Abstract
Introduction.The Ecotropic viral integration site 1 (EVI1) gene is located on chromosome band 3q26.2, and is composed of 16 exons. Upon diagnosis, EVI1 overexpression has been implicated in 10% of patients with acute myeloid leukemia (AML) and confers a poor prognosis. It occurs with or without chromosomal rearrangements involving 3q26.2 and is often associated with adverse cytogenetic abnormalities, but can also be found in normal karyotypes. At this point, expression of the EVI1 mRNA relative to the housekeeping gene Abelson (ABL) is determined by real time quantitative PCR (RQ-PCR) with ΔCt method. Although this technique gives good results upon diagnosis, it is not powerful enough to monitor patients in minimal residual disease (MRD). We therefore decided to develop a technique of absolute quantification of EVI1 expression in RQ-PCR with plasmid synthesis in order to determine overexpression thresholds in bone marrow (BM) and peripheral blood (PB), to follow up on changes in AML disease, and to look for differences of overexpression according to cytogenetics abnormalities.
Methods. This study focuses on 71 patients with AML overexpressing EVI1 included in the ALFA-0701 and ALFA-0702 trials for whom frozen material was available. BM and PB samples were collected at AML diagnosis (n=71) and during follow-up (n=152) as defined in the ALFA-0701 and ALFA-0702 trials. Threshold values in PB and BM were established by analyzing 31 normal PB and normal 19 BM samples. To achieve quantification, we cloned a sequence of EVI1 localized on exons 14 and 15, framing forward primer, reverse primer and TaqMan probe used by Groschel et al in 2010. The sequence was integrated into plasmids pCR2.1 TOPO (Invitrogen) which were used to determine the standard curve. RQ-PCR was performed on ABI7900 (Applied Biosystems). In parallel, the expression of the housekeeping gene ABL was evaluated by RQ-PCR for each sample. The results were expressed as a percentage of EVI1 copies by ABL copies. For every patient, Karyotype and follow-up data were confronted with the percentage of EVI1 expression.
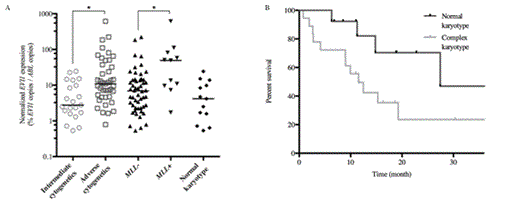
Results. To establish our overexpression threshold, we analyzed EVl1 expression in the population of negative witnesses. On PB, the median value of EVl1 expression was 0,11% [0,02 – 0,44], and the maximal value was 0,49%. On BM, the median value was 0,36% [0,01 – 0,96], and the maximal value was 0,96%. Thus, the overexpression threshold was fixed at 0,5% for PB and at 1% for BM. Upon diagnosis, we confirmed EVl1 overexpression for 62 samples out of 71. Mismatches were observed for 9 samples of PB which were ranked in weak positive in relative quantification. EVl1 median expression were 23,26% [4,29 – 516,1] on BM and 3,61% [0,63 – 34,3] on PB. We analyzed EVI1 expression upon diagnosis and MRD1 for 20 patients. In BM, expression varied from 21,61% [4,18 – 592,70] to 3,56% [0,48 – 85,63]. In PB, expression varied from 4% [0,61 – 58,71] to 0,22% [0,01 – 11,51]. Although reduction was significant (p < 0,001), it remained insufficient for MRD follow-up on this parameter.Otherwise, upon diagnosis, we found a statistically significant difference between patients with intermediate cytogenetic (n=21) and patients with adverse cytogenetic (n=41) (p < 0,01) with an EVl1 median expression of 2.75% [0,54 – 24,07] against 10,88% [1,63 – 218,7] (fig 1). Strongest EVI1 overexpression were found in the MLL rearrangement sub-group (n=11) (p < 0,01) : 49,18% [1,7 – 614,1] vs 6,93% [0,69 – 109,9].We did not reveal a strong link between rates and variations of EVI1 expression on the survival of patients, but a trend towards a less pejorative survival seems to appear when EVI1 overexpression is associated with a normal karyotype versus a complex karyotype (fig 2).
1) EVI1 expression in diagnosis 2) Overall survival according to cytogenetic.
Conclusions. We developed a model of absolute quantification of EVI1 expression based on plasmids containing a target sequence located in exons 14 and 15. Nevertheless, the basal expression of EVI1 is high and variations are weak between diagnosis and follow-up. However, we observed higher expression in patients with adverse cytogenetics abnormalities. But, it seems that cytogenetic abnormalities associated with EVI1 overexpression confer a poor prognosis, rather than overexpression in itself.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal