Key Points
Coexpression of NUP98/NSD1 and FLT3/ITD in AML is associated with very low complete remission rates and poor survival.
It is the interaction between NUP98/NSD1 and FLT3/ITD that determines poor outcome in NUP98/NSD1-associated AML.
Abstract
NUP98/NSD1 has recently been reported in association with poor outcome in acute myeloid leukemia (AML). Previous studies also observed a high overlap between NUP98/NSD1 and FLT3/ITD, raising the question as to whether the reported poor outcome is due to NUP98/NSD1 or caused by the co-occurrence of these 2 genetic lesions. We aimed to determine the prognostic significance of NUP98/NSD1 in the context of FLT3/ITD AML. A total of 1421 patients enrolled in 5 consecutive Children's Oncology Group/Children's Cancer Group and SWOG trials were evaluated. NUP98/NSD1 was found in 15% of FLT3/ITD and 7% of cytogenetically normal (CN)-AML. Those with dual FLT3/ITD and NUP98/NSD1 (82% of NUP98/NSD1 patients) had a complete remission rate of 27% vs 69% in FLT3/ITD without NUP98/NSD1 (P < .001). The corresponding 3-year overall survival was 31% vs 48% (P = .011), respectively. In CN-AML, patients with concomitant NUP98/NSD1 and FLT3/ITD had a worse outcome than those harboring NUP98/NSD1 only. In multivariate analysis, the dual NUP98/NSD1 and FLT3/ITD remained an independent predictor of poor outcome, and NUP98/NSD1 without FLT3/ITD lost its prognostic significance. Our study demonstrates that it is the interaction between NUP98/NSD1 and FLT3/ITD that determines the poor outcome of patients with NUP98/NSD1 disease.
Introduction
Acute myeloid leukemia (AML) is a genetically diverse disease.1,2 Analysis of recurrent chromosomal translocation has identified numerous genes important for malignant transformation and allowed the establishment of new disease classifications.3,4 Although cytogenetic abnormalities have long been appreciated as independent predictors of prognosis in AML patients, ∼25% and 50% of pediatric and adult patients, respectively, have no cytogenetic abnormalities.5,6 The advent of genome-wide studies has led to the discovery of several recurrent mutations in AML with prognostic significance.7-10 Such studies have revealed the novel fusion transcript NUP98/NSD1, which is created by the juxtaposition of Nucleoporin 98 (NUP98) and the nuclear receptor binding SET-domain protein 1 (NSD1) genes.11 Because both NUP98 and NSD1 are located in the telomeric region of chromosomes 11 and 5, respectively, fusions involving these 2 genes are cryptic by conventional karyotyping.12-14
FLT3/ITD is one of the most common molecular alterations in AML, occurring in ∼25% and 12% of adult and pediatric AML cases, respectively. Although the presence of FLT3/ITD does not affect complete remission (CR) rates, it is associated with decreased survival in both age groups due to increased relapse rate.15-18 In recent studies analyzing the prevalence and clinical significance of NUP98/NSD1 in adult and pediatric AML patients, the investigators found that patients with NUP98/NSD1 were more likely to also have FLT3/ITD.11,19-22 Despite the previous reported association between FLT3/ITD and NUP98/NSD1, the prevalence and clinical significance of NUP98/NSD1 in FLT3/ITD AML have not yet been studied.
Herein, we examined pediatric patients enrolled in 4 pediatric AML trials (2 Children's Cancer Group [CCG]2941/CCG2961 and 2 Children's Oncology Group [COG] trials: COGAAML03P1/COGAAML0531) and adult patients enrolled in SWOG trial S0106 providing the largest cohort of AML patients screened for NUP98/NSD1. In this study, we present a comprehensive evaluation of the prevalence and prognostic significance of NUP98/NSD1 in the context of other validated biologic, cytogenetic, and molecular risk factors in pediatric and adult patients with AML.
Patients and methods
Patient samples
Pediatric patients enrolled in the 4 consecutive pediatric AML protocols CCG-2941/2961, COG-AAML03P1, and -AAML0531 and adults enrolled in SWOG trial S0106 were eligible for this study. Acute promyelocytic leukemia (APL) patients enrolled in the COG trial AAML0631 were also screened. Collectively, these studies enrolled 3229 patients with newly diagnosed de novo AML. Details of these studies have been previously described.23-26 AAML03P1 (N = 350) and CCG-2941/2961 (N = 1106) enrolled patients 0 to 21 years of age, whereas AAML0531 (N = 1070) enrolled patients that were 0 to 30 years of age. S0106 (N = 595) enrolled patients 18 to 60 years of age. AAML0631 enrolled patients 2 to 21 years of age with APL (N = 108). A total of 1504 (N = 1267 in the COG and N = 237 in the SWOG) patients with diagnostic samples available were screened for NUP98/NSD1. Karyotyping was centrally reviewed by each study group. Consent was obtained from all study participants in accordance with the Declaration of Helsinki. Institutional review board approval was obtained from the Fred Hutchinson Cancer Research Center before mutation analysis, and this study was approved by the Myeloid Disease Biology Committees of the COG and SWOG.
Molecular genotyping and flow cytometric analysis
Genetic material was extracted from diagnostic marrow specimens using the AllPrep DNA/RNA Mini Kit via the QIAcube automated system (Qiagen, Valencia, CA). Presence of NUP98/NSD1 was determined by reverse transcriptase-polymerase chain reaction (RT-PCR) using cDNA prepared as per the standard protocol (Invitrogen, Carlsbad, CA). Primers and thermocycler conditions are presented in supplemental Table 1 available on the Blood Web site. The fusion transcripts were verified by Sanger sequencing. Real-time quantitative PCR (qPCR) primer probe sets are presented in supplemental Table 1. Expression analysis was performed on a StepOne Plus RT-PCR instrument, using the TaqMan system (Applied Biosystems, Foster City, CA) per the manufacturer’s instructions. Patient samples were run in duplicate, with the β-glucuronidase (GUSB) housekeeping gene as an internal control. The ΔCT method was used to determine the expression levels of NUP98/NSD1 relative to GUSB. Normal bone marrow was used as a control on each run.
Other relevant genes were assessed for frequently occurring mutations as previously described (ie, NPM1, FLT3/ITD, CEBPA, and WT1).17,27-29 The mutation analysis for CEBPA was not available for the SWOG cohort. Flow cytometric analysis for detection of minimal residual disease (MRD) was performed as previously described.30
Statistical methods
Overall survival (OS) was measured from the date of registration to date of death due to any cause, with patients last known to be alive censored at the date of last contact. Event-free survival (EFS) was measured from the date of registration to the date of the first of the following events: removal from protocol therapy without achieving a CR, progression, or death due to any cause; patients last known to be alive and progression free were censored at the date of last contact. Disease-free survival (DFS) was measured from the date of CR to the date of the first relapse or death due to any cause; patients last known to be alive and in CR were censored at the date of last contact.
The Kruskal-Wallis test was used to compare medians of quantitative variables, and Fisher’s exact test was used to compare categorical variables. Logistic regression was used to assess associations with CR. The Kaplan-Meier method was used to estimate OS, EFS, and DFS. Cox regression models were used to assess associations with these outcomes.
Results
Prevalence of NUP98/NSD1 and characteristics of the studied population
To determine the prevalence of NUP98/NSD1 in pediatric and adult patients and its association with FLT3/ITD in AML, we screened specimens for the presence of this fusion transcript from patients treated in 6 different COG and SWOG trials, which collectively enrolled 3229. We initially evaluated all available diagnostic specimens from patients treated during COG-AAML0531 (N = 683) and SWOG-S0106 (N = 237) for the presence of this fusion. NUP98/NSD1 was identified in 32 of 683 (5%) and 7 of 237 (3%) of the COG and SWOG patients, respectively; of those, all but 3 patients were either FLT3/ITD positive or cytogenetically normal (CN)-AML. Thus, the remaining evaluations in COG-AAML03P1 and CCG-2941/2961 were limited to patients with either FLT3/ITD or CN-AML.
In the 1421 patients of the combined COG and SWOG cohort with available specimens, there were 253 patients who harbored FLT3/ITD and 367 patients who were CN-AML. Of the patients with CN-AML, 35% (130 of 367) of them also harbored FLT3/ITD. Within the FLT3/ITD cohort, NUP98/NSD1 was identified in 15% (37 of 253) of the patients, whereas within the CN-AML cohort, this fusion was identified in 7% (26 of 367) of the patients. When evaluating all positive cases for NUP98/NSD1, 82% (37 of 45) of them also harbored FLT3/ITD, substantiating the significant overlap between the 2 genetic alterations.
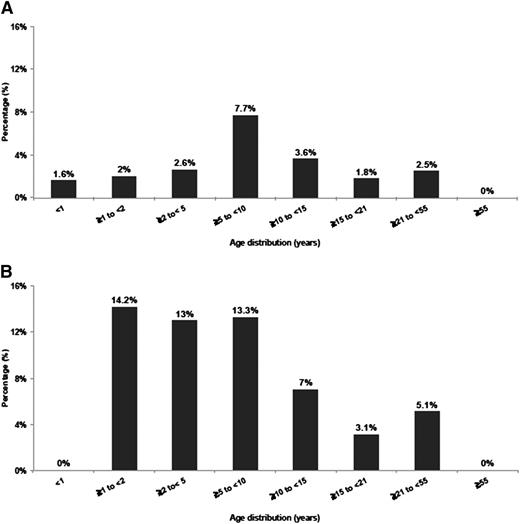
Age of patients with NUP98/NSD1 varied from <1 to 50 years (median, 10 years), indicating that NUP98/NSD1, although more frequent in younger age, is not restricted to pediatric patients (Figure 1A-B). In the COG cohort, NUP98/NSD1 was found in 16% (32 of 194) of the FLT3/ITD positive and in 8% (22 of 267) of the CN-AML patients. In the SWOG cohort, NUP98/NSD1 was identified in 8% (5 of 59) of the FLT3/ITD positive and in 4% (4 of 100) of CN-AML patients. All patients harbored the identical in-frame fusion of NUP98 exon 12 and NSD1 exon 6.
Histogram representing the age distribution of NUP98/NSD1 in AML. (A) Age distribution in all cytogenetic groups treated on protocols COG-AAML0531 and SWOG-S0106. (B) Age distribution in CN-AML treated on protocols CCG-2941/2961, COG-AAML03P1, AAML0531, and SWOG-S0106.
Histogram representing the age distribution of NUP98/NSD1 in AML. (A) Age distribution in all cytogenetic groups treated on protocols COG-AAML0531 and SWOG-S0106. (B) Age distribution in CN-AML treated on protocols CCG-2941/2961, COG-AAML03P1, AAML0531, and SWOG-S0106.
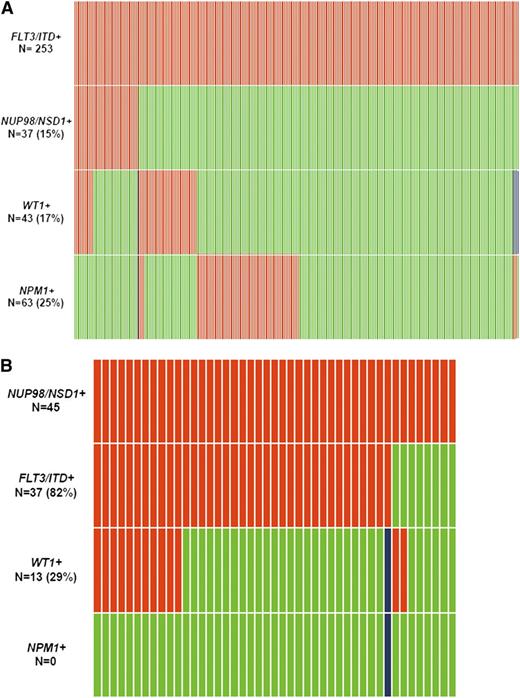
Demographics and disease characteristics in FLT3/ITD and CN-AML patients were compared between those with and without NUP98/NSD1 (Table 1). The molecular distribution of FLT3/ITD and NUP98/NSD1 is depicted in Figure 2A-B. NUP98/NSD1 was significantly associated with younger age, higher white blood count (WBC), and higher platelet count in both the FLT3/ITD and CN-AML subgroups. NPM1 mutations (NPM1+) were not detected in patients with NUP98/NSD1. In contrast, WT1 mutations (WT1+) were enriched among patients who harbored both FLT3/ITD and NUP98/NSD1 compared with those with FLT3/ITD only (30% vs 15%, P = .031).
Characteristics of patients with FLT3/ITD and CN-AML according to NUP98/NSD1 status
| Characteristics . | FLT3/ITD+ . | Normal cytogenetics . | ||||
|---|---|---|---|---|---|---|
| NUP98/NSD1− ( N = 216) . | NUP98/NSD1+ (N = 37) . | P value . | NUP98/NSD1− (N = 341) . | NUP98/NSD1+ (N = 26) . | P value . | |
| Age (years), median (range) | 15 (2-61) | 11 (3-51) | .003 | 15 (0-61) | 12 (1-51) | .022 |
| Study protocol, N (%) | .033 | .35 | ||||
| AAML03P1 | 21 (10) | 1 (3) | 28 (8) | 1 (4) | ||
| AAML0531 | 113 (52) | 20 (54) | 162 (48) | 15 (58) | ||
| CCG-2961 | 28 (13) | 11 (30) | 55 (16) | 6 (23) | ||
| S0106 | 54 (25) | 5 (14) | 96 (28) | 4 (15) | ||
| Male, N (%) | 117 (54) | 26 (70) | .075 | 177 (52) | 19 (73) | .042 |
| Cytogenetics, N (%) | .024 | NA | NA | NA | ||
| Favorable | 9 (4) | 0 (0) | ||||
| Intermediate | 133 (61) | 26 (70) | ||||
| Unfavorable | 25 (12) | 0 (0) | ||||
| Unknown risk | 15 (7) | 0 (0) | ||||
| Missing | 34 (16) | 11 (30) | ||||
| WBC × 109/L, median (range) | 44 (0-827) | 173 (3-860) | <.001 | 22 (0-827) | 79 (3-243) | <.001 |
| Marrow blast %, median (range) | 80 (3-100) | 86 (44-98) | .16 | 68 (0-100) | 85 (44-94) | .008 |
| Platelets × 109/L, median (range) | 48 (8-9300) | 85 (17-536) | <.001 | 52 (4-9300) | 72 (39-280) | .0075 |
| WT1+, N (%) | 32 (15) | 11 (30) | .031 | 26 (8) | 10 (38) | <.001 |
| NPM1+, N (%) | 63 (29) | 0 (0) | <.001 | 119 (35) | 0 (0) | <.001 |
| Characteristics . | FLT3/ITD+ . | Normal cytogenetics . | ||||
|---|---|---|---|---|---|---|
| NUP98/NSD1− ( N = 216) . | NUP98/NSD1+ (N = 37) . | P value . | NUP98/NSD1− (N = 341) . | NUP98/NSD1+ (N = 26) . | P value . | |
| Age (years), median (range) | 15 (2-61) | 11 (3-51) | .003 | 15 (0-61) | 12 (1-51) | .022 |
| Study protocol, N (%) | .033 | .35 | ||||
| AAML03P1 | 21 (10) | 1 (3) | 28 (8) | 1 (4) | ||
| AAML0531 | 113 (52) | 20 (54) | 162 (48) | 15 (58) | ||
| CCG-2961 | 28 (13) | 11 (30) | 55 (16) | 6 (23) | ||
| S0106 | 54 (25) | 5 (14) | 96 (28) | 4 (15) | ||
| Male, N (%) | 117 (54) | 26 (70) | .075 | 177 (52) | 19 (73) | .042 |
| Cytogenetics, N (%) | .024 | NA | NA | NA | ||
| Favorable | 9 (4) | 0 (0) | ||||
| Intermediate | 133 (61) | 26 (70) | ||||
| Unfavorable | 25 (12) | 0 (0) | ||||
| Unknown risk | 15 (7) | 0 (0) | ||||
| Missing | 34 (16) | 11 (30) | ||||
| WBC × 109/L, median (range) | 44 (0-827) | 173 (3-860) | <.001 | 22 (0-827) | 79 (3-243) | <.001 |
| Marrow blast %, median (range) | 80 (3-100) | 86 (44-98) | .16 | 68 (0-100) | 85 (44-94) | .008 |
| Platelets × 109/L, median (range) | 48 (8-9300) | 85 (17-536) | <.001 | 52 (4-9300) | 72 (39-280) | .0075 |
| WT1+, N (%) | 32 (15) | 11 (30) | .031 | 26 (8) | 10 (38) | <.001 |
| NPM1+, N (%) | 63 (29) | 0 (0) | <.001 | 119 (35) | 0 (0) | <.001 |
NA, not applicable.
Relationship between common mutations in patients with AML. (A) relationship between FLT3/ITD and other common mutations. (B) relationship between NUP98/NSD1 and other common mutations. Red indicates the presence of the specified mutation in the designated patients, green indicates the absence of the mutation, and blue indicates that data were not available for that sample. This analysis include patients treated on protocols CCG-2941/2961, COG-AAML03P1, AAML0531, and SWOG-S0106.
Relationship between common mutations in patients with AML. (A) relationship between FLT3/ITD and other common mutations. (B) relationship between NUP98/NSD1 and other common mutations. Red indicates the presence of the specified mutation in the designated patients, green indicates the absence of the mutation, and blue indicates that data were not available for that sample. This analysis include patients treated on protocols CCG-2941/2961, COG-AAML03P1, AAML0531, and SWOG-S0106.
Clinical outcome
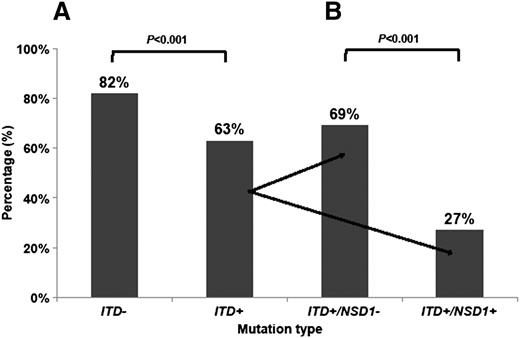
Given the association of FLT3/ITD and NUP98/NSD1, we next evaluated the prognostic impact of NUP98/NSD1 in patients with FLT3/ITD. The CR rate for patients with and without FLT3/ITD was 63% and 82%, respectively (P < .001). Within the FLT3/ITD cohort, the CR rate in patients with additional NUP98/NSD1 fusions was 27% (10 of 37) vs 69% (149 of 216) in those without NUP98/NSD1 (P < .001; Figure 3A-B). The 3-year OS for all patients with FLT3/ITD in this study was 45%. The 3-year OS for FLT3/ITD patients with and without NUP98/NSD1 was 31% and 48% (P = .011), respectively (Figure 4A). Similarly, the 3-year EFS for all patients with FLT3/ITD was 30%. The 3-year EFS for FLT3/ITD patients with and without NUP98/NSD1 was 14% and 33% (P < .001), respectively.
CR rate in AML patients. (A) CR rate according to FLT3/ITD status only and (B) CR rate according to FLT3/ITD and NUP98/NSD1 status. NSD1+ (NUP98/NSD1 present) NSD1− (NUP98/NSD1 absent); ITD+ (FLT3/ITD present); and ITD− (FLT3/ITD absent).
CR rate in AML patients. (A) CR rate according to FLT3/ITD status only and (B) CR rate according to FLT3/ITD and NUP98/NSD1 status. NSD1+ (NUP98/NSD1 present) NSD1− (NUP98/NSD1 absent); ITD+ (FLT3/ITD present); and ITD− (FLT3/ITD absent).
Kaplan-Meier estimates of OS. (A) OS according to FLT3/ITD and NUP98/NSD1 status and (B) OS in CN-AML according to FLT3/ITD and NUP98/NSD1 status. NUP98/NSD1+ (NUP98/NSD1 present), NUP98/NSD1− (NUP98/NSD1 absent), FLT3/ITD+ (FLT3/ITD present), and FLT3/ITD− (FLT3/ITD absent).
Kaplan-Meier estimates of OS. (A) OS according to FLT3/ITD and NUP98/NSD1 status and (B) OS in CN-AML according to FLT3/ITD and NUP98/NSD1 status. NUP98/NSD1+ (NUP98/NSD1 present), NUP98/NSD1− (NUP98/NSD1 absent), FLT3/ITD+ (FLT3/ITD present), and FLT3/ITD− (FLT3/ITD absent).
We previously demonstrated the clinical significance of the FLT3/ITD allelic ratio (AR).17 We evaluated the impact of FLT3/ITD AR on NUP98/NSD1 based on high (>0.4) and low (≤ 0.4) internal tandem duplication (ITD) AR. The median ITD AR was 0.51 and 0.48 (P = .63) in patients with and without NUP98/NSD1, respectively. In patients with FLT3/ITD AR > 0.4, the CR rate for those with and without NUP98/NSD1 was 26% (6 of 23) and 67% (84 of 126; P < .001), respectively. The corresponding 3-year OS was 32% vs 42% (P = .29). In patients with FLT3/ITD AR ≤ 0.4, the CR rate was 23% (3 of 13) vs 72% (64 of 89; P = .001), respectively. The corresponding 3-year OS was 31% vs 56% (P = .015) for those with and without NUP98/NSD1, respectively.
The prognostic significance of NUP98/NSD1 was also evaluated in CN-AML. The prevalence of FLT3/ITD in CN-AML patients with NUP98/NSD1 was 73% (19 of 26). The CR rate in the entire cohort of patients with CN-AML was 75% (276 of 367), whereas the CR rate for patients with and without NUP98/NSD1 was 50% (13 of 26) and 77% (263 of 341; P = .004), respectively. The 3-year OS and EFS for CN-AML with and without NUP98/NSD1 were 38% vs 63% (P = .029) and 19% vs 48% (P < .001), respectively. Within CN-AML patients harboring NUP98/NSD1, those with concomitant FLT3/ITD had a CR rate of 37% (7 of 19) vs 86% (6 of 7; P = .07) in patients with NUP98/NSD1 without FLT3/ITD. In CN-AML patients with NUP98/NSD1, the 3-year OS in CN-AML patients who also harbored FLT3/ITD was 28% vs 67% in those with NUP98/NSD1 without FLT3/ITD (P = .092; Figure 4B). Within the FLT3/ITD cohort, there were 130 patients who were CN-AML. In this analysis of CN-AML with FLT3/ITD, 37% (7 of 19) of the patients with additional NUP98/NSD1 achieved CR vs 67% (74 of 111) in those without NUP98/NSD1 (P = .019).
Among patients who harbored NUP98/NSD1, 26% (11 of 42) of them harbored simultaneously FLT3/ITD and WT1 mutations. The CR rate of patients harboring these 3 genetic alterations was 9% (1 of 11), whereas the CR rate of those harboring FLT3/ITD and NUP98/NSD1 without WT1 was 33% (8 of 24; P = .22).
In multivariate regression analysis including other known prognostic factors for AML, the dual alteration (FLT3/ITD and NUP98/NSD1) remained significantly associated worse CR rate (odds ratio [OR] = 0.1; 95% confidence interval [CI], 0.04-0.25; P < .001), OS (hazard ratio [HR] = 2.05; 95% CI, 1.19-3.53; P = .01), and EFS (HR = 2.34; 95% CI, 1.45-3.78; P < .001) compared with the dual negative patients (Table 2). Multivariate regression analysis for the CN-AML cohort only is shown in supplemental Table 2.
Multivariate regression analysis including AML patients with FLT3/ITD and normal cytogenetics for CR, OS, and EFS
| Covariate . | CR . | OS . | EFS . | |||
|---|---|---|---|---|---|---|
| OR, 95% CI . | P value . | HR, 95% CI . | P value . | HR, 95% CI . | P value . | |
| NUP98-NSD1+/FLT3/ITD+* | 0.1 (0.04, 0.25) | <.001 | 2.05 (1.19, 3.53) | .01 | 2.34 (1.45, 3.78) | <.001 |
| NUP98-NSD1−/FLT3-ITD+* | 0.48 (0.28, 0.81) | .0058 | 1.39 (0.99 1.95) | .055 | 1.43 (1.07, 1.91) | .015 |
| NUP98-NSD1+/FLT3-ITD−* | 1.76 (0.2, 15.76) | .61 | 0.34 (0.05, 2.5) | .29 | 1.16 (0.42, 3.22) | .77 |
| Age (years) | 1.01 (0.98, 1.04) | .5 | 1.02 (1, 1.04) | .12 | 1.01 (0.99, 1.02) | .58 |
| Male gender (ref = female) | 1.19 (0.75, 1.91) | .46 | 0.8 (0.59, 1.06) | .12 | 0.97 (0.76, 1.25) | .84 |
| WBC | 0.86 (0.67, 1.1) | .22 | 1.18 (1.06, 1.31) | .0034 | 1.24 (1.12, 1.37) | <.001 |
| Marrow blasts (10%) | 1.08 (0.97, 1.2) | .14 | 1.07 (1, 1.15) | .061 | 1.03 (0.97, 1.09) | .37 |
| Platelets | 1.02 (0.91, 1.15) | .68 | 0.98 (0.84, 1.14) | .82 | 1.02 (1, 1.04) | .11 |
| NPM1+ (ref = NPM1−) | 2.69 (1.45, 5) | .0016 | 0.67 (0.47, 0.96) | .031 | 0.55 (0.4, 0.75) | <.001 |
| WT1+ (ref = WT+) | 0.59 (0.3,1.16) | .13 | 1.88 (1.24, 2.83) | .0028 | 1.63 (1.12, 2.37) | .0099 |
| SWOG (ref = COG) | 0.43 (0.13, 1.41) | .16 | 1.35 (0.65, 2.83) | .42 | 2.02 (1.04, 3.92) | .037 |
| Covariate . | CR . | OS . | EFS . | |||
|---|---|---|---|---|---|---|
| OR, 95% CI . | P value . | HR, 95% CI . | P value . | HR, 95% CI . | P value . | |
| NUP98-NSD1+/FLT3/ITD+* | 0.1 (0.04, 0.25) | <.001 | 2.05 (1.19, 3.53) | .01 | 2.34 (1.45, 3.78) | <.001 |
| NUP98-NSD1−/FLT3-ITD+* | 0.48 (0.28, 0.81) | .0058 | 1.39 (0.99 1.95) | .055 | 1.43 (1.07, 1.91) | .015 |
| NUP98-NSD1+/FLT3-ITD−* | 1.76 (0.2, 15.76) | .61 | 0.34 (0.05, 2.5) | .29 | 1.16 (0.42, 3.22) | .77 |
| Age (years) | 1.01 (0.98, 1.04) | .5 | 1.02 (1, 1.04) | .12 | 1.01 (0.99, 1.02) | .58 |
| Male gender (ref = female) | 1.19 (0.75, 1.91) | .46 | 0.8 (0.59, 1.06) | .12 | 0.97 (0.76, 1.25) | .84 |
| WBC | 0.86 (0.67, 1.1) | .22 | 1.18 (1.06, 1.31) | .0034 | 1.24 (1.12, 1.37) | <.001 |
| Marrow blasts (10%) | 1.08 (0.97, 1.2) | .14 | 1.07 (1, 1.15) | .061 | 1.03 (0.97, 1.09) | .37 |
| Platelets | 1.02 (0.91, 1.15) | .68 | 0.98 (0.84, 1.14) | .82 | 1.02 (1, 1.04) | .11 |
| NPM1+ (ref = NPM1−) | 2.69 (1.45, 5) | .0016 | 0.67 (0.47, 0.96) | .031 | 0.55 (0.4, 0.75) | <.001 |
| WT1+ (ref = WT+) | 0.59 (0.3,1.16) | .13 | 1.88 (1.24, 2.83) | .0028 | 1.63 (1.12, 2.37) | .0099 |
| SWOG (ref = COG) | 0.43 (0.13, 1.41) | .16 | 1.35 (0.65, 2.83) | .42 | 2.02 (1.04, 3.92) | .037 |
ref, reference.
Reference = NUP98-NSD1−/FLT3-ITD−.
In a subset analysis limited to the COG patients, the prevalence of NUP98/NSD1 within FLT3/ITD cohort was 16%. WT1 mutations were enriched in patients with dual FLT3/ITD and NUP98/NSD1, with a prevalence of 31% vs 17% in those with FLT3/ITD only (P = .047). In contrast, NPM1 or CEBPA mutations were not observed in patients with dual FLT3/ITD and NUP98/NSD1 (supplemental Table 3). Similarly to the combined COG and SWOG cohorts, in the COG cohort only, the CR rate in FLT3/ITD positive patients with and without NUP98/NSD1 was 28% (9 of 32) and 69% (112 of 162; P = .0001) and 3-year OS was 33% and 55% (P = .0049), respectively (supplemental Figure 1A). Within the CN-AML cohort, those harboring both NUP98/NSD1 and FLT3/ITD had an adverse outcome, whereas patients with NUP98/NSD1 without FLT3/ITD had a more favorable CR rate of 86% vs 28% for those with the dual alteration, with a corresponding 3-year OS similar to the patients without FLT3/ITD, suggesting that both molecular lesions are required to bestow chemotherapy resistance to the leukemic clone (supplemental Figure 1B).
In the SWOG FLT3/ITD cohort, 8% (5 of 59) of the patients also harbored NUP98/NSD1. Of those 5 patients, 4 failed to achieve CR. The only patient with dual FLT3/ITD and NUP98/NSD1 who achieved CR relapsed after 354 days and died within a year after relapse. In the SWOG CN-AML cohort, 4% (4 of 100) of the patients harbored NUP98/NSD1. Of those, all but one also harbored FLT3/ITD. None of the patients with dual NUP98/NSD1 and FLT3/ITD achieved CR. The only patient who achieved CR and remained alive at 4.5 years of follow-up harbored NUP98/NSD1 but not FLT3/ITD.
Data on allogeneic stem cell transplantation (allo-SCT) were available for the COG patients only. Of the 32 patients with dual NUP98/NSD1 and FLT3/ITD, only 12 patients were evaluable for allo-SCT. The other 20 patients died, relapsed, failed induction, or electively withdrew from the study prior to allo-SCT. Of the 12 patients with dual NUP98/NSD1 and FLT3/ITD, 7 received allo-SCT and 5 were treated with chemotherapy only. The 3-year DFS for patients who received allo-STC was 43% vs 0% for those who did not undergo transplantation (P = .025).
NUP98/NSD1 and MRD
Data on MRD assessed by flow cytometry were available for the COG cohort only. MRD data were available for 117 patients with FLT3/ITD AML and were assessed at the end of cycle 1 of induction chemotherapy (EOI-1). The incidence of MRD in patients with FLT3/ITD who also harbored NUP98/NSD1 was 76% (13 of 17) compared with 37% (37 of 100) for those with FLT3/ITD without NUP98/NSD1 (P = .0032).
We further studied the molecular assay for NUP98/NSD1 and its ability to detect residual disease at the EOI-1. Specimen for this evaluation was available for the COG cohort only. Of the 39 patients with NUP98/NSD1 at diagnosis, 15 had samples available for analysis at the EOI-1 and were subjected to RT-qPCR for NUP98/NSD1. Of these 15 patients, concurrent MRD data assessed by flow cytometry were available in 13 patients. Three patients who were MRD negative by flow cytometry were positive for NUP98/NSD1 by RT-qPCR. Only 1 patient was MRD positive by flow cytometry but negative by RT-qPCR. This patient did not achieve CR and had persistent FLT3/ITD at EOI-1. Of the 15 patients who had samples available for analysis, 14 remained positive for NUP98/NSD1 at EOI-1. Of these 14 patients, 10 (71%) never achieved CR. Of the 4 patients who achieved CR, 2 relapsed and 2 are alive after receiving allo-SCT in the first CR.
Discussion
In this large collaborative study, we define the prevalence and prognostic significance of NUP98/NSD1 in children and adults with AML. We demonstrate that the co-occurrence of NUP98/NSD1 and FLT3/ITD leads to high induction failure and poor survival in AML. Our data suggest that the presence of only NUP98/NSD1 may not have a significant impact on outcome in the absence of other genomic lesions. It has been previously demonstrated that FLT3/ITD leads to a high incidence of postremission relapse, but it does not impact remission rate.15-18 In this study, we showed that FLT3/ITD patients who also harbor NUP98/NSD1 have a very low rate of remission induction, suggesting cooperation between these 2 genetic lesions to generate chemotherapy resistance to the leukemic clone.
Hollink et al recently showed that NUP98/NSD1 is associated with poor outcome in AML. The investigators also observed a high overlap between NUP98/NSD1 and FLT3/ITD, raising the question as to whether the reported poor outcome is due to NUP98/NSD1 or determined by the co-occurrence of these 2 genetic lesions.11 Our study specifically addresses this question and demonstrates that patients harboring both NUP98/NSD1 and FLT3/ITD have dismal CR rates, worse survival, and higher postinduction MRD than FLT3/ITD patients without NUP98/NSD1. This is also shown in CN-AML, where those with NUP98/NSD1 without FLT3/ITD had a more favorable outcome, and similar to patients who are dual negative for NUP98/NSD1 and FLT3/ITD. In multivariate analysis accounting for other known prognostic factors, the dual FLT3/ITD and NUP98/NSD1 remains an independent predictor of poor outcome, which is worse than FLT3/ITD alone, whereas NUP98/NSD1 in the absence of FLT3/ITD loses its prognostic significance.
In addition to the high overlap between FLT3/ITD and NUP98/NSD1, we also found a high simultaneous co-occurrence of WT1 in FLT3/ITD with NUP98/NSD1. Although the numbers are small, our data suggest that additional genetic lesions (ie, WT1+) might further impact response to therapy and outcome of patients harboring NUP98/NSD1 and FLT3/ITD. In our study, most patients remained positive for NUP98/NSD1 at EOI-1, suggesting that NUP98/NSD1 can be used as a potential tool for response evaluation or MRD detection either after chemotherapy or stem cell transplantation. The utility of NUP98/NSD1 as a marker of MRD has also been shown in a previous study.22 Finally, because there is a high prevalence of FLT3/ITD among patients with APL,31,32 we also screened these patients for NUP98/NSD1 and found that this fusion is not seen, at least, in pediatric APL patients.
ARs of FLT3/ITD have been shown to carry prognostic significance in AML.16,17 We previously showed that patients with ITD AR > 0.4 are at high risk for treatment failure, whereas the outcome of patients with ITD AR ≤ 0.4 is similar to FLT3 wild type. In the current study, NUP98/NSD1 was seen with similar frequency in patients with low and high ITD AR and the presence of this fusion resulted in a very high risk of treatment failure in patients with either low (≤0.4) or high (>0.4) AR, with CR rates of only 23% and 26%, respectively.
Although a high overlap between other NUP fusions (ie, NUP98/HOX9; NUP98/HOXD13, NUP214/DEK) and FLT3/ITD has also been demonstrated in several other studies, the cooperative mechanism between FLT3/ITD and Nucleoporin rearrangements in AML is unknown.11,33-38 Previous work has shown that FLT3/ITD induces a myeloproliferative neoplasm without leukemic phenotype in murine model.39 Nonetheless, when the FLT3/ITD murine model of myelodysplastic syndrome is combined with NUP98/HOXD13, the mice developed AML with 100% penetrance and short latency, suggesting an in vivo cooperating mechanism between Nucleoporin rearrangements and FLT3/ITD in AML.40
Our data raise questions about the therapeutic strategy that should be used for this high-risk group of patients with a dismal rate of remission induction. Given that patients with the dual genetic alterations (NUP98/NSD1 and FLT3/ITD) have such a poor remission induction rate (<30%), it might be justifiable to avoid unnecessary exposure to conventional chemotherapy and offer these patients alternative therapeutic approaches upfront. The next obvious question is whether the incorporation of FLT3 inhibitors into the treatment algorithm of patients with the dual genetic alteration could lead to improvement of their remission induction rate and overall outcome. As this fusion transcript was shown to alter the methyltransferase activity of NSD1,41 targeting methyltransferases may be another attractive treatment option for this high-risk patient population. Finally, upregulation of Hox genes and Meis1 is seen in AML harboring NUP98/NSD1.11,41 In a recent study, Daigle et al showed a concentration-dependent decrease in HoxA9 and Meis1 mRNA levels in cells harboring MLL rearrangements that were treated with the DOT1L inhibitor EPZ5676. Time course experiments revealed that full depletion of HoxA9 and Meis1 mRNA occurs in ∼8 days.42 The activity of this class of agents has not yet been investigated in AML harboring NUP98/NSD1, and it is conceivable that these compounds alone or in combination with FLT3 inhibitors may provide an attractive therapeutic option for patients harboring both genetic lesions.
Although the number of patients who underwent allo-SCT in our study is small, our results suggest that patients with NUP98/NSD1 and FLT3/ITD who achieved CR and underwent allo-SCT seem to have an improved 3-year DFS compared with those who were treated with chemotherapy only. Another smaller study also suggested a potential survival benefit of allo-SCT in this patient population.22 The role of allo-SCT in patients coexpressing NUP98/NSD1 and FLT3/ITD who achieve CR has not been determined and needs to be investigated in prospective clinical trials.
In summary, this is the largest study evaluating the prognostic significance of NUP98/NSD1 in FLT3/ITD AML in children and adult patients. We demonstrate that the co-occurrence of these 2 genetic lesions identifies a subgroup of patients at very high risk for induction failure. Our findings further support the upfront screening of NUP98/NSD1 fusion in AML, which requires the use of either FISH or RT-PCR, given that this fusion is cytogenetically cryptic. The high likelihood of induction failure suggests that alternative treatment regimens should be considered as frontline therapy in this group of patients. Nonetheless, as many of the patients coexpressing NUP98/NSD1 and FLT3/ITD present with high WBCs, which require rapid initiation of induction chemotherapy, the screening for this fusion transcript at diagnosis should be done expeditiously, otherwise alternative treatment may not be feasible. Finally, further insight into the role that NUP98/NSD1 plays in leukemogenesis and the mechanism of cooperation between NUP98/NSD1 and other genetic alterations (ie, FLT3/ITD and WT1) may eventually lead to the development of effective targeted therapies in AML.
Presented in part at the 55th Annual Meeting at the American Society of Hematology, New Orleans, LA, December 7-10, 2013.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health, National Cancer Institute grants K12 CA76930-15 and R01 CA114563, the Alex Lemonade Stand Foundation, and the Coltman Fellowship Award.
Authorship
Contribution: F.O. and S.M. designed and performed research, analyzed data, and wrote the manuscript; T.A.A., R.B.G., and M.O. served as statisticians, performed statistical analyses, and edited the manuscript; K.L.M. performed research and edited the manuscript; and M.L., S.C.R., B.A.H., B.J.L., S.P., J.R., F.R.A., and A.S.G. analyzed data and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fabiana Ostronoff, Clinical Research Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N (D2-140), Seattle, WA 98109; e-mail: fostrono@fhcrc.org.