Key Points
Recurrent hypomorphic cohesin defects and cohesin low expression were identified in a significant proportion of patients with MDS and AML.
Cohesin mutations likely represent secondary events in clonal hierarchy and contribute to clonal transformation.
Abstract
Somatic cohesin mutations have been reported in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). To account for the morphologic and cytogenetic diversity of these neoplasms, a well-annotated cohort of 1060 patients with myeloid malignancies including MDS (n = 386), myeloproliferative neoplasms (MPNs) (n = 55), MDS/MPNs (n = 169), and AML (n = 450) were analyzed for cohesin gene mutational status, gene expression, and therapeutic and survival outcomes. Somatic cohesin defects were detected in 12% of patients with myeloid malignancies, whereas low expression of these genes was present in an additional 15% of patients. Mutations of cohesin genes were mutually exclusive and mostly resulted in predicted loss of function. Patients with low cohesin gene expression showed similar expression signatures as those with somatic cohesin mutations. Cross-sectional deep-sequencing analysis for clonal hierarchy demonstrated STAG2, SMC3, and RAD21 mutations to be ancestral in 18%, 18%, and 47% of cases, respectively, and each expanded to clonal dominance concordant with disease transformation. Cohesin mutations were significantly associated with RUNX1, Ras-family oncogenes, and BCOR and ASXL1 mutations and were most prevalent in high-risk MDS and secondary AML. Cohesin defects were associated with poor overall survival (27.2 vs 40 months; P = .023), especially in STAG2 mutant MDS patients surviving >12 months (median survival 35 vs 50 months; P = .017).
Introduction
The use of high-throughput next-generation sequencing for somatic mutational profiling has led to the identification of recurrent somatic mutations within the cohesin complex in patients with acute myeloid leukemia (AML),1,2 myelodysplastic syndrome (MDS),3 glioblastoma multiforme,4 Ewing sarcoma,5 and colorectal6 and bladder carcinomas.7,8 Cohesin is a multiprotein complex that has an established role as an effector of sister-chromatid cohesin during metaphase. The cohesin complex is composed of 2 long structural maintenance proteins, SMC1A and SMC3, which heterodimerize at the hinge domain, forming a closed loop by binding at the α-klesin end to RAD21 and adapter proteins STAG1/STAG2.9 Despite the demonstrated role of cohesin in mitosis, the mechanism by which altered cohesin function leads to clonal expansion and malignant transformation has not been elucidated. Given the role of cohesin in alignment of sister chromatids during cellular replication,10 cohesin mutations could theoretically lead to chromosomal instability.5,6 However, in AML, cohesin mutations have not been associated with aneuploidy or complex cytogenetics,2 suggesting an alternate mechanism of transformation.11
Perturbation of other functions of cohesin complex genes may contribute to hematopoietic transformation, particularly through their role in regulating gene expression12,13 and DNA-loop formation.14,15 More recently, cohesin has been found to localize at superenhancers, which are locus-control regions with broad peaks of H3K27 acetylation. These sites regulate the expression of specific genes that govern physiological cell identity and pluripotency/self-renewal in malignancy and in embryonic stem cells.14,16 These data suggest the possibility that loss-of-function cohesin mutations alter transcriptional orchestration of differentiation/proliferation and may have a role in clonal evolution and tumorigenesis. Notably, germline cohesin mutations in NIBPL, SMC1A, SMC3, and HDAC8 occur in patients with Cornelia deLange syndrome,17 hallmarked by craniofacial abnormalities and cognitive deficits. Although no propensity to malignancy has been described, these children have a markedly abbreviated lifespan, abrogating the oncogenic propensity of these defects. Somatic loss of function of HDAC8 impairs SMC3 deacetylation, leading to decreased cohesin occupancy and altered transcriptional programs,18 underscoring a role for cohesin in gene regulation.14,19
Somatic cohesin mutations have been reported in several cohorts of patients with de novo AML.2,3 Given the molecular heterogeneity of myeloid neoplasms, precise establishment of the clinical impact of these leukemia disease alleles and their relevance to clinical phenotype, somatic genotype, and clinical outcome requires large patient cohorts with detailed clinical annotation. We therefore studied a cohort of 1060 patients with MDS, myeloproliferative neoplasms (MPNs), MDS/MPN overlap syndromes, and primary and secondary AML (pAML and sAML, respectively) to elucidate the clinical characteristics of patients with myeloid neoplasms harboring cohesin mutations with respect to disease phenotype, genetic background, therapeutic response, and clinical outcome.
Materials and methods
Patient population
The mutational status for cohesin complex genes (STAG2, STAG1, RAD21, SMC3, SMC1A, PDS5B, WAPAL, MAU2, REC8, NIPBL, SMC5, and ESCO2) were analyzed in bone marrow and blood specimens of 1060 patients with MDS (n = 386), MPNs (n = 55), MDS/MPNs (n = 169), and sAML (n = 149) that evolved from these conditions and pAML (n = 301), including 200 patients in The Cancer Genome Atlas (TCGA) AML cohort (see supplemental Table 1, available at the Blood Web site). MDS subtypes of refractory anemia (RA), RA with ringed sideroblasts, and refractory cytopenia with multilineage dysplasia were classified as low-risk MDS. RA with excess blasts (RAEB1 and RAEB2) cases were considered as high-risk MDS. Informed consent was obtained according to protocols approved by the institutional review boards and in accordance with the Declaration of Helsinki. Diagnosis was confirmed at each institution according to World Health Organization classification criteria. Clinical parameters studied included age, gender, overall survival (OS), and blood counts. All of these parameters were available for 780 patients in this cohort after excluding 200 pAML TCGA cases and 80 patients with incomplete clinical details. Metaphase cytogenetics details were retrieved from 892 patients. The median follow-up of the cohort was 20.4 months (range, 1-282 months).
Whole-exome sequencing and targeted multiamplicon deep sequencing
For whole-exome sequencing, the 50 Mb of protein coding sequences was enriched from total genomic DNA by liquid-phase hybridization using SureSelect (version 4) (Agilent Technology, Santa Clara, CA), followed by massively parallel sequencing with HiSequation 2000 (Illumina, San Diego, CA). Data were analyzed using our in-house pipeline for somatic mutation calling (Genomon) as previously described (http://genomon.hgc.jp/exome/en/index.html).20 To minimize false positives and focus on the most prevalent or relevant somatic events, we implemented a rational bioanalytic filtering approach and applied heuristic bioanalytic pipelines. We used 2 independent pipelines for identifying somatic and germline alterations. For confirmation of somatic mutations, we analyzed paired germline DNA from CD3+ lymphocytes. The selected observations were validated by targeted deep sequencing using MiSeq. Our sequence library for deep sequencing was generated by TruSeqCustom Amplicon (Illumina). Moreover, cohesin gene mutations were screened using whole-exome sequencing results available through TCGA (http://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp). To detect germline variants of cohesin genes, our strategy was to select nonsynonymous variants present in both somatic and germline samples that are possibly deleterious. Further, the putative candidates were prioritized based on population genotypic frequency. For the purpose of this study, nonsynonymous variants known to be pathogenic, rare, or with unknown genotypic frequency variants in general population are included. The mean coverage for cohesin genes (STAG2, SMC3, and RAD21) is 265× for the deep-sequencing samples.
Expression array and RNA sequencing
RNA sequencing from TCGA patients was downloaded from the TCGA repository. They were aligned to hg19 using STAR.21 Differential expression analysis for this data set was performed using the Bioconductor DESeq2 package.22 Patients with t(15;17) were excluded from any analyses. Cutoffs for low expression of STAG2 was considered the bottom 7.5% transcript levels, and high expressors were considered the top 25%.
Candidate genes identified were validated using 2 external validation cohorts. Affymetrix U133plus2 CEL files for 183 MDS patients and 17 healthy controls in Gene Expression Omnibus GSE19429 were pooled using Bioconductor’s gcrma tools for quantile normalization. Histograms of log2 expression values were then used to identify cutoffs that defined low expression of STAG2, RAD21, and SMC3 as compared with the cohort of healthy donors. Genes differentially expressed between these 2 groups were then identified using the Bioconductor package limma and Benjamini-Hochberg adjusted (ie, to correct for multiple hypothesis testing23 ) P values of ≤.001; specifically, we used the limma pipeline of voom() ->lmFit() ->eBayes(), which can be applied to both microarray and RNA-sequencing data.24 Further analyses were performed using oncomine (https://www.oncomine.org/).
Statistical analyses
Comparisons of proportions were performed by the χ2 test and by Fisher’s exact test, and differences in values and in ranks were assessed by Student t tests and Mann-Whitney U tests, respectively. Cox models were used to identify correlates with OS. Examination of log (−log) survival plots and partial residuals confirmed that the underlying assumption of proportional hazards was met except in the case wherein survival curves are equal during the first 12 months and separate drastically thereafter (Figure 3); we therefore dichotomized follow-up into before and after 12 months. Significant covariates were dichotomized at their medians, and resulting 2-group log-rank test differences were visualized using Kaplan-Meier methods. Throughout, 2-sided tests were used with significance defined as α <.05. These analyses were performed using JMP9 (SAS, Cary, NC) and R-statistical language.
Results
Cohesin gene family sequence alterations in myeloid neoplasms
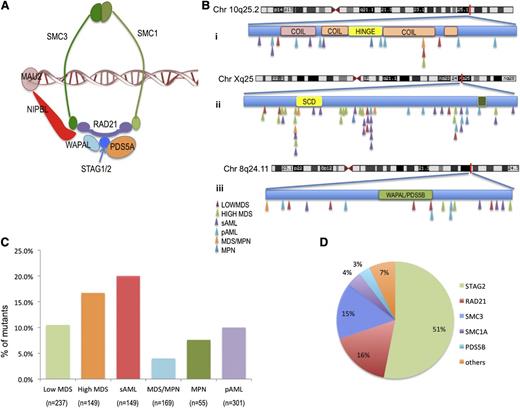
After excluding all known single-nucleotide polymorphisms and filtering out alterations present in paired germline samples, somatic alterations in cohesin complex genes were identified in 11.6% of patients, including STAG2 (5.9%), RAD21 (2%), and SMC3 (2%). Somatic mutations in other cohesin genes (SMC1A, PDS5B, WAPAL, MAU2, REC8, NIPBL, SMC5, and ESCO2) were present in <1% of our cohort (Figure 1 and supplemental Tables 2-4). We identified 2 cases with germline known rare nonsynonymous alterations, specifically a germline alteration in STAG1 (p.E760D) with population allelic frequency of 0.01, and a novel alteration in WAPAL (p.G230V; supplemental Table 4) All other sequence variants were confirmed to be somatic by sequencing paired flow-sorted CD3+ cells. Cohesin complex mutations were almost always mutually exclusive (Figure 2A). All cohesin mutations were heterozygous in nature except for mutations in STAG2 and SMC1A, which are located on chromosome X and thus result in the absence of expression of a wild-type (WT) allele. In STAG2, we detected mutations in 17 females (14 heterozygous and 3 homozygous) and in 44 males (hemizygous). In addition, we identified 8 deletions involving STAG2 (chromosome X) or RAD21 (chromosome 8) (supplemental Figure 3 and supplemental Table 3). Among STAG2 sequence alterations, 62 out of 63 predicted loss of function, including frameshift (n = 19), splice site (n = 10), and/or nonsense (n = 33) mutations. Similarly, 15 out of 20 RAD21 sequence alterations were nonsense (n = 2), frameshift (n = 11), or splice site (n = 2) mutations causing low expression of the gene; 5 were missense mutations. Among SMC3 alterations, 8 out of 19 were nonsense (n = 1), frameshift (n = 1), or splice site (n = 6) mutations, whereas 11 out of 19 were missense mutations.
Characterization of cohesin mutations in patients with myeloid disease. (A) Structure of the cohesin ring. (B) Position and disease subtype for identified cohesin mutations SMC3 (i), STAG2 (ii), and RAD21 (iii). No correlation was found with any structural motif, and no mutational hotspots were identified. (C) Frequency of cohesin family gene mutations in each myeloid malignancy. (D) Distribution of cohesin family gene mutations identified across the patient cohort.
Characterization of cohesin mutations in patients with myeloid disease. (A) Structure of the cohesin ring. (B) Position and disease subtype for identified cohesin mutations SMC3 (i), STAG2 (ii), and RAD21 (iii). No correlation was found with any structural motif, and no mutational hotspots were identified. (C) Frequency of cohesin family gene mutations in each myeloid malignancy. (D) Distribution of cohesin family gene mutations identified across the patient cohort.
Cooccurring somatic mutations in cohesin mutant patients. (A) Mutually exclusive cohesin gene mutations and cooccurring somatic genetic events. (B) Somatic mutational correlation in cohesin mutant/deletion cases.
Cooccurring somatic mutations in cohesin mutant patients. (A) Mutually exclusive cohesin gene mutations and cooccurring somatic genetic events. (B) Somatic mutational correlation in cohesin mutant/deletion cases.
Clinical features of cases harboring cohesin complex gene mutations
Cohesin defects were found in 11% of lower-risk MDS patients and 4% of MDS/MPN overlap patients (supplemental Table 4). A higher frequency of cohesin defects was found in sAML and high-risk MDS patients, with somatic alterations in 20% and 17% of these disease phenotypes, respectively (Figure 1C). STAG2 was associated with higher-risk disease (P = .0023), and RAD21 and SMC3 defects were most frequently identified in sAML (3.5% and 4%, respectively; supplemental Figure 1). We did not identify any other clinically relevant differences in the distribution of cohesin defects. Cohesin mutant MDS patients were more likely to be associated with high International Prognostic Scoring System (IPSS) scores (Table 1) and were more prevalent in patients who transformed to sAML.
Clinical characteristics of patients with cohesin family gene defects
| . | Mutants (n = 93) . | WT (n = 687) . | P . |
|---|---|---|---|
| Age | 65 (24-91) | 63 (18-91) | |
| >60 y | 72 | 504 | |
| <60 y | 21 | 183 | .45 |
| Sex | |||
| Male | 58 | 426 | |
| Female | 35 | 261 | 1 |
| Diagnosis | |||
| MDS-low | 22 | 190 | |
| MDS-high/sAML | 52 | 219 | .0036 |
| MDS/MPN | 7 | 147 | |
| MPN | 3 | 50 | |
| pAML | 9 | 93 | |
| IPSS* | |||
| Low | 8 | 99 | .05 |
| Int-1 | 20 | 106 | |
| Int-2 | 10 | 57 | |
| High | 4 | 19 | |
| Karyotype† | |||
| Normal | 56 (48%) | 351 (45%) | .1 |
| Abnormal | 60 (53%) | 425 (54%) | .1 |
| Complex | 11 (10%) | 104 (13%) | .36 |
| −5/del(5q) | 4 (4%) | 89 (11%) | .007 |
| −7/del(7q) | 4 (4%) | 99 (13%) | .002 |
| Trisomy 8 | 22 (19%) | 69 (9%) | .004 |
| −20/del(20q) | 6 (5%) | 45 (6%) | 1 |
| OS (mo) | 27.2 | 39.9 | .02 |
| . | Mutants (n = 93) . | WT (n = 687) . | P . |
|---|---|---|---|
| Age | 65 (24-91) | 63 (18-91) | |
| >60 y | 72 | 504 | |
| <60 y | 21 | 183 | .45 |
| Sex | |||
| Male | 58 | 426 | |
| Female | 35 | 261 | 1 |
| Diagnosis | |||
| MDS-low | 22 | 190 | |
| MDS-high/sAML | 52 | 219 | .0036 |
| MDS/MPN | 7 | 147 | |
| MPN | 3 | 50 | |
| pAML | 9 | 93 | |
| IPSS* | |||
| Low | 8 | 99 | .05 |
| Int-1 | 20 | 106 | |
| Int-2 | 10 | 57 | |
| High | 4 | 19 | |
| Karyotype† | |||
| Normal | 56 (48%) | 351 (45%) | .1 |
| Abnormal | 60 (53%) | 425 (54%) | .1 |
| Complex | 11 (10%) | 104 (13%) | .36 |
| −5/del(5q) | 4 (4%) | 89 (11%) | .007 |
| −7/del(7q) | 4 (4%) | 99 (13%) | .002 |
| Trisomy 8 | 22 (19%) | 69 (9%) | .004 |
| −20/del(20q) | 6 (5%) | 45 (6%) | 1 |
| OS (mo) | 27.2 | 39.9 | .02 |
n = 42 (mutants), 281 (WT).
n = 116 (mutants), 776 (WT).
Cytogenetic and molecular abnormalities associated with cohesin defects
To examine the potential association of cohesin complex mutations with other somatic defects, cases with mutated cohesin complex genes were compared with WT. Normal cytogenetics were present in 48% of mutant cases, whereas 10% had complex cytogenetics, a distribution comparable to WT patients. Among cases with an abnormal karyotype, trisomy 8 was more common in cohesin mutant patients (19% vs 9%; P = .004; supplemental Table 6), whereas −5/del(5q) and −7/del(7q) were less common (Table 1). When compared with the cohesin WT cohort, co-occurring mutations in RUNX1 (P = .0001), Ras-family oncogenes (P = .0001), BCOR (n = 15; P = .003), and ASXL1 (P = .03) were significantly enriched in patients with cohesin mutations (Figure 2 and supplemental Table 7). When stratifying by normal/abnormal karyotype, no particular genetic lesion was enriched based on karyotype, although 5 out of 9 patients (55%) with RUNX1 mutations and abnormal karyotype had trisomy 8.
Therapeutic response and clinical outcomes in patients with cohesin alterations
Within our cohort, we analyzed response to therapeutic interventions of lenalidomide (n = 92; supplemental Table 7) or the hypomethylating agents (HMAs) decitabine and azacitidine (n = 140; Table 2). Patients who received at least 4 cycles of treatment were assessed by modified International Working Group criteria.25 For this study, we considered complete remission, partial remission, and hematologic improvement in any cell lineage as response and stable disease, death, progression, or relapse during therapy as nonresponse. Patients with STAG2/RAD21 mutations were more likely to respond to treatment with HMAs compared with WT cases (79% vs 47%; P = .04) and were comparable in age and IPSS scores with the nonresponders. With respect to lenalidomide-treated patients (8 cohesin mutant and 84 WT), we did not observe a significant difference in response rate in cohesin mutant (50%) vs cohesin WT (52%) patients. No OS advantage was observed despite the increased response to HMAs. Patients with cohesin gene alterations had poor OS compared with patients without cohesin lesions (27.2 vs 39.9; P = .023). Further, we performed cox multivariable analysis of myeloid malignancy subtypes and identified that presence of the STAG2 mutation predicted poor survival independent of age, sex, and IPSS scores in high- and low-risk MDS patients (hazard ratio [HR] = 1.5 [0.9, 2.6]; P = .11) and in patients with MDS who survived beyond 12 months (HR = 2.1 [1.1, 3.8]; P = .017; Figure 3).
Clinical characteristics of patients treated with HMAs
| . | Cohesin mutant (n = 93) . | Cohesin WT (n = 627) . | P . |
|---|---|---|---|
| Patients treated with HMAs and response assessed | 16 | 124 | |
| Median age at start of therapy (y) | 71 | 69 | .98 |
| Median IPSS score at diagnosis | 1.5 | 1 | .7 |
| Cytogenetics at treatment | |||
| Normal | 8 | 53 | .79 |
| Abnormal karyotype | 8 | 67 | |
| Complex karyotype | 2 | 28 | .52 |
| −5/del(5q) | 1 | 19 | .46 |
| −7/del(7q) | 0 | 27 | .08 |
| Trisomy 8 | 3 | 8 | .22 |
| −20/del(20q) | 0 | 16 | .08 |
| Diagnosis at treatment | |||
| Low-risk MDS | 1 | 38 | .04 |
| High-risk MDS | 7 | 38 | .39 |
| sAML | 6 | 20 | .07 |
| MDS/MPN | 2 | 24 | .76 |
| MPN | 0 | 2 | 1 |
| pAML | 0 | 2 | 1 |
| Types of response per IWG criteria | |||
| Responder (CR, PR, and HI-E) | 13 | 59 | .02 |
| Nonresponder (SD and failure) | 3 | 65 | |
| Other somatic mutations | |||
| TET2 | 3 | 15 | .43 |
| TP53 | 0 | 5 | 1 |
| ASXL1 | 5 | 16 | .06 |
| DNMT3A | 1 | 14 | 1 |
| RUNX1 | 5 | 12 | .02 |
| IDH1 and IDH2 | 3 | 3 | .02 |
| . | Cohesin mutant (n = 93) . | Cohesin WT (n = 627) . | P . |
|---|---|---|---|
| Patients treated with HMAs and response assessed | 16 | 124 | |
| Median age at start of therapy (y) | 71 | 69 | .98 |
| Median IPSS score at diagnosis | 1.5 | 1 | .7 |
| Cytogenetics at treatment | |||
| Normal | 8 | 53 | .79 |
| Abnormal karyotype | 8 | 67 | |
| Complex karyotype | 2 | 28 | .52 |
| −5/del(5q) | 1 | 19 | .46 |
| −7/del(7q) | 0 | 27 | .08 |
| Trisomy 8 | 3 | 8 | .22 |
| −20/del(20q) | 0 | 16 | .08 |
| Diagnosis at treatment | |||
| Low-risk MDS | 1 | 38 | .04 |
| High-risk MDS | 7 | 38 | .39 |
| sAML | 6 | 20 | .07 |
| MDS/MPN | 2 | 24 | .76 |
| MPN | 0 | 2 | 1 |
| pAML | 0 | 2 | 1 |
| Types of response per IWG criteria | |||
| Responder (CR, PR, and HI-E) | 13 | 59 | .02 |
| Nonresponder (SD and failure) | 3 | 65 | |
| Other somatic mutations | |||
| TET2 | 3 | 15 | .43 |
| TP53 | 0 | 5 | 1 |
| ASXL1 | 5 | 16 | .06 |
| DNMT3A | 1 | 14 | 1 |
| RUNX1 | 5 | 12 | .02 |
| IDH1 and IDH2 | 3 | 3 | .02 |
CR, complete remission; HI-E, hematologic improvement (erythroid); IWG, International Working Group; PR, partial remission; SD, stable disease.
Effect of cohesin mutational status on OS in MDS. Kaplan-Meier curves for patients with MDS are shown for (A) STAG2 mutant patients and cohesin WT patients and (B) all cohesin mutant patients vs WT cohesin patients. Median OS of MDS patients with a STAG2 mutation was 35 months vs 50 months (P = .17; HR: 1.8 CI [1.1-3.1]), median survival for cohesin mutant MDS cases was inferior to WT MDS cases (34.2 vs 48.2; P = .21), and a significant survival difference was pronounced in patients with MDS who survived for at least 12 months (P = .01).
Effect of cohesin mutational status on OS in MDS. Kaplan-Meier curves for patients with MDS are shown for (A) STAG2 mutant patients and cohesin WT patients and (B) all cohesin mutant patients vs WT cohesin patients. Median OS of MDS patients with a STAG2 mutation was 35 months vs 50 months (P = .17; HR: 1.8 CI [1.1-3.1]), median survival for cohesin mutant MDS cases was inferior to WT MDS cases (34.2 vs 48.2; P = .21), and a significant survival difference was pronounced in patients with MDS who survived for at least 12 months (P = .01).
Clonal acquisition of cohesin complex gene mutations
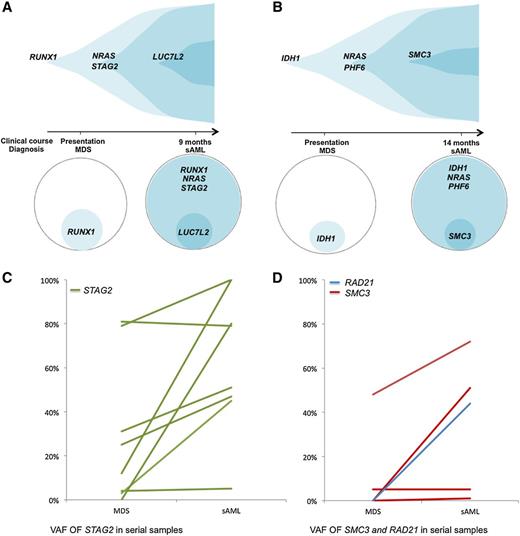
Clonal hierarchy analysis was performed to determine if cohesin complex gene variants were ancestral or secondary genetic events. This first involved variant allelic frequency (VAF) analyses of co-occurring mutations in samples, adjusted for copy number. The adjusted VAF of STAG2, RAD21, and SMC3 mutations were significantly lower than those for other concomitantly present TET2 gene mutations (P = .01). In 2 cases of sAML that evolved from RAEB, cohesin mutations were observed only after transformation to AML (Figure 4A-B), indicating these mutations can occur as a late event that is associated with AML transformation. In this cross-sectional analysis of the cohesin mutant cohort, STAG2 and SMC3 mutant clones were ancestral in only 18% of the cases. We also performed serial analyses for 12 cases with STAG2 and SMC3 mutations and 1 case with a mutation of RAD21 (Figure 4C-D). This enabled us to determine clonal architecture, and we observed that cohesin mutant clones expanded or remained stable in all 13 patients with a statistically significant increase in clone size (P = .0032). Findings were similar for each individual mutation, with clonal expansion in 7 of 8 patients with STAG2 mutations (P = .05), 2 of 4 SMC3 mutant patients, as well as in the single patient with a RAD21 mutation. In all, 10 of 13 patients had cohesin present in the dominant clone after transformation (supplemental Tables 10-11). Three AML cases with cohesin mutations (SMC3, STAG2, and RAD21) at diagnosis achieved remission with cytarabine and idarubicin treatment. Analysis for mutational burden after therapy demonstrated the STAG2 and SMC3 mutant clones in 2 cases were undetectable and the third case had persistence of a small RAD21 clone (44% to 7%).
Analysis of clonal hierarchy points toward cohesin as a secondary mutation. (A-B) Exemplary serial samples illustrating clonal architecture of cohesin family genes. (C) VAFs of STAG2 mutations by deep sequencing in serial samples. For each STAG2 mutant patient with available serial samples, the VAF in each sampling is shown. Although only 2 patients were found to harbor a STAG2 mutation in the dominant clone at the first time point, 7 out of 8 patients (87.5%) who were initially found to have a subclonal STAG2 mutation underwent clonal expansion, becoming the dominant clone at the time of transformation (P = .05). (D) VAFs of SMC3 and RAD21 mutations by deep sequencing in serial samples. For 4 patients with SMC3 mutations and the 1 patient with a RAD21 mutation with available serial samples, the VAF in each sampling is shown. All of these 5 patients harbored subclonal cohesin mutations at the first time point, with 2 out of 4 (50%) of the SMC3 mutant patients and the 1 out of 1 RAD21 mutant patient undergoing clonal expansion, becoming the dominant clone at the time of transformation.
Analysis of clonal hierarchy points toward cohesin as a secondary mutation. (A-B) Exemplary serial samples illustrating clonal architecture of cohesin family genes. (C) VAFs of STAG2 mutations by deep sequencing in serial samples. For each STAG2 mutant patient with available serial samples, the VAF in each sampling is shown. Although only 2 patients were found to harbor a STAG2 mutation in the dominant clone at the first time point, 7 out of 8 patients (87.5%) who were initially found to have a subclonal STAG2 mutation underwent clonal expansion, becoming the dominant clone at the time of transformation (P = .05). (D) VAFs of SMC3 and RAD21 mutations by deep sequencing in serial samples. For 4 patients with SMC3 mutations and the 1 patient with a RAD21 mutation with available serial samples, the VAF in each sampling is shown. All of these 5 patients harbored subclonal cohesin mutations at the first time point, with 2 out of 4 (50%) of the SMC3 mutant patients and the 1 out of 1 RAD21 mutant patient undergoing clonal expansion, becoming the dominant clone at the time of transformation.
Expression signatures associated with cohesin defects
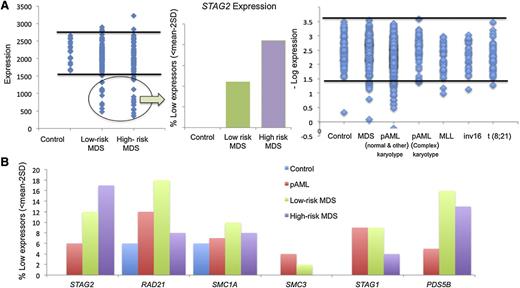
The presence of hypomorphic mutations suggests that somatic alterations of cohesin lead to loss of function. It is thus possible that functionally decreased expression may produce a clinical phenocopy of cohesin mutations. When dichotomized using a threshold expression of <mean − 2 standard deviations (95% confidence interval) as a cutoff value, 10% to 20% of cases showed decreased cohesin expression (Figure 5A-B). A subset of these patients had mutations in cohesin complex members and reduced expression of STAG2/RAD21/SMC3; indeed, we noted that most patients with cohesin mutations had reduced expression of the entire cohesin complex. However, we also noted reduced expression of cohesin genes in patients without mutations, including 15% of lower- and higher-risk MDS patients with reduced expression of STAG2 and RAD21. We also observed a subset of AML patients with marked reduction in STAG2 and PDS5B expression in the absence of cohesin mutation. We also investigated expression signatures of genes associated with cohesin mutations or with reduced expression of STAG2/RAD21/SMC3. Our analysis showed a similar expression profile in patients with STAG2 mutations or patients with reduced expression of cohesin complex members (Figure 6A). We observed decreased expression of DCK and increased expression of UCK in patients with cohesin mutations or with nonmutational suppression of cohesin expression, indicating common alterations of downstream pathways (supplemental Table 12). Notably, expression of the entire cohesin complex was reduced in patients with STAG2 mutations in a manner similar to that seen in patients with reduced cohesin expression in the absence of mutations (Figure 6B). When comparing patients with reduced expression or mutation of STAG2 to patients with intact expression of WT STAG2, we identified 28 genes, which were differentially expressed in patients with STAG2 alterations regardless of whether this was due to somatic mutations or transcriptional downregulation (supplemental Table 13). This includes decreased expression of signaling effectors (NRAS, JAK1, and CBL) and reduced expression of HIF1A, suggesting a novel mechanism of transformation for patients with myeloid malignancies characterized by alterations in the cohesin complex. Previous work in a zebrafish model identified runx dependence on rad21 expression,26 and given the high association of RUNX1 mutations in cohesin mutant myeloid disease, we queried if this relationship would be evident in our patient cohort. We were unable to find any correlation between RAD21 and RUNX1 expression or between mutational status and expression in our patient cohorts, suggesting that this interaction in humans may be subject to pathway redundancy.
Analysis of cohesin expression identifies a discrete subset of low expressors. (A) Expression of STAG2 in MDS and pAML subtypes. A discrete subset of patients exhibit low STAG2 expression defined as <2 standard deviations below the mean as compared with 17 healthy donors. This was more prevalent in patients with high-risk MDS and non–core binding factor AML without complex cytogenetics. (B) Percent of low expressors in cohesin genes. The frequency of cohesin low expressors for each cohesin complex gene (STAG2, RAD21, SMC1A, SMC3, STAG1, and PDS5B) is shown by disease subtype. STAG2 was most commonly underexpressed in high-risk MDS, whereas RAD21 was most commonly underexpressed in the remaining myeloid malignancies. SD, standard deviation.
Analysis of cohesin expression identifies a discrete subset of low expressors. (A) Expression of STAG2 in MDS and pAML subtypes. A discrete subset of patients exhibit low STAG2 expression defined as <2 standard deviations below the mean as compared with 17 healthy donors. This was more prevalent in patients with high-risk MDS and non–core binding factor AML without complex cytogenetics. (B) Percent of low expressors in cohesin genes. The frequency of cohesin low expressors for each cohesin complex gene (STAG2, RAD21, SMC1A, SMC3, STAG1, and PDS5B) is shown by disease subtype. STAG2 was most commonly underexpressed in high-risk MDS, whereas RAD21 was most commonly underexpressed in the remaining myeloid malignancies. SD, standard deviation.
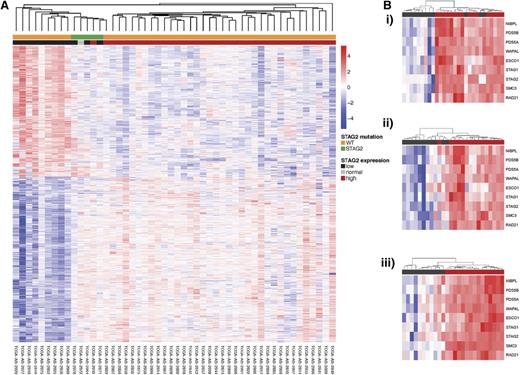
Unsupervised hierarchical clustering of gene expression with annotation by cohesin mutational status and cohesin expression. (A) Unsupervised hierarchical clustering of TCGA AML patients with annotation of STAG2 low expression and STAG2 mutation. Patients with STAG2 mutations and low STAG2 expression cluster show a unique expression profile and cluster separately from patients with WT STAG2 and high expression. (B) Cohesin complex expression in STAG2 (i), RAD21 (ii), and SMC3 (iii) low expressors. Within each group of cohesin low expressors, the entire cohesin family gene complex has similarly decreased expression.
Unsupervised hierarchical clustering of gene expression with annotation by cohesin mutational status and cohesin expression. (A) Unsupervised hierarchical clustering of TCGA AML patients with annotation of STAG2 low expression and STAG2 mutation. Patients with STAG2 mutations and low STAG2 expression cluster show a unique expression profile and cluster separately from patients with WT STAG2 and high expression. (B) Cohesin complex expression in STAG2 (i), RAD21 (ii), and SMC3 (iii) low expressors. Within each group of cohesin low expressors, the entire cohesin family gene complex has similarly decreased expression.
Discussion
We have demonstrated that somatic alterations in cohesin complex genes occur in 11.6% of patients with myeloid neoplasms in our cohort, consistent with recent reports in smaller patient cohorts.1,2 The observation that most mutations are nonsense and frameshift types suggests that cohesin mutations lead to decreased function of the cohesin complex. As low expression of these genes may have functionally equivalent consequences, we defined a subset of patients with altered expression of the cohesin complex in an additional 15% of patients. Such nonmutated cohesin low expressors have been described in both AML and Ewing sarcoma cell lines.3,5 Additionally, this study expands the current knowledge of cohesin mutations across the spectrum of myeloid diseases, identifying cohesin mutation frequencies in different disease subtypes, most prominently in sAML (20%). Clonal hierarchy analysis rarely identified cohesin mutations in founding clones; rather, cohesins were present in subclones that underwent clonal expansion over time. This subclonal architecture occurs in the setting of co-occurring genetic and molecular abnormalities, including mutations in chromatin modifiers (ASXL1) and key transcriptional regulators (RUNX1, Ras family genes, and BCOR). Our data further suggest that cohesin has a cooperative role in epigenetic regulation and transcriptional programming by virtue of the higher response rate in cohesin mutant MDS patients treated with HMAs.
In line with other studies, cohesin mutations were not associated with an increase in the frequency of complex cytogenetic abnormalities. Despite cohesin’s role in alignment of sister chromatids during metaphase, these data provide further evidence that the leukemogenic consequences of cohesin defects do not involve disrupted mitosis and global chromosomal instability. The expanded sample size of our cohort also enabled precise analysis of clinical associations. Cohesin mutations did not alter the IPSS cytogenetic risk of MDS patients (the average IPSS cytogenetics score was 0.55 points for cohesin mutant cohort and 0.60 points for WT). Nonetheless, overall IPSS risk was higher in cohesin-mutated patients, driven by high blast count, suggesting a stem cell defect in the mutant clone.
The vast majority of mutations identified across cohesin proteins were heterozygous nonsense and frameshift events, which would be anticipated to cause expression of a truncated protein. In addition to clearly hypomorphic mutations, splice site mutations were also identified (eg, STAG2 5′ of exon 4 and 8). Splice site mutations can cause activation of a nearby cryptic splice site (upstream or downstream of the normal site), omission of the whole exon, or intron retention, any of which can lead to frameshifts and premature stop codons that will trigger nonsense-mediated decay and haploinsufficiency. Cohesin haploinsufficiency is thus the operative pathophysiologic mechanism, a notion further supported by the mutual exclusivity of cohesin mutations: haploinsufficiency for one cohesin complex member may be sufficient to initiate the myeloid transformation pathway, whereas a double hit would be deleterious. Recent studies of cohesin dynamics at super enhancer sites are in line with these conclusions: cohesin localizes to sites of high mediator occupancy at the enhancer-promoter interaction sites in lymphoma and myeloma. Treatment with the BRD4 inhibitor JQ.1 leads to a 2-fold decrease in cohesin occupancy at superenhancer sites and normalization of the expression of oncogenic drivers.16,27 Taken together, these data suggest that cohesin function is integral to expression of key cell-identity genes and that reduction in gene dose of cohesin alters cellular identity.
Analysis of clonal dynamics in patients in whom serial samples are obtained at various time points along their disease course provides important insight into the timing and effect of cohesin loss of function on leukemia initiation and progression. Using deep-sequencing and cross-sectional and serial analyses, we found that cohesin mutations were not commonly present in the founder clone but rather promoted clonal expansion and transformation to more aggressive disease. In 10 of 13 cohesin mutant patients with serial samples, the cohesin mutant clone achieved clonal dominance at the time of transformation. Cohesin mutations co-occur with other mutations known to be drivers of clonal evolution. The strong association with ASXL1 indicates that epigenetic dysregulation may be synergistic in the mechanism of action exerted by cohesin haploinsufficiency. Furthermore, activated Ras signaling may alter the expression of key identity genes, which are then further dysregulated by cohesin inactivity.
High numbers of informative patients (those with mutations) are needed to establish correlations with response to specific drugs. In exploring the effects of cohesin downmodulation/mutations on expression pattern in cohort, we noted a consistent downmodulation of DCK in patients with cohesin complex alterations. Expression of this enzyme has been reported to be reciprocal to the levels of UCK, a rate-limiting enzyme for azacitidine metabolism. Thus, azacitidine would be expected to have greater efficacy in cohesin-deficient patients. Indeed, we observed a higher response rate in patients with cohesin mutations treated with azacitidine.
In summary, cohesin mutations, which likely result in loss of function, are recurrent molecular genetic events in both solid and hematopoietic tumors. Mechanistic studies to understand their functional and pathophysiological role are currently underway, but our genetic data suggest that cohesin mutations do not contribute to hematopoietic transformation through altered chromosomal instability. The high frequency with which cohesin mutations occur in myeloid disease and their presence in expanding subclones during disease progression suggests that the cohesin complex may represent an attractive therapeutic target for future preclinical and clinical studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by National Institutes of Health grants R01 HL118281-01 (National Heart, Lung, and Blood Institute), R01 CA138858-05, (National Cancer Institute), K24 HL077522-07 (National Heart, Lung, and Blood Institute) (J.P.M.); and R01 CA169784 (National Cancer Institute) (R.L.L.), as well as a grant from the Robert Duggan Research Fund (J.P.M.), a grant from the Mortimer J. Lacher fellowship of the Lymphoma Foundation (A.D.V.), and a grant from the Edward P. Evan's CRC (M.S.).
Authorship
Contribution: A.V., S.T., H.M., R.L., and J.M. designed research; A.V., S.T., B.S., and B.P. performed research; A.V., S.T., H.M., B.S., B.P., and T.R. analyzed data; and A.V., S.T., H.M., B.S., B.P., T.R., M.S., R.L., and J.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw Maciejewski, Translational Hematology & Oncology Research, Taussig Cancer Institute R/40, Cleveland Clinic, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: maciejj@ccf.org.
References
Author notes
S.T. and A.D.V. contributed equally to this study.


![Figure 3. Effect of cohesin mutational status on OS in MDS. Kaplan-Meier curves for patients with MDS are shown for (A) STAG2 mutant patients and cohesin WT patients and (B) all cohesin mutant patients vs WT cohesin patients. Median OS of MDS patients with a STAG2 mutation was 35 months vs 50 months (P = .17; HR: 1.8 CI [1.1-3.1]), median survival for cohesin mutant MDS cases was inferior to WT MDS cases (34.2 vs 48.2; P = .21), and a significant survival difference was pronounced in patients with MDS who survived for at least 12 months (P = .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/11/10.1182_blood-2014-04-567057/4/m_1790f3.jpeg?Expires=1769085186&Signature=OS7I2bxwdtFg07OBmxIC5dH-JMvz6F0v8zLodOxzUI9gdbkf5XndwydYfBGapjbrj6hebOw-7zMOEHo4ee09Wlv-puSjOJwPjAx2syfsYeIqyCOAKT0j8IWQi3FYDlXLZiq1x2J8rFGSqBTLP5ZaNPSBcuTdQUfEBgQaPXS6Tf8Wet0FFv9TGHGZiWvbLsY97fxc~qdxmKMjea9j5Nv9Z9kfGtZnOscJMEG0wtG7Rlec97c1vRwiyloc8vcCCmjIB0~4iA~F0mYlu5OaXM7TqDq6H16sZTs4dF34FFsSyp5y7gNHxsO6oCVgAR0avDGSMyL4nMO-jE-qhY1WZkETFQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal