Abstract
Background: Additional sex combs like transcription factor 1 (ASXL1) is a member of the polycomb group protein. ASXL1 mutation has been implicated in myeloid malignancy transformation. It is hypothesized that mutated ASXL1 leads to the loss of polycomb repressive complex 2 (PRC2) mediated gene repression and subsequent transforming events. Recent studies identify ASXL1 mutation as a poor prognostic marker in patients (pts) with de novo acute myeloid leukemia (AML) who present with intermediate–risk cytogenetic lesions (Patel, NEJM 2012; Schnittger, Leukemia2013). To study the impact of ASXL1 mutations in an unselected AML population, we analyzed clinical and molecular characteristics of patients with untreated AML who express ASXL1 mutation at presentation.
Methods: Using next generation sequencing, 254 adult patients with AML seen at the Hospital of the University of Pennsylvania were analyzed for mutations, including ASXL1, using a 33-gene hematologic malignancy panel. Clinical characteristics were obtained from retrospective chart review. Kaplan-Meier estimates were used to calculate overall survival (OS) from time of diagnosis. Living patients were censored at date last seen.
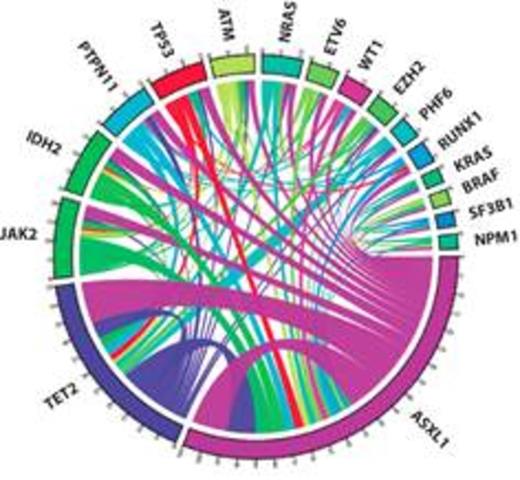
Results: ASXL1 mutations were detected in 36/254 (14%) AML pts. There were 29 known pathologic mutations, 1 benign, 1 probable pathologic, and 9 variants of unknown clinical significance (VUS). In 6/36 (16.7%) pts, ASXL1 was the sole mutation identified. Of the 30 pts with additional mutations (Figure 1), 6/30 (20%) pts harbored 2 independent ASXL1 mutations. When the 27 patients with pathologic ASCL mutations were analyzed for co-mutations, TET2 (13/27, 48%) was the most frequent ASXL1 co-mutation. FLT3 (0/27, 0%) and NPM1 (1/27, 3.7%) were notable for their absence.
Median age of pts at diagnosis was 69 years (range 23-80). Prior myelodysplastic syndrome (MDS) or myeloproliferative neoplasm (MPN) was noted in 9/36 (25%) and 11/36 (30.6%) pts, respectively. Four pts (11.1%) had received chemotherapy and/or radiation therapy for a prior non-myeloid neoplasm. Karyotype was normal in 18/36 (50%) pts, and 7 additional pts had intermediate cytogenetic lesions. There were 7 pts (19.4%) with unfavorable cytogenetics (complex karyotype (3 pts), 7q- (3 pts), and 5q- (1 pt)). Four pts (11.1%) had a favorable karyotype, with t(8;21) in 3 pts and t(15;17) in 1 pt.
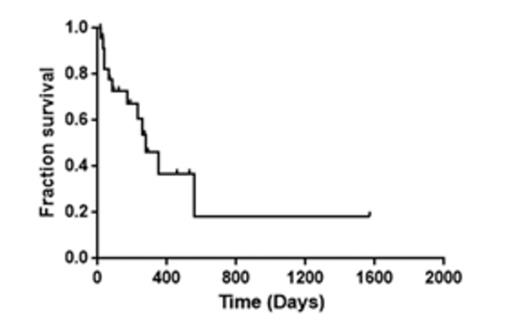
At presentation, median white blood cell count (WBC) was 6.4x103/uL (1.0 x -103). In pts whose AML transformed from prior MPN, median WBC was 50 X103/uL (3.3-140). Standard induction chemotherapy with an anthracycline and cytarabine was given to 17/36 (47%) pts. An additional 3/36 (8.3%) pts underwent induction therapy with clofarabine. Complete remission (CR) was documented in 14/20 (70%) evaluable pts. Of the remaining pts, 11 received a hypomethylating agent, and 5 received other therapies. Thirty-day treatment mortality for all 36 pts and for 27 pts with known ASXL1 pathologic mutation was 13.4% and 18.5% respectively. Kaplan-Meier estimate showed a median overall survival of 349 days (median follow up of 107 days (range 15-1570)). For the 27 pts with a pathologic ASXL1 mutation, the OS was 276 days (Figure 2, median follow up of 145 days (range 18-1570)).
Conclusion: ASXL1 mutations in de novo AML with intermediate-risk cytogenetics is associated with poor clinical outcome in cooperative group trials. Strikingly we demonstrate in a single institution, retrospective analysis that 66.7% of pts who present with ASXL1 mutations in the setting of previously untreated AML had documented MDS, MPN and/or prior chemotherapy/radiation. Further studies are necessary to evaluate if ASXL1 mutation has independent prognostic significance in AML or if it is primarily a marker for secondary leukemia.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal