Key Points
A Cdk6 R31C knock-in mutation resistant to INK4 inhibitors cooperates with Cdk4 hyperactivity in the development of hematopoietic tumors.
In Cdk6 R31C cells, p16INK4a increasingly binds and inhibits Cdk4, suggesting that both kinases cooperate in sequestering INK4 proteins in cancer.
Abstract
Cdk4 and Cdk6 are related protein kinases that bind d-type cyclins and regulate cell-cycle progression. Cdk4/6 inhibitors are currently being used in advanced clinical trials and show great promise against many types of tumors. Cdk4 and Cdk6 are inhibited by INK4 proteins, which exert tumor-suppressing functions. To test the significance of this inhibitory mechanism, we generated knock-in mice that express a Cdk6 mutant (Cdk6 R31C) insensitive to INK4-mediated inhibition. Cdk6R/R mice display altered development of the hematopoietic system without enhanced tumor susceptibility, either in the presence or absence of p53. Unexpectedly, Cdk6 R31C impairs the potential of hematopoietic progenitors to repopulate upon adoptive transfer or after 5-fluorouracil–induced damage. The defects are overcome by eliminating sensitivity of cells to INK4 inhibitors by introducing the INK4-insensitive Cdk4 R24C allele, and INK4-resistant mice are more susceptible to hematopoietic and endocrine tumors. In BCR-ABL–transformed hematopoietic cells, Cdk6 R31C causes increased binding of p16INK4a to wild-type Cdk4, whereas cells harboring Cdk4 R24C and Cdk6 R31C are fully insensitive to INK4 inhibitors, resulting in accelerated disease onset. Our observations reveal that Cdk4 and Cdk6 cooperate in hematopoietic tumor development and suggest a role for Cdk6 in sequestering INK4 proteins away from Cdk4.
Introduction
Cyclin-dependent kinases (Cdks) regulate multiple cellular processes including cell-cycle progression, transcription, or neuron biology.1 Cdk4 and Cdk6 are highly related (∼70% homology) cell-cycle kinases that phosphorylate retinoblastoma protein (pRb) family members and regulate progression through G1 and entry into the DNA synthesis (S) phase. Cdk4 and Cdk6 bind D-type cyclins (D1, D2, and D3), which are transcriptionally induced by mitogenic signals, leading to Cdk4/6 activation and pRb phosphorylation. Cdk-dependent phosphorylation of pRb ultimately results in its inactivation and allows for the subsequent transcriptional induction of several genes required for S-phase or mitosis.2 These 2 kinases are thought to be at least partially redundant in regulating G1 transition when coordinately expressed. In vivo, lack of either of these kinases causes specific defects in a cell type–specific manner. Cdk4 deficiency results in defective proliferation of postnatal pancreatic β cells and pituitary cells,3,4 whereas lack of Cdk6 provokes specific defects in hematopoietic cells.5,6 Concomitant ablation of both Cdk4 and Cdk6 results in embryonic lethality from defective erythropoiesis.5
Most tumor types display functional upregulation of these kinases through overexpression (in some cases from amplification or translocation) of D-type cyclins or inactivation of the cyclin D-Cdk4/6–specific inhibitors: the INK4 proteins.7 This family is composed of 4 inhibitors—p16INK4a, p15INK4b, p18INK4c, and p19INK4d—which specifically inactivate Cdk4 and Cdk6 by provoking structural changes that avoid binding and activation by D-type cyclins.8 At least 3 of them, p16INK4a, p15INK4b, and p18INK4c, are inactivated in tumors by deletion, mutation, or promoter hypermethylation, and in vivo models suggest that concomitant inactivation of these inhibitors results in a stronger oncogenic effect.9,10 In particular, the 9p21 chromosomal region encoding p16INK4a and p15INK4b is one of the most commonly mutated loci in human cancer.7,11 Interestingly, a few tumor-associated point mutations have been found in p16INK4a and p18INK4c that avoid binding and therefore inhibition of Cdk4/Cdk6.12,13 On the other hand, a specific point mutation in Cdk4 (Arg24 to Cys; R24C) prevents binding of the INK4 inhibitors in melanoma patients with sporadic or hereditary disease.14,15 Mice carrying this mutation in the endogenous Cdk4 locus develop a spectrum of tumors and are more susceptible to melanoma development.16,17 Lack of Cdk4 inhibition by INK4 proteins in Cdk4R24C/R24C mice results in a stronger phenotype than lack of the individual INK inhibitors,9,10,18 as expected from the compensatory roles among these proteins. However, INK4 proteins may still be partially functional in Cdk4R24C/R24C animals by inhibiting Cdk6.
To understand the unique or combined effect of lack of inhibition of Cdk4 and Cdk6 by INK4 inhibitors, we have generated knock-in mice expressing a Cdk6 mutant allele in which Arg31 (homologous to Arg24 in Cdk4) has been replaced by Cys (Cdk6 R31C or Cdk6R). Preventing inhibition of Cdk6 by INK4 proteins has only a minor impact on mouse development or survival. In Cdk6R/R cells, more p16INK4a binds to Cdk4, potentially accounting for the limited oncogenic potential of the Cdk6 R31C mutation. The inhibitory function of INK4 proteins is completely abrogated in Cdk4R/R; Cdk6R/R double-mutant mice display a significant increase in hematopoietic tumors. These results suggest that the precise modulation of Cdk4/6 activity by INK4 proteins contributes to maintaining the homeostasis of proliferation by sequestering INK4 inhibitors.
Methods
Mice colony and pathological analysis
To generate the targeting vector, we subcloned 5 kb of Cdk6 DNA (supplemental Figure 1 and supplemental Methods on the Blood Web site) into pBH48,3 and the GCCCGC (Ala30-Arg31) sequence located in exon 1 was replaced by GCATGC (Ala30-Cys31). Heterozygous mice carrying this allele were bred to CMV-Cre mice19 to generate “knock-in” Cdk6+/R31C (Cdk6+/R) animals. Cdk4 R24C3,17 and p53-null20 mice were reported previously. All of these animals were maintained at the Spanish National Cancer Research Centre Animal Facility, and all international guidelines were followed for animals used in research after approval by the corresponding ethics committees and conformed to Austrian laws (license 68.205/0218-II/3b/2012).
Flow cytometry
Splenic single-cell suspensions were prepared using a 70-µm cell strainer followed by red blood cell lysis using Red Blood Cell Lysis Buffer (Sigma-Aldrich). Femur and tibia were used to prepare single-cell suspensions from the bone marrow. Peripheral blood was obtained by vena facialis puncture, and red blood cells were lysed as described previously. The following antibodies were used: Gr-1 (Ly6-G, Ly6-C)-FITC, CD11b-PerCP-Cy5.5, CD3-PE, CD4-PE-Cy7, CD8-APC, CD19-PacificBlue, B220-PerCP-Cy5.5, CD43-PE, BP1-biotinylated, Streptavidin-APC-Cy7, IgM-FITC, IgD-APC, B220-PacificBlue, Ter119-PacificBlue, Gr (Ly6-G, Ly6-C)-PacificBlue, CD11b-PacificBlue, CD3-PacificBlue, Sca-1 (Ly6A/E)-PE-Cy7, Sca1-PerCP-Cy5.5 c-kit (CD117)-PE-Cy5, CD34-FITC, CD150-APC, CD48-PE, and CD25-biotinylated; CD135-biotinylated, Ly5.1 (CD45.1)-FITC, Ly5.2 (CD45.2)-APC-Cy7, Ly5.1 (CD45.1)-APC, Ly5.2 (CD45.2)-PacificBlue, and aCD16/CD32 were used as Fc receptor blocks (all from eBioscience). CD3ε (2C11), CD11b (M1/70), CD45R/B220 (RA3-6B2), Ter119, Gr1 (RB6-8C5), and anti-c-Kit-APC-H7 (2B8) were from Pharmingen (BD Biosciences, San Jose CA). For the analysis of erythropoiesis, samples from bone marrow and spleen were stained using anti-Ter 119 PE (Ter-119) and anti-CD71 FITC (C2) all from Pharmingen. Samples were analyzed using a FACSCanto II flow cytometer (BD Biosciences, Heidelberg, DE) and calculated with FACSDiva software version 6.1.2 (BD Biosciences) or using FlowJo v9.3.1 (Treestar, OR).
Adoptive transfer studies and 5-FU treatment
For transplant experiments 6- to 8-week-old recipient mice (C57BL/6-CD45.1) were used after whole-body irradiation with 9.0 Gy. Single-cell suspensions of whole bone marrow were obtained from Ly5.2 animals and transplanted by tail vein injection. The total cell number injected was normalized to the number of hematopoietic stem cells (HSCs) (population A: Lin− Sca1+ CD117+ CD150+ CD48− cells) of wild-type within 4 × 106 bone marrow cells. Sixteen weeks later, recipient mice were euthanized and Ly5.2 donor cell contribution was analyzed using flow cytometry. Treatment was performed as described previously using 5-fluorouracil (5-FU; F 6627, Sigma-Aldrich).21
Cell culture and tumor cell implants
Virus preparation, infection of hematopoietic cells, and establishment of cell lines were performed as described previously.22 Cell lines were characterized performing [3H]-thymidine incorporation assay and cell-cycle analysis as described previously.23 To analyze leukemogenesis, 1 × 105 cells from independently derived p185BCR-ABL-transformed cell lines were intravenously injected into NSG mice. To study subcutaneous tumor formation, 1 × 106 cells were injected into the flank of NSG mice. Mice were euthanized on day 10 and tumor weights were analyzed. To analyze the response to the Cdk4/6 inhibitor PD-0332991 (Pfizer), cells were exposed to different doses of the compound and proliferation was quantified using a CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions.
Transcriptional profiling
Total RNA was extracted from asynchronous cultures using Trizol (Invitrogen). The quality of the RNA obtained was evaluated using the Laboratory-Chip technique (Agilent Bioanalyzer). Samples were then fluorescent-labeled by transcription in vitro using commercial Two-Color Microarray-Based Gene Expression Analysis (Agilent), and the Mouse Gene Expression G3 60K (Agilent) containing ∼56 000 60-mer probes was used. Images were acquired and quantified by means of confocal scanner and software (Agilent G2505C and Feature Extraction). The expression levels were processed using standard methods of normalization, significance analysis, multiple testing correction (Benjamini-Hochberg), and analysis of pathways using Ingenuity Pathways Analysis, Database for Annotation, Visualization and Integrated Discovery,24 and PASTAA.25
Biochemical analysis
Cells were lysed in egg lysis buffer composed of 0.1% NP-40, 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 250 mM NaCl, and 5 mM EDTA in the presence of a protease inhibitor cocktail (Roche). Protein concentrations were determined using the Roti-Quant kit (Roth). Immunoprecipitations were performed overnight at 4°C with 6 µg antibody or 7 µL of serum against Cdk6 (a gift from Mariano Barbacid) and 25 µL Pierce Protein A/G Agarose beads (50% volume to volume ratio; Thermo Scientific). Proteins were separated using a 12% sodium dodecyl sulfate -polyacrylamide gel electrophoresis and blotted onto Whatman nitrocellulose membranes (GE Healthcare). Equal amounts of protein (50 µg) from input, supernatant fractions, and from immunoprecipitation corresponding to 1.5 mg of input cell lysate were loaded per lane, respectively. The following antibodies were used for detection: Cdk4, Cdk6, p16INK4a, p15INK4b, p18INK4c, p19INK4d, insulin-like growth factor-1 (IGF-1), Traf2, and Hsc70 (all from Santa Cruz Biotechnology), nuclear factor-κB (NF-κB) p65 (from Merck Millipore), and phospho-pRb S780 and phospho-pRb S208/811 (from Cell Signaling).
Statistical and imaging analyses
Statistical analyses were performed using analysis of variance (ANOVA) or the Student t or log-rank tests (GraphPad Prism 5). All data are shown as mean ± standard deviation or ± standard error of the mean. Probabilities of P < .05 were considered significant. Images were quantified using ImageJ (National Institutes of Health, Bethesda, MD).
Results
Cdk6 R31C and Cdk4 R24C cooperate in tumor development
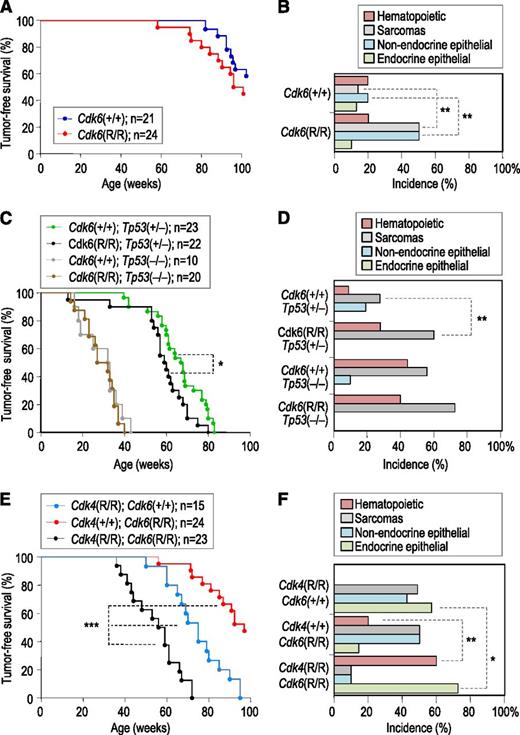
We targeted the mouse Cdk6 locus in mouse embryonic stem cells by homologous recombination with a targeting vector carrying the first exon of Cdk6 and a loxP site-flanked PGK-neo cassette for selection (supplemental Figure 1). The GCCCGC (Ala30-Arg31) sequence located in exon 1 of Cdk6 was replaced by GCATGC, resulting in a Cdk6 protein in which the arginine residue (R31) is replaced by a cysteine (R31C mutation), known to prevent binding to INK4 inhibitors.26,27 Cdk6R/R-homozygous mutants were born at the expected ratio, were fertile, and developed normally during the first months of life. These mutant mice displayed a slightly reduced lifespan when compared with wild-type littermates, which was mostly attributed to a higher incidence of angiosarcomas and epithelial tumors at a greater age (Figure 1A,B). No significant changes in survival or overall disease incidence were observed when maintaining Cdk6R/R mice on a p53-null background, and all animals died within 40 weeks bearing sarcomas and hematopoietic tumors (Figure 1C). In contrast, the presence of p53 heterozygosity unmasked alterations in tumorigenesis resulting from the Cdk6 R31C mutation, as detected by the increased incidence of both hematopoietic tumors and sarcomas in Cdk6R/R; Tp53+/− when compared with Cdk6+/+; Tp53+/− mice or Cdk6R/R; and Tp53+/− mice (Figure 1C-D; supplemental Table 1).
Genetic analysis of Cdk6 R31C mice. (A) Tumor-free survival of wild-type and Cdk6R/R mice. (B) Incidence of tumors in the indicated genotypes. (C) Tumor-free survival of Cdk6 R31C mice in p53-heterozygous or null backgrounds. (D) Incidence of tumors of different origins in the indicated genotypes. The presence of the Cdk6 R31C allele results in increased incidence of sarcomas and hematopoietic tumors only in a p53-heterozygous background. See supplemental Table 1 for details. (E) Tumor-free survival of Cdk4R/R; Cdk6R/R or double Cdk4R/R; Cdk6R/R mutant mice. (F) Tumor distribution in mutant mice with the indicated genotypes. See Table 1 for details. Long-rank test; *P < .05; **P < .01; ***P < .001.
Genetic analysis of Cdk6 R31C mice. (A) Tumor-free survival of wild-type and Cdk6R/R mice. (B) Incidence of tumors in the indicated genotypes. (C) Tumor-free survival of Cdk6 R31C mice in p53-heterozygous or null backgrounds. (D) Incidence of tumors of different origins in the indicated genotypes. The presence of the Cdk6 R31C allele results in increased incidence of sarcomas and hematopoietic tumors only in a p53-heterozygous background. See supplemental Table 1 for details. (E) Tumor-free survival of Cdk4R/R; Cdk6R/R or double Cdk4R/R; Cdk6R/R mutant mice. (F) Tumor distribution in mutant mice with the indicated genotypes. See Table 1 for details. Long-rank test; *P < .05; **P < .01; ***P < .001.
Because INK4 proteins function by inhibiting both Cdk4 and Cdk6, we next generated mice fully insensitive to INK4-mediated inhibition by combining the new Cdk6 R31C allele with the Cdk4 R24C allele.3 Cdk4R/R mice develop a variety of tumors, with angiosarcomas and tumors of endocrine origin being dominant (Figure 1E).16 The combination of Cdk4 R24C and Cdk6 R31C alleles did not alter embryonic development or the proliferation index of normal tissues (not shown) in young animals. However, the survival of Cdk4R/R; Cdk6R/R mice significantly shortened when compared with the single mutants (Figure 1E). This reduction was primarily caused by hematopoietic malignancies that occurred with reduced latency and enhanced incidence compared with Cdk6R/R mice. In addition, we found an increased incidence of endocrine tumors, including neoplasias of the pituitary gland (Figure 1F, Table 1; supplemental Figure 2). The histopathological examination of diseased animals also revealed other pathologies caused by the combination of these 2 alleles, including hypertrophy of ventricular walls in the heart, abundant cysts in the kidney, and altered megakaryocyte morphology and number (Table 1; supplemental Figure 2).
Pathologies in Cdk4 R24C; Cdk6 R31C compound mice
| Pathology . | Cdk4+/+;Cdk6+/+ . | Cdk4R/R; Cdk6+/+ . | Cdk4+/+;Cdk6R/R . | Cdk4R/R; Cdk6R/R . |
|---|---|---|---|---|
| n = 20 . | n = 12 . | n = 16 . | n = 18 . | |
| Tumoral or pretumoral phenotypes | ||||
| Acinar cell adenoma (pancreas) | 5% (103) | 0% | 0% | 0% |
| Angiosarcoma | 0% | 58% (68) | 38% (89) | 11% (72) |
| Bronchial adenocarcinoma | 5% (118) | 33% (71) | 38% (114) | 0% |
| Fibrosarcoma | 5% (80) | 8% (64) | 0% | 0% |
| Histiocytic sarcoma | 5% (86) | 0% | 0% | 22% (54) |
| Leydig cell hyperplasia | 0% | 17% (71) | 0% | 50% (52) |
| Leydig cell tumor | 0% | 50% (69) | 0% | 50% (61) |
| Lung tumors | 0% | 0% | 0% | 11% (61) |
| Lymphoma | 10% (88) | 0% | 19% (83) | 39% (62) |
| Mammary gland hyperplasia | 10% (92) | 17% (83) | 19% (83) | 39% (59) |
| Pancreatic endocrine hyperplasia | 20% (112) | 33% (65) | 19% (84) | 50% (54) |
| Pancreatic endocrine tumor | 0% | 25% (70) | 13% (97) | 11% (72) |
| Pituitary gland hyperplasia | 0% | 17% (54) | 13% (90) | 22% (49) |
| Pituitary gland tumors | 10% (92) | 25% (76) | 13% (77) | 72% (61) |
| T-cell hyperplasia | 5% (80) | 17% (65) | 50% (114) | 0% |
| Rhabdomyosarcoma | 0% | 0% | 13% (139) | 0% |
| Total incidence | 50% | 94% | 88% | 83% |
| Average latency | 97.3 | 69 | 99 | 58.3 |
| Nontumoral phenotypes | ||||
| Bronchus-associated lymphoid tissue | 0% | 0% | 31% (101) | 0% |
| Cardiac defects | 20% (102) | 0% | 13% (85) | 39% (55) |
| Chronic hepatitis | 5% (107) | 0% | 13% (70) | 0% |
| Chronic pancreatitis | 30% (78) | 0% | 13% (85) | 10% (36) |
| Extramedullary hematopoiesis | 5% (103) | 58% (71) | 19% (114) | 22% (50) |
| Kidney disease | 20% (96) | 17% (66) | 19% (104) | 89% (57) |
| Liver steatosis | 20% (91) | 0% | 0% | 11% (44) |
| Megakaryocyte malformation | 0% | 0% | 0% | 78% (60) |
| Splenic atrophy | 0% | 0% | 13% (97) | 0% |
| Vasculitis | 5% (81) | 0% | 19% (90) | 0% |
| Pathology . | Cdk4+/+;Cdk6+/+ . | Cdk4R/R; Cdk6+/+ . | Cdk4+/+;Cdk6R/R . | Cdk4R/R; Cdk6R/R . |
|---|---|---|---|---|
| n = 20 . | n = 12 . | n = 16 . | n = 18 . | |
| Tumoral or pretumoral phenotypes | ||||
| Acinar cell adenoma (pancreas) | 5% (103) | 0% | 0% | 0% |
| Angiosarcoma | 0% | 58% (68) | 38% (89) | 11% (72) |
| Bronchial adenocarcinoma | 5% (118) | 33% (71) | 38% (114) | 0% |
| Fibrosarcoma | 5% (80) | 8% (64) | 0% | 0% |
| Histiocytic sarcoma | 5% (86) | 0% | 0% | 22% (54) |
| Leydig cell hyperplasia | 0% | 17% (71) | 0% | 50% (52) |
| Leydig cell tumor | 0% | 50% (69) | 0% | 50% (61) |
| Lung tumors | 0% | 0% | 0% | 11% (61) |
| Lymphoma | 10% (88) | 0% | 19% (83) | 39% (62) |
| Mammary gland hyperplasia | 10% (92) | 17% (83) | 19% (83) | 39% (59) |
| Pancreatic endocrine hyperplasia | 20% (112) | 33% (65) | 19% (84) | 50% (54) |
| Pancreatic endocrine tumor | 0% | 25% (70) | 13% (97) | 11% (72) |
| Pituitary gland hyperplasia | 0% | 17% (54) | 13% (90) | 22% (49) |
| Pituitary gland tumors | 10% (92) | 25% (76) | 13% (77) | 72% (61) |
| T-cell hyperplasia | 5% (80) | 17% (65) | 50% (114) | 0% |
| Rhabdomyosarcoma | 0% | 0% | 13% (139) | 0% |
| Total incidence | 50% | 94% | 88% | 83% |
| Average latency | 97.3 | 69 | 99 | 58.3 |
| Nontumoral phenotypes | ||||
| Bronchus-associated lymphoid tissue | 0% | 0% | 31% (101) | 0% |
| Cardiac defects | 20% (102) | 0% | 13% (85) | 39% (55) |
| Chronic hepatitis | 5% (107) | 0% | 13% (70) | 0% |
| Chronic pancreatitis | 30% (78) | 0% | 13% (85) | 10% (36) |
| Extramedullary hematopoiesis | 5% (103) | 58% (71) | 19% (114) | 22% (50) |
| Kidney disease | 20% (96) | 17% (66) | 19% (104) | 89% (57) |
| Liver steatosis | 20% (91) | 0% | 0% | 11% (44) |
| Megakaryocyte malformation | 0% | 0% | 0% | 78% (60) |
| Splenic atrophy | 0% | 0% | 13% (97) | 0% |
| Vasculitis | 5% (81) | 0% | 19% (90) | 0% |
Bold-faced type indicates major differences compared with control groups.
Cdk6 R31C mice display defective progenitor potential
We next focused our attention at the hematopoietic system because Cdk6 is known to modulate differentiation and proliferation in different hematopoietic compartments,1,5,6,27 and its activation in double-mutant mice resulted in hematopoietic neoplasias that are otherwise infrequent in Cdk4 R24C mice (Figure 1F; Table 1). Total cell numbers in spleen and bone marrow were comparable in all 4 genotypes (supplemental Figure 2). Young (8 week old) Cdk6R/R mice displayed reduced numbers of T cells and granulocytes in the spleen, which was not evident in Cdk4R/R mice (Figure 2A; supplemental Figure 3). On the other hand, single-mutant Cdk6R/R mice did not show significant alterations in total numbers of B cells (CD3− CD19+), whereas the percentage of these cells was significantly increased in double Cdk4R/R; Cdk6R/R mice, accompanied by differences along the B-cell development as analyzed by determining Hardy fractions. Remarkably, the presence of INK4-insensitive Cdks seemed to affect individual developmental steps differentially; the biggest differences were observed between Hardy fraction B and C (CD19+ BP1− to CD19+ BP1+; Figure 2B).
Altered hematopoiesis in Cdk4 R24C and Cdk6 R31C mice. (A) Analysis of B-, T-, and myeloid cells in the spleen (n = 5 per genotype). (B) Analysis of B-cell development in the bone marrow determining Hardy fractions. (C) Analysis of the LSK fraction in the bone marrow. (D) Analysis of HSCs and multipotent progenitor cells (MPPs). For all experiments described in panels c-d, 5 Cdk4R/R single-mutant mice, 7 Cdk6R/R single mutants and wild-type mice, and 5 double-mutant mice were analyzed and compared. All data are given as the percentage of living cells. Significant differences are indicated as follows: *P < .05; **P < .01; ***P < .001. One-way ANOVA including Bonferroni’s multiple comparison test.
Altered hematopoiesis in Cdk4 R24C and Cdk6 R31C mice. (A) Analysis of B-, T-, and myeloid cells in the spleen (n = 5 per genotype). (B) Analysis of B-cell development in the bone marrow determining Hardy fractions. (C) Analysis of the LSK fraction in the bone marrow. (D) Analysis of HSCs and multipotent progenitor cells (MPPs). For all experiments described in panels c-d, 5 Cdk4R/R single-mutant mice, 7 Cdk6R/R single mutants and wild-type mice, and 5 double-mutant mice were analyzed and compared. All data are given as the percentage of living cells. Significant differences are indicated as follows: *P < .05; **P < .01; ***P < .001. One-way ANOVA including Bonferroni’s multiple comparison test.
The relative percentage of hematopoietic progenitors of the bone marrow was also altered in both Cdk4R/R and Cdk6R/R single mutants. Despite the comparable cellularity in the bone marrow, the percentage of LSK cells was significantly reduced in 8-week-old Cdk4R/R and Cdk6R/R single-mutant mice (Figure 2C and supplemental Figure 3). The combination of both alleles in Cdk4R/R; Cdk6R/R double-mutant mice resulted in a partial recovery in the numbers of these progenitor cells when compared with single mutants (Figure 2C). Similarly, the ratio of HSCs (CD150+ CD48− CD34−), containing the most quiescent HSCs, was reduced in single and double mutants. Again, the combination of the Cdk4 R24C and Cdk6 R31C alleles partially rescued this defect as well as the reduced numbers in multipotent progenitors observed in single-mutant mice (Figure 2D).
We next performed bone marrow transplantation studies into lethally irradiated Ly5.1 mice. Ly5.2 Cdk6R/R progenitors displayed a reduced ability to repopulate as evident from the decreased numbers of LSK and HSC cells 16 weeks after transplantation (Figure 3A). In line with this, reduced numbers of donor-derived T cells, B cells, and granulocytes were found in bone marrow, spleen, and peripheral blood of recipient mice upon transplantation of Cdk6R/R bone marrow (Figure 3B). The concomitant presence of the Cdk4 R24C allele in a Cdk6 R31C background rescued these defects (Figure 3A-B). We also tested the potential of mutant hematopoietic progenitors on 5-FU treatment, which induces death of actively cycling but not quiescent stem cells. The percentage of LSK cells was significantly reduced both in Cdk6R/R and Cdk4R/R mice after 5-FU, whereas double mutants displayed a robust recovery both in LSK number and cellularity in the bone marrow (Figure 3C). A similar rescue in the erythroid lineage was observed after staining for Ter119 in the spleen (supplemental Figure 4). Finally, 7 days after treatment with 5-FU, Cdk6R/R, but not Cdk4R/R, mice displayed a significant defect in the recovery of red blood cells and hematocrit as well as white blood cells and lymphocytes in peripheral blood. These defects were also partially rescued in Cdk4R/R; Cdk6R/R double-mutant mice (supplemental Figure 4), suggesting that the alterations observed in Cdk6 R31C mice could be mediated by the effect of INK4 inhibitors on Cdk4. In summary, these studies show cell intrinsic defects in progenitors in Cdk6 R31C mice that are compensated for by the concomitant presence of a Cdk4 R24C protein.
The Cdk4 R24C allele rescues defective progenitor potential in Cdk6 R31C mice. (A) Analysis of the donor cells’ contribution within the LSK fraction 16 weeks after bone marrow transplantation (n = 5 for wild-type, Cdk4R/R or Cdk6R/R single mutants; n = 4 double-mutant mice). (B) Analysis of the donor contribution to individual lineages in bone marrow, spleen, and peripheral blood (n = 5 for wild-type, Cdk4R/R or Cdk6R/R single mutants; n = 4 double-mutant mice). (C) Analysis of bone marrow repopulation after 5-FU treatment. Flow cytometry analysis of hematopoietic progenitors (LSK cells) 7 days after treatment with 5-FU (n = 3 Cdk4R/R or Cdk6R/R single mutants; n = 6 wild-type mice; n = 7 double-mutant mice). Representative hematoxylin and eosin images of the bone marrow from untreated mice or mice treated with 5-FU mice are included. DMSO, dimethylsulfoxide. Scale bars, 50 μm. *P < .05; **P < .01; ***P < .001. 1-way ANOVA including Bonferroni’s multiple comparison test.
The Cdk4 R24C allele rescues defective progenitor potential in Cdk6 R31C mice. (A) Analysis of the donor cells’ contribution within the LSK fraction 16 weeks after bone marrow transplantation (n = 5 for wild-type, Cdk4R/R or Cdk6R/R single mutants; n = 4 double-mutant mice). (B) Analysis of the donor contribution to individual lineages in bone marrow, spleen, and peripheral blood (n = 5 for wild-type, Cdk4R/R or Cdk6R/R single mutants; n = 4 double-mutant mice). (C) Analysis of bone marrow repopulation after 5-FU treatment. Flow cytometry analysis of hematopoietic progenitors (LSK cells) 7 days after treatment with 5-FU (n = 3 Cdk4R/R or Cdk6R/R single mutants; n = 6 wild-type mice; n = 7 double-mutant mice). Representative hematoxylin and eosin images of the bone marrow from untreated mice or mice treated with 5-FU mice are included. DMSO, dimethylsulfoxide. Scale bars, 50 μm. *P < .05; **P < .01; ***P < .001. 1-way ANOVA including Bonferroni’s multiple comparison test.
Cdk4 R24C and Cdk6 R31C cooperate in increasing proliferation and preventing cell death
To gain further insights in the mechanism underlying these observations and their relevance in cancer, we decided to make use of a well-established model in which bone marrow progenitors are transformed by the BCR-ABL1 oncogene.28,29 Bone marrow progenitors were derived from wild-type, Cdk4R/R; Cdk6R/R, or double Cdk4R/R; Cdk6R/R knock-in mice and transduced by viruses expressing the p185 form of the BCR-ABL1 oncogene. The presence of the Cdk4 R24C or Cdk6 R31C alleles enhanced DNA replication in these cell lines, especially when grown in low-serum conditions (Figure 4A). To analyze their effect during leukemia development, several independent cell lines were injected intravenously into recipient mice. As shown in Figure 4B, leukemia evolved rapidly, and the transformed cells infiltrated bone marrow, spleen, and liver (data not shown). The disease latency of mice injected with cells carrying either Cdk4R/R or Cdk6R/R single knock-in alleles was significantly reduced when compared with mice injected with wild-type tumors. Tumor cells harboring both the Cdk4R/R and Cdk6R/R alleles killed the animals even faster (Figure 4B). In line, Cdk4R/R; Cdk6R/R tumor cells were able to form significantly larger tumors when implanted subcutaneously into recipient mice (supplemental Figure 5). In this experimental system, the effect of Cdk6 R31C was more pronounced, in line with the relevance of this kinase in the hematopoietic system. Tumors derived from double-mutant clones displayed enhanced mitotic index (as monitored by phosphorylation of histone H3) and decreased number of apoptotic cells (as scored by antibodies against the active form of caspase 3; supplemental Figure 5). Thus, in these 2 independent assays, the concomitant presence of the 2 mutant Cdks was able to significantly accelerate tumor growth and progression.
Analysis of BCR-ABL1–induced leukemogenesis. (A) [3H]-thymidine incorporation in low-serum cultures of bone marrow–derived p185BCR-ABL-transformed cells (mean ± standard deviation [SD]; n = 4 clones, Cdk4R/R; Cdk6R/R and n = 3 clones, all other genotypes; 1-way ANOVA including Bonferroni’s multiple comparison test). (B) Survival of NSG mice after intravenous injection of p185BCR-ABL–transformed cell lines. Data from 3 independent experiments are presented; log-rank test). (C) Analysis of the cell-cycle profile in the absence or presence of 255 nM PD-0332991 (mean ± SD; n = 4 clones for all genotypes). (D) Dose response curves for PD-0332991 after 48 hours’ treatment (mean ± SD; n = 4 clones for all genotypes). (E) A summary of transcripts up- (red) or down- (green) regulated (fold-change >2) in Cdk4R/R; Cdk6R/R compared with wild-type clones (n = 3 clones per genotype). (F) Transcripts deregulated in BCR-ABL1–transformed Cdk4R/R; Cdk6R/R cells displayed promoter sequences significantly enriched compared with BCR-ABL1–transformed wild-type cultures) in recognition sites for the indicated transcription factors. (G) Validation of the differential expression of specific proteins, predicted in the microarray analysis, using immunoblot (p65 NF-kB, IGF-1 Traf2) or flow cytometry (interleukin-2Ra [IL-2Ra]) analysis using 4 individual cell clones per genotype; 1-way ANOVA including Bonferroni’s multiple comparison test; *P < .05; **P < .01).
Analysis of BCR-ABL1–induced leukemogenesis. (A) [3H]-thymidine incorporation in low-serum cultures of bone marrow–derived p185BCR-ABL-transformed cells (mean ± standard deviation [SD]; n = 4 clones, Cdk4R/R; Cdk6R/R and n = 3 clones, all other genotypes; 1-way ANOVA including Bonferroni’s multiple comparison test). (B) Survival of NSG mice after intravenous injection of p185BCR-ABL–transformed cell lines. Data from 3 independent experiments are presented; log-rank test). (C) Analysis of the cell-cycle profile in the absence or presence of 255 nM PD-0332991 (mean ± SD; n = 4 clones for all genotypes). (D) Dose response curves for PD-0332991 after 48 hours’ treatment (mean ± SD; n = 4 clones for all genotypes). (E) A summary of transcripts up- (red) or down- (green) regulated (fold-change >2) in Cdk4R/R; Cdk6R/R compared with wild-type clones (n = 3 clones per genotype). (F) Transcripts deregulated in BCR-ABL1–transformed Cdk4R/R; Cdk6R/R cells displayed promoter sequences significantly enriched compared with BCR-ABL1–transformed wild-type cultures) in recognition sites for the indicated transcription factors. (G) Validation of the differential expression of specific proteins, predicted in the microarray analysis, using immunoblot (p65 NF-kB, IGF-1 Traf2) or flow cytometry (interleukin-2Ra [IL-2Ra]) analysis using 4 individual cell clones per genotype; 1-way ANOVA including Bonferroni’s multiple comparison test; *P < .05; **P < .01).
The ability of Cdk4R/R; Cdk6R/R BCR-ABL1–transformed cell lines to grow in vitro allowed us to determine cell-cycle kinetics and revealed a reduced G0/G1 phase and high ratio of S/G2/M cells in the absence of a significant level of apoptosis when compared with wild-type or single-mutant mice (Figure 4C). Double knock-in cells were also more resistant than the single mutants or wild-type cells to the Cdk4/6 inhibitor PD-0332991 (Figure 4D), indicating the relevance of Cdk4/6 kinase activity for the growth of these leukemic cells.
We next compared the transcriptional profiles of asynchronous Cdk4R/R; Cdk6R/R to control cultures. Genes deregulated in these mutant cells (723 transcripts upregulated and 986 downregulated; fold-change >2; Figure 4E; supplemental Table 2) were enriched in targets of the transcription factors Egr1/2, Zf5, Rfx1, Atf4, Ctf1/2, Lrf, E12, or E2f, among others (Figure 4F). Gene ontology (GO) analysis of deregulated genes also indicated control of apoptosis as the major cellular process deregulated in double-mutant cultures (Table 2; supplemental Table 3). These INK4-insensitive cells also displayed significant differences in molecular pathways involved in cell adhesion and motion or regulation of cytokine signaling (Table 2; supplemental Table 3). The deregulation of some critical molecules involved in apoptosis or other major pathways of relevance in B-cell leukemias was validated using immunoblot (p65NFkB, IGF-1, Traf2) or cytometry analysis (interleukin-2Ra; Figure 4G). Altogether, these data suggest the involvement of multiple signaling pathways, in addition to the retinoblastoma/E2F route, in the response to INK4 insensitivity in Cdk4R/R; Cdk6R/R leukemic cells.
GO analysis of transcripts deregulated in Cdk4R/R; Cdk6R/R BCR-ABL1–transformed cells
| Annotation clusters . | Description . | Representative genes upregulated . | Representative genes downregulated . |
|---|---|---|---|
| Apoptosis | GO:0043065, positive regulation of apoptosis; GO:0043067, regulation of programmed cell death; GO:0006917, induction of apoptosis; GO:0006916, anti-apoptosis; GO:0043066, negative regulation of apoptosis | AIPL1, ANK2, CDK5R1, CDKN2A, CHST11, DFFA, EAF2, FGFR3, IFIH1, IGF1, IL2RB, NOTCH2, NR4A1, PHLDA3, SPP1, TCF7, TGM2, TRAF2 | BBC3, BCL2A1D, BCL2A1C, BCL2L11, BCL3, BNIP3L, BTG2, CARD11, CD74, CDH1, CEBPB, CUL7, DDIT3, DMRT2, FAS, FCGR1, GCH1, IL18, IL2RA, LST1, LTB, MAPK8IP1, NLRC4, NUPR1, PROK2, RRM2B, SPHK1, TRP63, TSC22D3, UACA, VDR |
| Cell adhesion | GO:0007155, cell adhesion; PIR: cell adhesion; GO:0016337, cell–cell adhesion | ALCAM, APP, CD33, CD97, CDK5R1, CELSR1, ESAM, F11R, FBLN7, HSPG2, IBSP, ITGAM, L1CAM, LGALS3BP, NCAM2, NRP2, PCDH17, PCDHB16, PCDHB17, PCDHB20, PGM5, SPP1, SSPO | ARHGAP6, BCL2L11, CD36, CDH1, CLDN12, COL6A6, DSG2, FAT1, FNDC3A, ITGB4, LMO7, MEGF10, MYBPC2, NLGN2, PCDH11X, PVRL2, ROBO2, SELL, THY1, TRO |
| Cell projection and motion | GO:0007411, axon guidance; GO:0030030, cell projection organization; GO:0048858, cell projection morphogenesis; GO:0032989, cellular component morphogenesis; GO:0006928, cell motion | ALCAM, ANK3, APP, CDK5R1, DNAJA1, EFHD1, GM4776, ITGAM, L1CAM, NR2F1, NRP2, SEMA3A, SHROOM2, SOX6, TBR1, TNNT2, TNS3, VAV3, WWTR1 | ARID5B, BBS4, CHRNB2, EFNA5, EGR2, ETV1, FAT1, FEZF2, FGD3, ITGB4, LHX2, LPIN1, LST1, NEURL2, PDGFA, PLEKHG5, PTK2, RHOX5, ROBO2, ROBO3, RYK, TTC8, VANGL2 |
| Regulation of neurological processes | GO:0051969, regulation of transmission of nerve impulse; GO:0044057, regulation of system process; GO:0031644, regulation of neurological system process; GO:0019226, transmission of nerve impulse | APP, CELSR1, DLG2, IRX5, KCNMA1, MAOA, PCDHB16, SEMA3A, TNNT2, TRF | CHRNB2, COLQ, DRD5, EGR2, FKBP1B, GRIA2, GRIK5, GRM8, HEXA, LST1, LTB, NLGN2, P2RX3, PARK2, PROK2, PTK2 |
| Regulation of cytokine signaling | GO:0019955, cytokine binding; GO:0004896, cytokine receptor activity; GO:0019221, cytokine-mediated signaling pathway | IL1R1, IL1RAP, IL2RB | ACVR2B, CCR2, CD74, CXCR3, CXCR5, EBI3, IL1R2, IL12RB2, LIFR |
| Annotation clusters . | Description . | Representative genes upregulated . | Representative genes downregulated . |
|---|---|---|---|
| Apoptosis | GO:0043065, positive regulation of apoptosis; GO:0043067, regulation of programmed cell death; GO:0006917, induction of apoptosis; GO:0006916, anti-apoptosis; GO:0043066, negative regulation of apoptosis | AIPL1, ANK2, CDK5R1, CDKN2A, CHST11, DFFA, EAF2, FGFR3, IFIH1, IGF1, IL2RB, NOTCH2, NR4A1, PHLDA3, SPP1, TCF7, TGM2, TRAF2 | BBC3, BCL2A1D, BCL2A1C, BCL2L11, BCL3, BNIP3L, BTG2, CARD11, CD74, CDH1, CEBPB, CUL7, DDIT3, DMRT2, FAS, FCGR1, GCH1, IL18, IL2RA, LST1, LTB, MAPK8IP1, NLRC4, NUPR1, PROK2, RRM2B, SPHK1, TRP63, TSC22D3, UACA, VDR |
| Cell adhesion | GO:0007155, cell adhesion; PIR: cell adhesion; GO:0016337, cell–cell adhesion | ALCAM, APP, CD33, CD97, CDK5R1, CELSR1, ESAM, F11R, FBLN7, HSPG2, IBSP, ITGAM, L1CAM, LGALS3BP, NCAM2, NRP2, PCDH17, PCDHB16, PCDHB17, PCDHB20, PGM5, SPP1, SSPO | ARHGAP6, BCL2L11, CD36, CDH1, CLDN12, COL6A6, DSG2, FAT1, FNDC3A, ITGB4, LMO7, MEGF10, MYBPC2, NLGN2, PCDH11X, PVRL2, ROBO2, SELL, THY1, TRO |
| Cell projection and motion | GO:0007411, axon guidance; GO:0030030, cell projection organization; GO:0048858, cell projection morphogenesis; GO:0032989, cellular component morphogenesis; GO:0006928, cell motion | ALCAM, ANK3, APP, CDK5R1, DNAJA1, EFHD1, GM4776, ITGAM, L1CAM, NR2F1, NRP2, SEMA3A, SHROOM2, SOX6, TBR1, TNNT2, TNS3, VAV3, WWTR1 | ARID5B, BBS4, CHRNB2, EFNA5, EGR2, ETV1, FAT1, FEZF2, FGD3, ITGB4, LHX2, LPIN1, LST1, NEURL2, PDGFA, PLEKHG5, PTK2, RHOX5, ROBO2, ROBO3, RYK, TTC8, VANGL2 |
| Regulation of neurological processes | GO:0051969, regulation of transmission of nerve impulse; GO:0044057, regulation of system process; GO:0031644, regulation of neurological system process; GO:0019226, transmission of nerve impulse | APP, CELSR1, DLG2, IRX5, KCNMA1, MAOA, PCDHB16, SEMA3A, TNNT2, TRF | CHRNB2, COLQ, DRD5, EGR2, FKBP1B, GRIA2, GRIK5, GRM8, HEXA, LST1, LTB, NLGN2, P2RX3, PARK2, PROK2, PTK2 |
| Regulation of cytokine signaling | GO:0019955, cytokine binding; GO:0004896, cytokine receptor activity; GO:0019221, cytokine-mediated signaling pathway | IL1R1, IL1RAP, IL2RB | ACVR2B, CCR2, CD74, CXCR3, CXCR5, EBI3, IL1R2, IL12RB2, LIFR |
Most significantly deregulated functional pathways and their GO codes are shown first. See supplemental Table 2 for further details.
Cdk6 sequesters p16INK4a inhibitors away from Cdk4
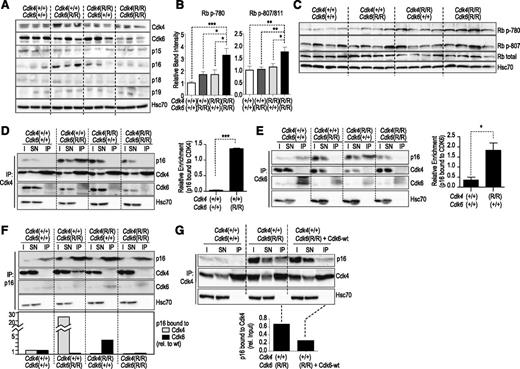
The availability of BCR-ABL1–transformed cell lines carrying the different knock-in alleles allowed us to investigate and compare the levels of cell-cycle regulatory proteins side by side. The level of expression of Cdk4 R24C or Cdk6 R31C mutant proteins from the corresponding alleles was similar to the wild-type isoforms, although Cdk6 R31C levels were significantly reduced in double Cdk4R/R; Cdk6R/R knock-in cells. The levels of p16INK4a were increased in single Cdk4R/R or Cdk6R/R tumor cells (Figure 5A and supplemental Figure 5), in agreement with previous reports showing that p16INK4a is induced as a consequence of ectopic Cdk4 or Cdk6 activity.30,31 Importantly, the concomitant presence of the Cdk4 R24C and Cdk6 R31C alleles resulted in a significant increase in the phosphorylation of the Cdk4/6 target retinoblastoma protein (Rb; Figure 5B,C).
Biochemistry in p185BCR-ABL-transformed cell lines. (A) Expression levels of Cdk4, Cdk6, and INK family members in 3 individually derived cell lines of each genotype as indicated. (B) Phosphorylation of Rb at Ser780 or Ser807/811 is significantly increased in Cdk4R/R; Cdk6R/R as compared with single-mutant or control cultures. Depicted is the summary from 3 independent western blots using 3 or 4 independent p185BCR-ABL–transformed cell lines. *P < .05; **P < .01; ***P < .001. One-way ANOVA including Bonferroni’s multiple comparison test. (C) Representative blots of Rb phosphorylation in the indicated residues in 4 independent clones per genotype. (D) Protein levels of p16INK4a, Cdk4, and Cdk6 after immunoprecipitation (IP) with antibodies against Cdk4. I and SN indicate the input lysate or supernatant after IP. The relative enrichment of p16INK4a complexed with Cdk4 calculated from 2 individual IP experiments is shown in the histogram. Student t test; ***P < .001. (E) Protein levels of p16INK4a, Cdk4, and Cdk6 after IP with antibodies against Cdk6 (IP). I and SN indicate the input lysate or supernatant after IP. The histogram depicts the relative enrichment of p16INK4a complexed with Cdk6 calculated from 3 individual IP experiments. Student t test; *P < .05. (F) Protein levels of p16INK4a, Cdk4, and Cdk6 after IP with antibodies against p16INK4a. I and SN indicate the input lysate or supernatant after IP. The histogram shows the quantification (relative to wild-type cells) of the amount of p16INK4a bound to Cdk4 or Cdk6 in the different clones. (G) Overexpression of wild-type Cdk6 (Cdk6-wt) reduces the levels of Cdk4-bound p16 in Cdk4+/+; Cdk6R/R cells. In all of these assays, Hsc70 was used as a loading control.
Biochemistry in p185BCR-ABL-transformed cell lines. (A) Expression levels of Cdk4, Cdk6, and INK family members in 3 individually derived cell lines of each genotype as indicated. (B) Phosphorylation of Rb at Ser780 or Ser807/811 is significantly increased in Cdk4R/R; Cdk6R/R as compared with single-mutant or control cultures. Depicted is the summary from 3 independent western blots using 3 or 4 independent p185BCR-ABL–transformed cell lines. *P < .05; **P < .01; ***P < .001. One-way ANOVA including Bonferroni’s multiple comparison test. (C) Representative blots of Rb phosphorylation in the indicated residues in 4 independent clones per genotype. (D) Protein levels of p16INK4a, Cdk4, and Cdk6 after immunoprecipitation (IP) with antibodies against Cdk4. I and SN indicate the input lysate or supernatant after IP. The relative enrichment of p16INK4a complexed with Cdk4 calculated from 2 individual IP experiments is shown in the histogram. Student t test; ***P < .001. (E) Protein levels of p16INK4a, Cdk4, and Cdk6 after IP with antibodies against Cdk6 (IP). I and SN indicate the input lysate or supernatant after IP. The histogram depicts the relative enrichment of p16INK4a complexed with Cdk6 calculated from 3 individual IP experiments. Student t test; *P < .05. (F) Protein levels of p16INK4a, Cdk4, and Cdk6 after IP with antibodies against p16INK4a. I and SN indicate the input lysate or supernatant after IP. The histogram shows the quantification (relative to wild-type cells) of the amount of p16INK4a bound to Cdk4 or Cdk6 in the different clones. (G) Overexpression of wild-type Cdk6 (Cdk6-wt) reduces the levels of Cdk4-bound p16 in Cdk4+/+; Cdk6R/R cells. In all of these assays, Hsc70 was used as a loading control.
We next asked whether p16INK4a, in the presence of the Cdk6 R31C mutant allele, would increasingly bind wild-type Cdk4. As shown in Figure 5D, whereas p16INK4a was almost undetectable in Cdk4 complexes in wild-type BCR-ABL1 cells as well as in Cdk4R/R; Cdk6+/+ or Cdk4R/R; Cdk6R/R clones, it efficiently bound Cdk4 in Cdk4+/+; Cdk6R/R cells. In control Cdk4+/+; Cdk6+/+ BCR-ABL1–transformed clones, Cdk6 complexes contained p16INK4a (Figure 5E), whereas, as expected, this inhibitor was not present in clones carrying the Cdk6 R31C mutant. Again, the amount of p16INK4a bound to Cdk6 increased in Cdk4R/R; Cdk6+/+ clones. Similar results were obtained in experiments in which we performed direct immunoprecipitation of p16INK4a and analyzed attached kinases (Figure 5F). Whereas p16INK4a did not form complexes with Cdk4 in control cells, it was readily detected in Cdk4 complexes in Cdk4+/+; Cdk6R/R cells. Vice versa, we observed an enrichment in Cdk6-p16INK4a complexes in Cdk4R/R; Cdk6+/+ cells. The absence of p16INK4a in Cdk4 or Cdk6 complexes in Cdk4R/R; Cdk6R/R cells correlated with increased phosphorylation of Rb (Figure 5B,C), the major substrate of these kinases. Overexpression of exogenous wild-type Cdk6 in Cdk4+/+; Cdk6R/R cells was sufficient to reduce the levels of Cdk4-bound p16INK4a (Figure 5G), suggesting the relevance of Cdk4/6 protein levels in counteracting the activity of INK4 inhibitors. Taken together, our findings reveal that expression of single Cdk4 R24C or Cdk6 R31C mutants suffices to shift binding of the cell-cycle inhibitor p16INK4a to the remaining wild-type Cdk6 or Cdk4, respectively, exerting a compensatory inhibitor effect. Only the presence of both INK4a-insensitive alleles enables the spontaneous development of hematopoietic tumors by completely preventing the tumor suppressor activity of INK4 proteins.
Discussion
Cdk4 and Cdk6 are 2 highly related Cdks activated by D-type cyclins and inhibited by INK4 proteins.1 Despite their similarity, specific functions have been proposed in different cell types, such as astrocytes,32 osteoblasts,33 or prostate cells.34 In vivo, whereas genetic ablation of Cdk4 results in specific deficiencies in pancreatic and pituitary endocrine cells,3,4 disruption of the Cdk6 gene results in reduced red blood cells and altered proliferation of T cells,5 suggesting differential functions for this kinase in the hematopoietic system. Cdk6 and Cdk4 also have separate activities in erythroblast differentiation, with nuclear activity of Cdk6 and cytoplasmic activity of Cdk4. Cdk6 blocks erythroid differentiation,35 but as cells commit to differentiation and Cdk6 declines, increasing amounts of Cdk4 appear in the nucleus,36 suggesting that Cdk6 controls proliferation of erythroblasts, whereas Cdk4 controls cell division in differentiated cells. Cdk6 is highly expressed in thymocytes, and Cdk6-cyclin D2 complexes are proposed to be involved in the acquisition of the competent state in human T lymphocytes.6,37 Cdk6 depletion causes reduced thymocyte cell numbers with a specific block in DN3 cells,6 a phenotype reminiscent of that observed in cyclin D3-null mice.38 Moreover, Cdk6 is also highly expressed in the cytoplasm of CD8 memory cells, favoring rapid division and expansion of these mature cells.39
Cdk4 and Cdk6 are frequently deregulated in human cancer through inactivation of INK4 inhibitors,2,7,11 and several INK4 family members are codeleted in specific tumor types, suggesting cooperation between family members.40,41 In the mouse, individual genetic ablation of INK4 inhibitors display limited effect in tumor formation, and codeletion of 2 family members results in some additive effects, at least in sarcomas and endocrine tumors.9,10,18 However, these assays have been limited by the complexity of combining mutant alleles for all the INK4 genes. The analysis of Cdk4R/R; Cdk6R/R mice (Figure 1; Table 1) suggests additional compensatory roles among the INK4 family, at least in hematopoietic and endocrine tumors.
Whereas specific functions for Cdk4 or Cdk6 have been clearly established in normal tissues, their separate or synergistic functions during tumor development are less understood. Cdk4 is mostly amplified in sarcomas,42-44 whereas the Cdk4 R24C mutation is specifically found in melanoma.14,15 Cdk6, on the other hand, is translocated and amplified in multiple hematopoietic neoplasms such as T- and B-cell lymphomas and leukemias45-50 and acute myeloid leukemias.51 Our study uncovers an oncogenic role for Cdk6 R31C in a Cdk4R/R background for hematopoietic tumors (Figures 1 and 4). As suggested recently, Cdk6 may have specific roles in these hematopoietic neoplasias because this kinase functions as a member of transcription complexes that stimulate different pathways, including angiogenesis.31,52 Yet, many human tumors including lymphoblastic leukemias overexpress both kinases (supplemental Figure 6). Our study provides the rationale for concomitant overexpression of both Cdk4 and Cdk6 in hematopoietic tumors, especially in those cases in which at least an INK4 protein is still present. In the presence of the Cdk6 R31C allele, p16INK4a is increasingly bound to Cdk4, leading to limited phosphorylation of pRb (Figure 5). Concomitant expression of the INK4-insensitive allele Cdk4 R24C results in enhanced phosphorylation of pRb and proliferation of hematopoietic precursors and tumoral cells (Figures 2-5), suggesting that these 2 cyclin D–dependent kinases cooperate not only by phosphorylating common or specific substrates but also by maintaining inhibitors away from the other family member.
The relevance of Cdk4/6 activity in cancer therapy has been recently highlighted by the success of Cdk4/6 inhibitors in phase 2 clinical trials in hormone-positive breast cancer.53 In fact, research in the past decade using mouse models has suggested the possible benefit of inhibiting Cdk4/6 kinases in breast, lung, and hematopoietic tumors, among others.54 Whereas Cdk4/6 inhibition results in the induction of senescence in epithelial tumors, hematopoietic cells are more susceptible to apoptotic cell death. Our expression data in Cdk4R/R; Cdk6R/R BCR-ABL1–transformed cells suggests that Cdk4/6 activity controls several apoptotic routes in addition to other molecular pathways (Table 2 and Figure 4). Further work on these pathways may provide new biomarkers of clinical use and will be instrumental to understand the effect of these cell-cycle kinases in leukemogenesis.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE59024).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank O. Domínguez, Gonzalo Gómez, and David G. Pisano (Spanish National Cancer Research Centre) for help with microarray and bioinformatics analysis and D. Santamaría and M. Barbacid for reagents.
This work was supported by grants from the Austrian Science Foundation (FWF) (SFB47 and P24297) (V.S.), fellowships from the Spanish Ministerio de Economía y Competitividad (MINECO) (E.R.-D., V.Q.), and grants from MINECO (SAF2012-38215), Fundación Ramón Areces, the OncoCycle Programme (S2010/BMD-2470) from the Comunidad de Madrid, the OncoBIO Consolider-Ingenio 2010 Programme (CSD2007-00017) from MINECO, and the European Union Seventh Framework Programme (MitoSys project; HEALTH-F5-2010-241548) (M.M.).
Authorship
Contribution: E.R.-D., V.Q., F.B., and M.P.-M. performed most of the experiments; M.E., M.G.d.C., and D.M. helped with cytometry analysis and the analysis of mouse models; D.P. contributed to the microarray and histological studies; P.D. and M.C. performed the histopathological analysis; and V.S. and M.M. designed and supervised the project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Veronika Sexl, Institute of Pharmacology and Toxicology, Veterinary University of Vienna, Veterinaerplatz 1, A-1210, Vienna, Austria; e-mail: veronika.sexl@vetmeduni.ac.at; and Marcos Malumbres, Cell Division and Cancer Group, Spanish National Cancer Research Centre, Melchor Fernandez Almagro 3, E-28029, Madrid, Spain; e-mail: malumbres@cnio.es.
References
Author notes
E.R.-D., V.Q., F.B., and M.P.-M. contributed equally to this work.



![Figure 4. Analysis of BCR-ABL1–induced leukemogenesis. (A) [3H]-thymidine incorporation in low-serum cultures of bone marrow–derived p185BCR-ABL-transformed cells (mean ± standard deviation [SD]; n = 4 clones, Cdk4R/R; Cdk6R/R and n = 3 clones, all other genotypes; 1-way ANOVA including Bonferroni’s multiple comparison test). (B) Survival of NSG mice after intravenous injection of p185BCR-ABL–transformed cell lines. Data from 3 independent experiments are presented; log-rank test). (C) Analysis of the cell-cycle profile in the absence or presence of 255 nM PD-0332991 (mean ± SD; n = 4 clones for all genotypes). (D) Dose response curves for PD-0332991 after 48 hours’ treatment (mean ± SD; n = 4 clones for all genotypes). (E) A summary of transcripts up- (red) or down- (green) regulated (fold-change >2) in Cdk4R/R; Cdk6R/R compared with wild-type clones (n = 3 clones per genotype). (F) Transcripts deregulated in BCR-ABL1–transformed Cdk4R/R; Cdk6R/R cells displayed promoter sequences significantly enriched compared with BCR-ABL1–transformed wild-type cultures) in recognition sites for the indicated transcription factors. (G) Validation of the differential expression of specific proteins, predicted in the microarray analysis, using immunoblot (p65 NF-kB, IGF-1 Traf2) or flow cytometry (interleukin-2Ra [IL-2Ra]) analysis using 4 individual cell clones per genotype; 1-way ANOVA including Bonferroni’s multiple comparison test; *P < .05; **P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/15/10.1182_blood-2014-02-555292/4/m_2380f4.jpeg?Expires=1763462941&Signature=xalj5Ob-~Wjq-bSELS1ghXTca-No7i6CzVzQSLgG4TWVCr4H~wORJexDnhmglkNOr3q84eFJDm8INABwtBaowUWnSpS8JwRWWHPrPudeRh9ZvIAsC~3Fue-KChNdHDkS2hgfY9pcjYWIlHsCaAlv2MBoF4-NXPn9kHqwWtQaE731KUv25AejPV8mUDSh~SgyhUCOqhnR1PnY5aqQOrbaqa5-HOAPpqsSOVBNwTbJUTx4RWjK2fl9cP2qVN0JWVqCCZXzGrPXv0e7PTGaSETM1MU74CD3XrtN7LbZ-cl51gJHFlOX6nBWZr9NyFuLAIm5HK0fPJqrM7OrgQQ2zyQyvA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal