Key Points
HOXB4 induces stable gene expression changes in transplanted HSCs that drive balanced self-renewal and differentiation divisions.
Marked downregulation of Prdm16 occurs concurrently with HOXB4-mediated HSC expansion and functions to prevent leukemia in vivo.
Abstract
Overexpression of HOXB4 in hematopoietic stem cells (HSCs) leads to increased self-renewal without causing hematopoietic malignancies in transplanted mice. The molecular basis of HOXB4-mediated benign HSC expansion in vivo is not well understood. To gain further insight into the molecular events underlying HOXB4-mediated HSC expansion, we analyzed gene expression changes at multiple time points in Lin−Sca1+c-kit+ cells from mice transplanted with bone marrow cells transduced with a MSCV-HOXB4-ires-YFP vector. A distinct HOXB4 transcriptional program was reproducibly induced and stabilized by 12 weeks after transplant. Dynamic expression changes were observed in genes critical for HSC self-renewal as well as in genes involved in myeloid and B-cell differentiation. Prdm16, a transcription factor associated with human acute myeloid leukemia, was markedly repressed by HOXB4 but upregulated by HOXA9 and HOXA10, suggesting that Prdm16 downregulation was involved in preventing leukemia in HOXB4 transplanted mice. Functional evidence to support this mechanism was obtained by enforcing coexpression of sPrdm16 and HOXB4, which led to enhanced self-renewal, myeloid expansion, and leukemia. Altogether, these studies define the transcriptional pathways involved in HOXB4 HSC expansion in vivo and identify repression of Prdm16 transcription as a mechanism by which expanding HSCs avoid leukemic transformation.
Introduction
In mammals, members of the homeobox transcription factor (Hox) family are important regulators of both embryonic development and hematopoiesis.1 Deregulated expression of certain Hox genes causes hematopoietic abnormalities including myeloproliferative disorders and acute leukemias.2 For instance, enforced overexpression of either HOXA9 or HOXA10 causes acute myeloid leukemia that is mediated by enhancement of progenitor cell proliferation and inhibition of normal hematopoietic differentiation.3,4 HOXA9 expression is frequently deregulated in human leukemias as demonstrated by expression array analysis.5 Human leukemia can also be caused by chromosomal rearrangements involving HOX genes6,7 or the mixed-lineage leukemia gene, a positive regulator of Hox gene expression in hematopoietic cells.8
HOXB4 is an important hematopoietic transcription factor that does not have the oncogenic potential of other HOX factors. HOXB4 is normally expressed in mouse hematopoietic stem cells (HSCs) and in aorta-gonad-mesonephros–derived CD45+CD144+ cells.9 Germline deletion of Hoxb4 has a mild phenotype likely due to redundant function from the other group 4 paralogs.10 HOXB4 overexpression in murine HSCs leads to a dramatic enhancement of HSC self-renewal and an accompanying increase in HSC numbers.11,12 Ectopic HOXB4 also confers expansion in human HSCs,13 although this effect is somewhat species specific.14 In mice, HOXB4-transduced HSCs rapidly increase in number in the bone marrow (BM) after transplantation, but this expansion is self-limited; HSC numbers remain normal, and no abnormalities in hematopoiesis are seen.15,16 It is not known how HOXB4-mediated HSC amplification avoids leukemic transformation, but one possibility is that HOXB4 could specifically downregulate proto-oncogenes that are required for leukemogenesis.
Previous studies have identified target genes altered by ectopic HOXB4 expression in primary myeloid progenitors17,18 and in a hematopoietic cell lines.19 More recent studies have identified HOXB4 target genes in embryonic stem cells.20,21 There was only a limited number of shared targets identified in these various studies, suggesting that HOXB4 target genes differ based on cell type and context. It would be ideal to identify downstream effectors and targets directly in HOXB4-transduced HSCs in transplanted animals. Furthermore, the transcriptional program induced by HOXB4 could change dynamically over time during the posttransplant reconstitution phase.
We now describe gene expression profiling experiments in HSC-enriched populations derived directly from mice transplanted with HOXB4-transduced BM cells. In these studies, Lin−Sca1+c-kit+ (LSK) cells were purified from transplanted mice at varying times after transplant, and extracted messenger RNA (mRNA) was analyzed by microarray analysis and reverse-transcription quantitative real-time PCR (qRT-PCR). These experiments identified dynamic transcriptional changes induced by HOXB4 during HSC reconstitution involving both upregulation of stem cell genes as well as several counterregulatory networks that may physiologically limit HSC expansion and/or prevent leukemic transformation. In particular, we show that HOXB4 leads to marked downregulation of Prdm16 expression, a proto-oncogene necessary for self-renewal and maintenance of murine HSCs.22,23
Materials and methods
Vector cloning and production
The retroviral vectors MSCV-ires-GFP and MSCV-HOXB4-ires-YFP/GFP have been described previously.17 MSCV-HA-HOXA9-ires-GFP and MSCV-HOXA10-ires-GFP retroviral vectors were generously provided by Dr R. Keith Humphries. Murine sPrdm16 complementary DNA (cDNA) (nucleotides 664–3822 of Mus musculus Prdm16 transcript, NM_00117795) was generated by polymerase chain reaction (PCR) using a mouse Prdm16 cDNA (clone ID 6409778; Thermo Scientific) as template. The sPrdm16 cDNA was inserted into the CL20 MSCV-ires-mCherry lentiviral vector to generate the CL20 MSCV-sPrdm16-ires-mCherry vector.24 The CL20i4r-EF1α-HOXB4-P2A-GFP lentiviral vector was generated by PCR using the MSCV-HOXB4-ires-YFP retroviral vector as template. An EcoRI-HOXB4-P2A-GFP-NotI fragment was generated by PCR and was digested with EcoRI and NotI and used to replace the hgcOPT fragment in the CL20i4r-EF1α-hgcOPT lentiviral vector.25 Lentiviral vectors were produced by transient transfection of the 293T cell line with helper plasmids and then further concentrated by ultracentrifugation.
BM cell transduction, transplantation, and cell separation
All animal experiments were carried out according to protocols approved by the St. Jude Institutional Animal Care and Use Committee. 5-Fluorouracil treatment of donor mice, BM cell harvest, cell processing and transplant procedures were carried out as described previously.17 BM cells were transduced with concentrated supernatant from GPE+86-derived producer cells containing MSCV-HOXB4-ires-YFP or MSCV-HA-HOXA9-ires-GFP or MSCV-HOXA10-ires-GFP or MSCV-ires-GFP retroviral vectors in 6-well plates preloaded with 25 μg/mL retronectin (Takara). Transduced cells were injected into the lateral tail vain of lethally irradiated (1100 cGy) female C57Bl/6L mice. BM cells were isolated from transplanted mice 6, 12, and 20 weeks posttransplantation and stained using commercially available lineage-specific antibodies.
In experiments to co-overexpress sPrdm16 and HOXB4, lineage-depleted BM cells from 6- to 8-week old female C57Bl/6L mice were transduced with CL20i4r-EF1α-HOXB4-P2A-GFP and CL20-MSCV-sPrdm16-ires-mCherry vectors concurrently for 4 continuous days. Fluorescence-activated cell sorting was used to isolate green fluorescent protein (GFP) and mCherry double-positive cells, 500 of which were plated into methylcellulose (STEMCELL Technologies) for the colony-forming unit in culture (CFU-C) assay, and 5 × 105 or 1 × 106 were transplanted into lethally irradiated female B6.SJL-PtprcaPepcb/BoyJ mice (The Jackson Laboratory).
RNA extraction, microarray expression analyses, and real-time RT-PCR
Total RNA from sorted cells was extracted using the RNAqueous-Micro kit (Life Technologies). For microarray, approximately 20 ng total RNA was processed using NuGEN WTA pico v2 and Encore Biotin labeling modules, and 6 μg biotin-labeled cDNA was hybridized overnight to HT_MG-430_PM microarray chips (Affymetrix). Hybridized arrays were scanned using the Gene Titan system, and signals were summarized using the RMA method (Partek Genomics Suite v6.5). Differentially expressed transcripts between GFP+ LSK and HOXB4+ LSK cells were identified from triplicate, independently derived from pools of cells at various time points using a 2-factor ANOVA model. The Benjamini-Hochberg method26 was used to estimate the false discovery rate (FDR). Pathways activated or repressed by HOXB4 expression were identified by gene set enrichment analysis (GSEA)27 using curated pathways obtained from the MolSigDB (Broad Institute) and Ingenuity Pathways (Ingenuity). MetaCore software (GeneGo, Philadelphia, PA) was also used to identify significant biological pathways defined by specific differentially expressed genes. Our microarray data are available through the Gene Expression Omnibus using accession ID GSE59804.
For real-time qRT-PCR, separate RNA samples from independent cohorts of transplanted mice or cultured cells were reverse transcribed using the SuperScript VILO cDNA Synthesis Kit (Life Technologies). Resulting cDNAs were quantitated by qRT-PCR assay using Power SYBR Green PCR Master Mix (Life Technologies). The relative expression levels for specific transcripts were calculated using the 2−ΔΔCT method normalized to the internal control gene Gapdh.
Characterization of retroviral integration sites in leukemic cells
The retroviral integration sites in leukemic cells that developed in mice transplanted with marrow transduced with the MSCV-HOXB4-ires-GFP vector were identified using the splinkerette-PCR technique as described previously.28 Restriction endonuclease–digested DNA was ligated to the splinkerette linker, and then the DNA was PCR amplified using nested primers corresponding to the splinkerette linker and the murine stem cell virus (MSCV) long terminal repeat. PCR fragments were separated by agarose gel electrophoresis and subjected to automated sequencing.
Western blot
Total protein lysate extraction of HOXB4- or sPrdm16-transduced NIH-3T3 cells and western blot were carried out as described previously.29 HOXB4 protein was probed using 0.1 μg/mL I12 rat anti-mouse HOXB4 antibody (Developmental Studies Hybridoma Bank, University of Iowa) followed by horseradish peroxidase–conjugated goat anti-rat secondary antibody (Santa Cruz Biotechnology), and sPrdm16 protein was probed with 0.1 μg/mL sheep anti-mouse Prdm16 polyclonal antibody (R&D Systems) followed by horseradish peroxidase–conjugated donkey anti-sheep secondary antibody (R&D Systems).
Results
Global gene expression changes in HOXB4-transduced LSK cells at various time points after transplant
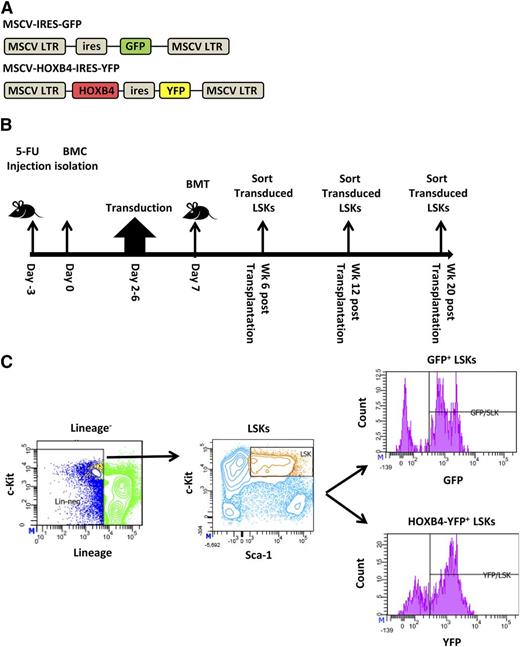
To identify both direct and indirect transcriptional target genes of HOXB4 in HSCs, we generated the MSCV-HOXB4-ires-YFP vector and a control GFP vector based on the MSCV γ-retroviral vector (Figure 1A). Groups of mice were transplanted with transduced BM cells and then killed at 6, 12, and 20 weeks after transplant for gene expression analysis (Figure 1B). Gene expression profiling was performed on HOXB4-YFP– or control GFP-expressing BM LSK cells isolated by cell sorting (Figure 1C and supplemental Table 1, available at the Blood Web site). For all time points, 3 independent transplant experiments and cell-isolation procedures were performed to establish reproducibility.
Experimental design of HOXB4 target gene analysis in transplanted murine HSCs. (A) Schematic diagram of HOXB4 retroviral and GFP control vectors used to transduce 5-fluorouracil (5-FU)–treated BM cells. (B) Timeline of transduction and mouse transplantation assay. C57Bl/6J mice were treated with 5-FU for 3 days prior to isolation of BM cells. After 2 days of prestimulation with a cytokine cocktail, cells were transduced for 4 days with either the GFP or HOXB4 retroviral vector. A total of 1 to 2 million cells were transplanted into lethally irradiated recipient C57Bl/6J mice. Vector-expressing LSK cells were isolated from the BM at 6, 12, and 20 weeks posttransplantation and were processed by flow sorting prior to total RNA isolation. (C) Flow cytometry sorting strategy for transduced LSKs. BM cells isolated from recipients were first gated for lineage-negative cells, then for cKit+ Sca-1+ marker status, and then for vector expression based on GFP or yellow fluorescent protein (YFP) expression. Sorted GFP/YFP+ LSK cells were used for subsequent gene expression analyses. BMC, BM cells; BMT, BM transplant.
Experimental design of HOXB4 target gene analysis in transplanted murine HSCs. (A) Schematic diagram of HOXB4 retroviral and GFP control vectors used to transduce 5-fluorouracil (5-FU)–treated BM cells. (B) Timeline of transduction and mouse transplantation assay. C57Bl/6J mice were treated with 5-FU for 3 days prior to isolation of BM cells. After 2 days of prestimulation with a cytokine cocktail, cells were transduced for 4 days with either the GFP or HOXB4 retroviral vector. A total of 1 to 2 million cells were transplanted into lethally irradiated recipient C57Bl/6J mice. Vector-expressing LSK cells were isolated from the BM at 6, 12, and 20 weeks posttransplantation and were processed by flow sorting prior to total RNA isolation. (C) Flow cytometry sorting strategy for transduced LSKs. BM cells isolated from recipients were first gated for lineage-negative cells, then for cKit+ Sca-1+ marker status, and then for vector expression based on GFP or yellow fluorescent protein (YFP) expression. Sorted GFP/YFP+ LSK cells were used for subsequent gene expression analyses. BMC, BM cells; BMT, BM transplant.
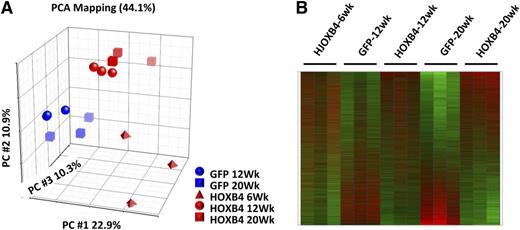
Principle component analysis (PCA) revealed significant clustering within experimental groups (Figure 2A). The GFP samples and the HOXB4 samples separated across the first principal component, indicating the most significant source of expression variation was associated with the presence or absence of HOXB4 expression. Within the GFP treatment arm, the 12- and 20-week samples grouped closely together. Within the HOXB4 treatment arm, the 12 and 20-week samples also grouped together and were statistically distinct from the GFP-control groups (Figure 2A). The 6-week samples from the HOXB4 treatment arm were separated from the 12- and 20-week HOXB4 samples, demonstrating distinct transcriptional profiles exist at these 2 time points. A heatmap representation of all expression profiles shows reproducible gene expression changes in HOXB4-transduced LSK cells that evolve between 6 and 20 weeks after transplant (Figure 2B). These gene expression patterns were reproducible in triplicate samples from both groups.
PCA and heatmap representation of HOXB4-induced expression profiles in BM LSK cells. (A) PCA using all probe sets, where each symbol represents a separate experiment as indicated by the legend, with red symbols denoting HOXB4 samples and blue symbols the GFP controls. The GFP+ LSK cells and HOXB4+ LSK cells segregate into statistically separate groups across the PC1 axis. HOXB4-LSK cells harvested at 6 weeks are distinct from 12-week and 20-week HOXB4-LSKs across the PC2 axis. (B) Heatmap of gene expression alterations seen across all probe sets, with triplicate samples per experimental arm and time points as indicated by the labels. Red indicates overexpressed probe sets relative to GFP controls, whereas green indicates underexpressed probe sets. PC, principal component.
PCA and heatmap representation of HOXB4-induced expression profiles in BM LSK cells. (A) PCA using all probe sets, where each symbol represents a separate experiment as indicated by the legend, with red symbols denoting HOXB4 samples and blue symbols the GFP controls. The GFP+ LSK cells and HOXB4+ LSK cells segregate into statistically separate groups across the PC1 axis. HOXB4-LSK cells harvested at 6 weeks are distinct from 12-week and 20-week HOXB4-LSKs across the PC2 axis. (B) Heatmap of gene expression alterations seen across all probe sets, with triplicate samples per experimental arm and time points as indicated by the labels. Red indicates overexpressed probe sets relative to GFP controls, whereas green indicates underexpressed probe sets. PC, principal component.
Identification of specific changes in gene expression in HOXB4-transduced LSK cells
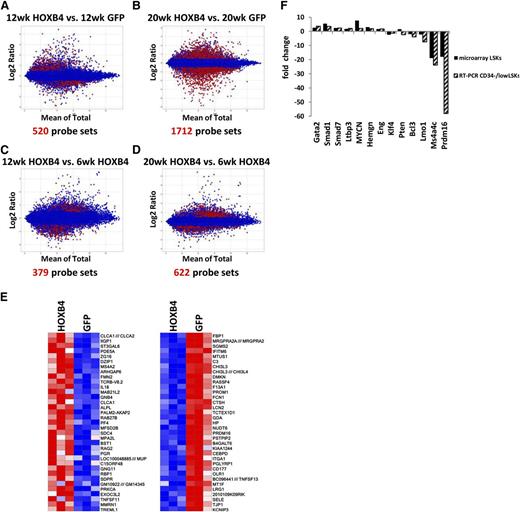
To identify either direct or indirect HOXB4 target genes, we applied a 2-factor ANOVA model to identify probe sets with >0.5-log difference and with a FDR of <0.05 in the HOXB4 vs the GFP groups at given time points. In the HOXB4-LSK cells, a total 520 and 1712 probe sets were identified with significant differential expression at 12 weeks and 20 weeks when compared with control GFP-LSK cells (Figure 3A-B). When 6-week HOXB4 samples were compared with the 12- and 20-week HOXB4 samples, 379 and 622 probe sets showed >0.5 log deference and FDR <0.05, respectively (Figure 3C-D). However, no significant differential gene expression was noted between 12 and 20 weeks in HOXB4+ LSK cells, demonstrating that the major transcriptional events in HOXB4-transduced cells occur by 12 weeks after transplantation. Heatmaps of the top 35 over- and underexpressed genes at 20 weeks show reproducible and significant differences in HOXB4 vs GFP samples in 3 independent transplant experiments (Figure 3E).
Identification and validation of HOXB4 target genes. Bland-Altman plots showing comparative gene expression between (A) 12-week HOXB4-LSK cells and 12-week GFP-LSK cells, (B) 20-week HOXB4-LSKs and 20-week GFP-LSKs, (C) 12-week HOXB4-LSKs and 6-week HOXB-LSKs, and (D) 20-week HOXB4-LSKs and 6-week HOXB4-LSKs. The y-axis indicates the magnitude of change for a given probe set and the x-axis the average of the signal observed for a given probe set on the Mouse HT_MG_430_PM array. The red dots represent differentially expressed probe sets with a >0.5-log differential expression and FDR < 0.05. (E) Heatmap of the top upregulated and downregulated genes from 20-week HOXB4-LSKs compared with 20-week GFP-LSKs. Each column contains data from an independent replicate of the designated experimental group. Red represents upregulated genes, and blue boxes represent downregulated genes. (F) Quantitative RT-PCR measurement of differential gene expression for 13 HOXB4-regulated genes in CD34−/low LSK cells harvested at 20 weeks posttransplantation. This is compared with the expression microarray results of the same genes. The fold change is indicated for HOXB4-transduced cells relative to GFP control cells.
Identification and validation of HOXB4 target genes. Bland-Altman plots showing comparative gene expression between (A) 12-week HOXB4-LSK cells and 12-week GFP-LSK cells, (B) 20-week HOXB4-LSKs and 20-week GFP-LSKs, (C) 12-week HOXB4-LSKs and 6-week HOXB-LSKs, and (D) 20-week HOXB4-LSKs and 6-week HOXB4-LSKs. The y-axis indicates the magnitude of change for a given probe set and the x-axis the average of the signal observed for a given probe set on the Mouse HT_MG_430_PM array. The red dots represent differentially expressed probe sets with a >0.5-log differential expression and FDR < 0.05. (E) Heatmap of the top upregulated and downregulated genes from 20-week HOXB4-LSKs compared with 20-week GFP-LSKs. Each column contains data from an independent replicate of the designated experimental group. Red represents upregulated genes, and blue boxes represent downregulated genes. (F) Quantitative RT-PCR measurement of differential gene expression for 13 HOXB4-regulated genes in CD34−/low LSK cells harvested at 20 weeks posttransplantation. This is compared with the expression microarray results of the same genes. The fold change is indicated for HOXB4-transduced cells relative to GFP control cells.
To validate that the alterations seen in the LSK population are also present in a more highly enriched HSC population, CD34−/low LSK cells were isolated 20 weeks after transplant. This population is a relatively pure subset of reconstituting HSCs.30 Reverse qRT-PCR was used to measure mRNA levels for a subset of differentially expressed transcripts potentially involved in HSC regulation (Figure 3F). Gata2 is a zinc-finger transcription factor that is required for adult HSC development31 and was increased 3.6-fold in HOXB4-transduced CD34− HSCs. Hemgn was identified as a HOXB4 direct target in HOXB4-transduced hematopoietic progenitor cells in our previous study17 and was increased about 2.0-fold in both LSK bulk and CD34− fractions (Figure 3F). Transforming growth factor β signaling is a negative regulator of HSC proliferation and utilizes Smad proteins as downstream effector molecules.32 Expression of Smad1 and Smad7 was 3.6- and 2.4-fold upregulated by qRT-PCR in HOXB4-transduced CD34−/low LSK cells (Figure 3F).
A 3- to 18-fold downregulation of Prdm16 was noted in all 4 microarray probes from the HOXB4 samples at 20 weeks. Prdm16 is SET-domain–containing protein associated with epigenetic remodeling and is required for brown fat determination33-35 and adult murine HSC and neural stem cell maintenance.22 We noted a 58.1-fold downregulation of Prdm16 mRNA by qRT-PCR in HOXB4 CD34− LSKs at 20 weeks (Figure 3F). Altogether, these results show a good general correlation between gene expression changes seen in LSK and the more highly purified CD34− subpopulation.
HOXB4 overexpression results in gene expression changes associated with HSC self-renewal and lymphoid-myeloid differentiation
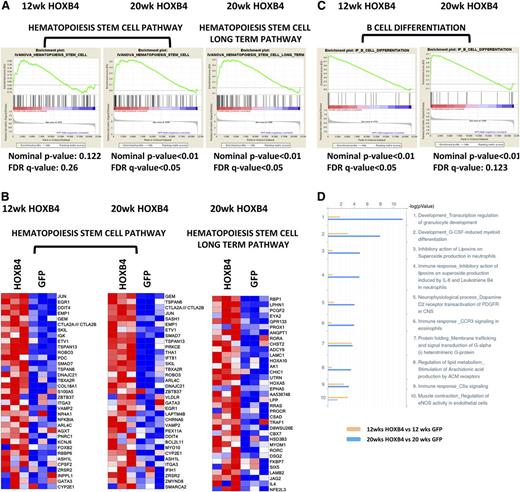
To further define the regulatory pathways underlying HOXB4-dependent HSC expansion, we performed GSEA with curated gene sets associated with signaling, metabolic, and differentiation pathways. HOXB4 expression at 20 weeks after transplant produced significant activation of genes associated with HSC function36 (Figure 4A). Activation of genes associated with long-term HSC potential was also observed in HOXB4-transduced LSK cells at 20 weeks, but not at 12 weeks (FDR < 0.05). The highest overexpressed genes in the HSC pathways included Jun, Evt1, Smad7, Angpt1, Laptm4b, Hoxa5, and Hoxa10 (Figure 4B).
GSEA and MetaCore pathway analysis of HOXB4 responsive genes. (A) GSEA analyses are shown for HOXB4 regulated genes identified for 2 different HSC pathways. LSK cell samples were obtained from transplanted mice at 12 and 20 weeks after transplant. For each GSEA, the nominal P value and FDR q-value are shown below each pathway graph. Statistical significance was validated for both pathways in 20-week samples. (B) Heatmap gene expression changes in individual gene probe sets within the Hematopoiesis Stem Cell pathway and Hematopoiesis Stem Cell Long Term pathway in LSK cells analyzed at 12 and 20 weeks after transplant. Each column represents an independent experiment within the indicated group. Red boxes represent upregulated genes and blue boxes represent downregulated genes. (C) GSEA analysis B-cell differentiation pathway genes altered by HOXB4 expression in 12- and 20-week LSK samples. Statistical parameters are listed below each analysis. (D) Pathway analysis of differentially expressed probe sets using MetaCore program analysis showing the top 10 pathways regulated by HOXB4 at 12 (yellow) and 20 weeks (blue) posttransplantation. The x-axis scale shows the −10 log (P value) of each pathway. The Development_Transcription regulation of granulocyte development pathway and Development_G-CSF-induced myeloid differentiation pathway are the top 2 pathways activated by HOXB4 at 20 weeks posttransplantation.
GSEA and MetaCore pathway analysis of HOXB4 responsive genes. (A) GSEA analyses are shown for HOXB4 regulated genes identified for 2 different HSC pathways. LSK cell samples were obtained from transplanted mice at 12 and 20 weeks after transplant. For each GSEA, the nominal P value and FDR q-value are shown below each pathway graph. Statistical significance was validated for both pathways in 20-week samples. (B) Heatmap gene expression changes in individual gene probe sets within the Hematopoiesis Stem Cell pathway and Hematopoiesis Stem Cell Long Term pathway in LSK cells analyzed at 12 and 20 weeks after transplant. Each column represents an independent experiment within the indicated group. Red boxes represent upregulated genes and blue boxes represent downregulated genes. (C) GSEA analysis B-cell differentiation pathway genes altered by HOXB4 expression in 12- and 20-week LSK samples. Statistical parameters are listed below each analysis. (D) Pathway analysis of differentially expressed probe sets using MetaCore program analysis showing the top 10 pathways regulated by HOXB4 at 12 (yellow) and 20 weeks (blue) posttransplantation. The x-axis scale shows the −10 log (P value) of each pathway. The Development_Transcription regulation of granulocyte development pathway and Development_G-CSF-induced myeloid differentiation pathway are the top 2 pathways activated by HOXB4 at 20 weeks posttransplantation.
The top canonical pathways activated and repressed in HOXB4-LSK cells at 20 weeks (supplemental Table 2) include multiple signaling, differentiation, and metabolic pathways. In particular, a distinct B-cell differentiation pathway (Ingenuity) was activated in HOXB4+ LSK cells at both 12 and 20 weeks (Figure 4C). Genes important for B-cell differentiation were activated in HOXB4+ LSK cells (supplemental Figure 1A). Pathway analysis of differentially expressed transcripts using MetaCore software identified several myeloid differentiation pathways as being activated by HOXB4 in LSK cells (Figure 4D). These included Development_Transcription regulation of granulocyte development and Development_G-CSF–induced myeloid differentiation pathways (Figure 4D and supplemental Figure 1B). Altogether, these results suggest HOXB4’s function may also include lineage-specific priming events and increased numbers of committed cells in the heterogenous LSK population.
Insertional activation of proto-oncogenes, including Prdm16, cooperates with HOXB4 to induce acute myeloid leukemia in transplanted mice
In an independent experiment to those described above, 54 mice were transplanted with MSCV-HOXB4-ires-GFP–transduced BM cells and followed over time for the development of hematopoietic abnormalities. Ninety MSCV-ires-GFP transplanted mice served as controls. One of 90 GFP recipients developed leukemia, which was late onset and GFP negative. Two out of the 54 HOXB4 recipients (#1869 and #2050) in the HOXB4 group developed GFP+ acute myelogenous leukemia (AML) at 4 and 9.5 months, respectively, following transplantation.
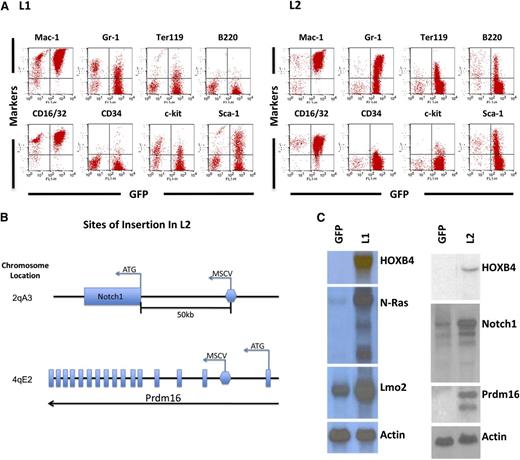
Leukemia #1869 (L1) was associated with marked leukocytosis, mild thrombocytopenia, and mild anemia (supplemental Table 3). Morphologic and immunophenotyping analyses of BM cells were consistent with AML and showed expression of Mac-1 and Gr-1 myeloid markers as well as CD16/32 and Sca1 (Figure 5A). Identification of vector insertion sites revealed 10 separate vector integration sites in L1 (supplemental Table 4), with integrations in or near the cellular proto-oncogenes Lmo2 and N-ras (supplemental Figure 2). Vector-induced gene activation was confirmed by northern blot analysis of tumor cells, showing a 15- to 20-fold increase in N-Ras mRNA and a twofold increase in Lmo2 relative to control GFP cells (Figure 5C).
HOXB4-mediated leukemia in transplanted mice due to vector insertion site-mediated oncogene activation. (A) Immunophenotyping of BM cells from recipients displaying leukemia cases L1 and L2. The y-axis shows the indicated cell surface markers identified by antibody staining, and the x-axis shows GFP expression associated with the HOXB4 vector. Expression of monomyeloid cell surface markers (Mac-1, CD16/32, Gr-1) and also erythroid (Ter119) and B-lymphoid (B220) markers are shown. A smaller percentage of the leukemic cells also expressed the stem cell markers c-kit and Sca1. (B) Retroviral insertion sites in L2 were identified by splinkerette PCR. The location of the 2 insertion sites in the adjacent to Notch1 and within Prdm16 and shown schematically along with the orientation of the vector at the insertion site. (C) Northern blot analysis of relevant oncogenic mRNAs from control and HOXB4 leukemic cells from both the L1 and L2 cases. The control cells consisted of BM cells of mice transplanted with normal BM cells transduced with the “empty” MSCV/GFP vector (lane labeled GFP). There is a markedly increased level of N-Ras mRNA (15- to 20-fold) and a mild increase (∼twofold) of Lmo2 mRNA in L1 BM cells when compared with control BM cells. There is a markedly increased level of Prdm16 mRNA and a mild increase (∼twofold) of Notch1 mRNA in L2 BM cells compared with control BM cells. Actin serves as an internal loading control.
HOXB4-mediated leukemia in transplanted mice due to vector insertion site-mediated oncogene activation. (A) Immunophenotyping of BM cells from recipients displaying leukemia cases L1 and L2. The y-axis shows the indicated cell surface markers identified by antibody staining, and the x-axis shows GFP expression associated with the HOXB4 vector. Expression of monomyeloid cell surface markers (Mac-1, CD16/32, Gr-1) and also erythroid (Ter119) and B-lymphoid (B220) markers are shown. A smaller percentage of the leukemic cells also expressed the stem cell markers c-kit and Sca1. (B) Retroviral insertion sites in L2 were identified by splinkerette PCR. The location of the 2 insertion sites in the adjacent to Notch1 and within Prdm16 and shown schematically along with the orientation of the vector at the insertion site. (C) Northern blot analysis of relevant oncogenic mRNAs from control and HOXB4 leukemic cells from both the L1 and L2 cases. The control cells consisted of BM cells of mice transplanted with normal BM cells transduced with the “empty” MSCV/GFP vector (lane labeled GFP). There is a markedly increased level of N-Ras mRNA (15- to 20-fold) and a mild increase (∼twofold) of Lmo2 mRNA in L1 BM cells when compared with control BM cells. There is a markedly increased level of Prdm16 mRNA and a mild increase (∼twofold) of Notch1 mRNA in L2 BM cells compared with control BM cells. Actin serves as an internal loading control.
The second leukemia case (L2) also showed marked leukocytosis and significant anemia (supplemental Table 3). Morphologic analysis revealed a myelomonocytic phenotype, and flow cytometry analysis demonstrated expression of myeloid (Gr-1, Mac-1), erythroid (Ter119), and B-lymphoid (B220) markers (Figure 5A). Retroviral integration site analysis in L2 revealed an intragenic integration in intron 1 of the Prdm16 gene and another integration event approximately 50 kb upstream of Notch1 gene (Figure 5B and supplemental Table 4). Increased levels of Prdm16 and Notch 1 mRNA were confirmed in L2 leukemia cells (Figure 5C). The 2 bands seen in the Prdm16 northern blot are consistent with expression of the short form of Prdm1637 from the larger mRNA. The smaller mRNA was most likely due to a noncanonical polyadenylation.
These leukemia cases illustrate that deregulated expression of specific proto-oncogenes is required in order for HOXB4 to induce AML. Induction of sPrdm16 expression by vector insertion in HOXB4-transduced hematopoietic cells has been previously reported in other transplant-induced leukemias in dogs and monkeys,38 suggesting that this event is recurrent and selected for in vivo. The fact that Prdm16 was never seen to cause insertional leukemia in the MSCV-GFP control group suggests a functional cooperation between Prdm16 and HOXB4 expression in leukemogenesis.
Repression of Prdm16 in HSCs is specific for HOXB4 and not seen with other leukemogenic Hox factors
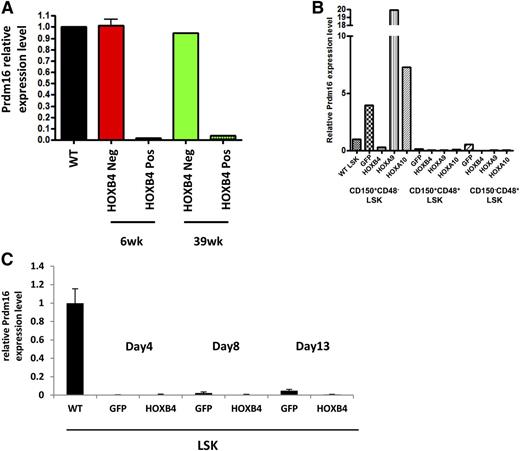
A time course of Prdm16 mRNA expression showed large decreases in Prdm16 transcripts in HOXB4-LSK cells relative to control LSK cells at both 6 weeks and 39 weeks after transplant (Figure 6A), demonstrating that downregulation of Prdm16 occurs early after transplant. The observations that sPrdm16 expression is activated in HOXB4-associated AML yet repressed in healthy HOXB4 transplanted mice suggests that repression of Prdm16 expression may explain why HOXB4 lacks the leukemogenic potential seen with other oncogenic HOX factors such as HOXA9 and HOXA10.
HOXB4 specific repression of Prdm16 in HSCs in vivo. (A) Quantitative RT-PCR for expression of Prdm16 mRNA in LSK cells harvested from transplanted mice at 6 weeks and 39 weeks posttransplantation. Cells were sorted based on expression of the HOXB4-YFP vector with negative cells serving as the control population. (B) Quantitative RT-PCR for expression of Prdm16 mRNA in GFP-, HOXB4-, HOXA9-, or HOXA10-transduced CD150+CD48−LSK cells, CD150+CD48+LSK cells, and CD150−CD48+ LSK cells from 6-week transplanted mice. Downregulation of Prdm16 by HOXB4 and upregulation of Prdm16 by HOXA9 or HOXA10 were detected in HSCs only. Low or no Prdm16 expression was detected in restricted HPCs compared with wild-type LSK cells. (C) Quantitative RT-PCR for expression of Prdm16 mRNA in GFP- and HOXB4-transduced LSKs 4 days, 8 days, and 13 days posttransduction. No Prdm16 expression was detected either GFP- or HOXB4-transduced LSK cells at any time point compared with wild-type LSK cells. In all panels, the expression level of Prdm16 in wild-type LSK cells was set to a control value of 1.0 and expression of Gapdh mRNA was used as internal control. WT, wild-type.
HOXB4 specific repression of Prdm16 in HSCs in vivo. (A) Quantitative RT-PCR for expression of Prdm16 mRNA in LSK cells harvested from transplanted mice at 6 weeks and 39 weeks posttransplantation. Cells were sorted based on expression of the HOXB4-YFP vector with negative cells serving as the control population. (B) Quantitative RT-PCR for expression of Prdm16 mRNA in GFP-, HOXB4-, HOXA9-, or HOXA10-transduced CD150+CD48−LSK cells, CD150+CD48+LSK cells, and CD150−CD48+ LSK cells from 6-week transplanted mice. Downregulation of Prdm16 by HOXB4 and upregulation of Prdm16 by HOXA9 or HOXA10 were detected in HSCs only. Low or no Prdm16 expression was detected in restricted HPCs compared with wild-type LSK cells. (C) Quantitative RT-PCR for expression of Prdm16 mRNA in GFP- and HOXB4-transduced LSKs 4 days, 8 days, and 13 days posttransduction. No Prdm16 expression was detected either GFP- or HOXB4-transduced LSK cells at any time point compared with wild-type LSK cells. In all panels, the expression level of Prdm16 in wild-type LSK cells was set to a control value of 1.0 and expression of Gapdh mRNA was used as internal control. WT, wild-type.
To test whether Prdm16 repression was specific to HOXB4, we compared Prdm16 transcript levels in HOXB4-, HOXA9-, and HOXA10-expressing HSCs (CD150+CD48−LSK cells), as well as restricted HPCs (CD150+CD48+LSK and CD150−CD48+ LSK cells), obtained from mice 6 weeks after transplant. Prdm16 was 13.5-fold downregulated in HOXB4-HSCs compared with GFP+ transduced control HSCs (Figure 6B). In contrast, Prdm16 expression was significantly upregulated by HOXA9 (5.0-fold) and HOXA10 (1.8-fold) compared with the GFP+ control. Therefore, the Prdm16 transcript levels in HOXA9 or HOXA10 HSCs were 67.5 times or 24.6 times higher than HOXB4 HSCs in vivo. In contrast, Prdm16 mRNA was very low to undetectable in all the HPC groups. The specific downregulation of Prdm16 in HOXB4-transduced HSCs in vivo may explain the specific lack of leukogenic activity with HOXB4 overexpression.
We next tested whether repression of Prdm16 by HOXB4 was also present in cultured LSK cells given that HSC expansion proceeds in a relatively unlimited fashion when HOXB4-transduced HSCs are grown in appropriate culture conditions.12 Compared with fresh wild-type LSK cells, Prdm16 expression was very low to undetectable in both GFP and HOXB4-transduced LSK cells at 4 days, 8 days, and 13 days posttransduction (Figure 6C). One possible explanation for this finding is that the LSK phenotype in cultured cells does not necessarily identify an enriched population of HSCs due to changes in cell surface marker expression in cultured cells.39,40 However, the very low levels detected in these populations are consistent with the possibility that downregulation of Prdm16 expression may not necessarily be required for maintaining HSC numbers within physiologic limits in vivo.
Enforced coexpression of sPrdm16 and HOXB4 causes preleukemia in mice
To define the normal endogenous Prdm16 expression pattern in HSCs and developing progenitors, we used qRT-PCR to measure mRNA expression levels at various stages of hematopoietic development in normal, untransplanted mice. Endogenous Prdm16 mRNA levels were 120-fold higher in long-term HSCs than in total BM cells (Figure 7A). Prdm16 mRNA was consistently downregulated in myeloid and lymphoid progenitors at various stages of development as well as in mature blood cell lineages. These results confirm prior studies showing that Prdm16 is selectively expressed at high levels in HSCs.22,23
Enforced coexpression of HOXB4 and sPrdm16 enhances HPC self-renewal in vitro and induces myeloid preleukemic transformation in transplanted mice. (A) Quantitative real-time PCR of endogenous Prdm16 mRNA levels in various sorted progenitor and stem cell populations. Gapdh mRNA was used as a loading control, and the level of Prdm16 mRNA expression in whole BM was set at a relative value of 1.0. (B) Schematic representation of lentiviral vectors to overexpress HOXB4 and sPrdm16. The promoters and fluorescent reporter genes used are indicated. The 400-bp chromatin insulator (i4r) was used as indicated to protect from position effects. (C) Western blot analysis of HOXB4 and sPrdm16 expression in NIH-3T3 cells transduced with the indicated vectors. Note the lower background band seen on the sPrmd16 blot is also present with the “empty” vector control. (D) Secondary colony numbers by myeloid CFU-C assay on sorted transduced cells as a measure of self-renewal capacity. Cells were transduced with control vectors or mock transduced or with single HOXB4 or sPrdm16 vectors or cotransduced with both HOXB4 and sPrdm16 vectors as indicated. After a primary CFU-C assay, cells were replated and colonies scored based on 10 000 cell inputs. Error bars show the standard deviation for multiple experiments, and statistical comparisons are indicated above the histograms. (E) Flow cytometry analyses of HOXB4 and sPrdm16 double-positive cells in peripheral blood (PB) and BMC of 2 primary recipients, # 890 and # 894. The top row shows staining for the donor background CD45.1, the middle row shows expression of the sPrdm16 vector marker (mCherry) and the HOXB4 vector marker (GFP), and the bottom row shows expression of the Mac-1 and Gr-1 myeloid markers. Note the expansion of double-positive myeloid cells in each of these cases. (F) BM cytospin photomicrographs of secondary recipients (#10037 and #10040) derived by transplantation from donor #890. Insets show higher magnification of the myeloid blasts seen in these cases, which comprised 30% to 50% of the cells on these slides. Slides were stained with Romanowsky stain, and images were acquired digitally on a Nikon ECLIPSE E800 microscope (Nikon, Japan) equipped with a Nikon DXM1200 camera and Nikon S Flour 40×/1.30 oil lens using NIS Elements software (Nikon). WT, wild-type.
Enforced coexpression of HOXB4 and sPrdm16 enhances HPC self-renewal in vitro and induces myeloid preleukemic transformation in transplanted mice. (A) Quantitative real-time PCR of endogenous Prdm16 mRNA levels in various sorted progenitor and stem cell populations. Gapdh mRNA was used as a loading control, and the level of Prdm16 mRNA expression in whole BM was set at a relative value of 1.0. (B) Schematic representation of lentiviral vectors to overexpress HOXB4 and sPrdm16. The promoters and fluorescent reporter genes used are indicated. The 400-bp chromatin insulator (i4r) was used as indicated to protect from position effects. (C) Western blot analysis of HOXB4 and sPrdm16 expression in NIH-3T3 cells transduced with the indicated vectors. Note the lower background band seen on the sPrmd16 blot is also present with the “empty” vector control. (D) Secondary colony numbers by myeloid CFU-C assay on sorted transduced cells as a measure of self-renewal capacity. Cells were transduced with control vectors or mock transduced or with single HOXB4 or sPrdm16 vectors or cotransduced with both HOXB4 and sPrdm16 vectors as indicated. After a primary CFU-C assay, cells were replated and colonies scored based on 10 000 cell inputs. Error bars show the standard deviation for multiple experiments, and statistical comparisons are indicated above the histograms. (E) Flow cytometry analyses of HOXB4 and sPrdm16 double-positive cells in peripheral blood (PB) and BMC of 2 primary recipients, # 890 and # 894. The top row shows staining for the donor background CD45.1, the middle row shows expression of the sPrdm16 vector marker (mCherry) and the HOXB4 vector marker (GFP), and the bottom row shows expression of the Mac-1 and Gr-1 myeloid markers. Note the expansion of double-positive myeloid cells in each of these cases. (F) BM cytospin photomicrographs of secondary recipients (#10037 and #10040) derived by transplantation from donor #890. Insets show higher magnification of the myeloid blasts seen in these cases, which comprised 30% to 50% of the cells on these slides. Slides were stained with Romanowsky stain, and images were acquired digitally on a Nikon ECLIPSE E800 microscope (Nikon, Japan) equipped with a Nikon DXM1200 camera and Nikon S Flour 40×/1.30 oil lens using NIS Elements software (Nikon). WT, wild-type.
We next sought to determine if enforced coexpression of HOXB4 and sPrdm16 was sufficient to cause leukemia. We generated lentiviral vectors that express HOXB4 with a GFP reporter gene or express sPrdm16 with a linked mCherry gene (Figure 7B). Western blot analysis showed robust expression of HOXB4 and sPrdm16 protein in transduced NIH-3T3 target cells (Figure 7C). Lin− BM cells were cotransduced with various combinations of these vectors, resulting in low proportions of cells expressing both the sPrmd16 and HOXB4 vector (supplemental Figure 3). Double-transduced cells were isolated and analyzed for myeloid progenitor self-renewal using a serial CFU-C replating assay. Cells transduced with the HOXB4 vector alone formed about 160 secondary colonies per 10 000 cells plated, whereas sPrdm16-transduced cells formed about 70 secondary colonies (Figure 7D). The HOXB4 and sPrdm16 double-positive group formed over 300 secondary colonies, significantly more than with the HOXB4 only group (P < .01). This result demonstrates that HOXB4 and sPrdm16 synergize to enhance self-renewal of hematopoietic progenitor cells.
We also transplanted bulk cotransduced BM cells into lethally irradiated recipient mice. Reconstitution with cells expressing the sPrdm16 vector was generally low, reflecting the low titer of this vector and/or direct toxic effects of enforced sPrdm16 expression. In most animals, no expansion of sPrdm16+ cells or hematopoietic abnormality was detected in both peripheral blood and BM. However, a large number of HOXB4 and sPrdm16 double-positive cells were detected in the peripheral blood in 2 of 12 recipients at 12 weeks after transplantation (cases #890 and #894). Expansion of HOXB4 and sPrdm16 double-positive blood cells occurred as early as 6 weeks after transplantation and further increased over time. When mouse #894 was killed at 12 weeks posttransplantation, 52.8% of peripheral blood cells and 97.1% of BM cells coexpressed the HOXB4 and sPrdm16 as determined by flow cytometry (Figure 7E). These double-positive cells expressed the Gr-1+, Mac-1+ myeloid phenotype.
BM cells from these 2 mice were used for secondary transplantation experiments. Secondary recipients from case #894 did not engraft with donor cells. In contrast, secondary recipients generated from #890 (#10037, #10038, and #10040) were all repopulated with HOXB4- and sPrdm16-coexpressing cells in peripheral blood and BM. At 12 and 16 weeks after transplant, pathological examination revealed prominent myeloid dysplasia with increased myeloid blasts in the BM, consistent with myelodysplasia and preleukemia (Figure 7F). Taken together, this experiment shows that HOXB4 and sPrdm16 overexpression can collaborate to promote preleukemia in at least some secondary transplant recipients.
Discussion
Enforced expression of HOXB4 causes murine HSC expansion in vivo without leading to hematopoietic malignancies. The genes that control this expansion have not been previously identified and are likely relevant for balancing proliferation and differentiation of HSCs. Prior gene expression profiling studies have identified various HOXB4 target genes in different systems,17-21 yet these studies show little overlap in identified target genes. None of these previous studies included analyses of primary adult HSCs derived directly from animals transplanted with HOXB4-transduced cells. Even less is known about how HOXB4 transcriptional events are dynamically regulated at various times during HSC reconstitution. This study addresses these questions by defining the HOXB4 transcriptional program directly in HSC-enriched populations derived from the BM of transplanted mice at multiple time points.
The HOXB4 gene expression pattern in transplanted LSK cells was fully established by 12 weeks after transplant. This is consistent with the known kinetics of HOXB4-induced HSC expansion in which HSC numbers in the BM were normal and stable by 12 weeks after transplant, but not at 8 weeks.15,16 The fact that the kinetics of the gene expression changes correlate with the plateauing of HSC numbers further establish that HOXB4-mediated HSC effects are transcriptionally mediated. GSEA showed that both short- and long-term HSC expression profiles were activated by HOXB4 expression and involved regulation of a number of genes known to be important for HSC function and self-renewal.
In addition to these HSC signatures, network analyses revealed enrichments in gene sets associated with B-cell and myeloid differentiation programs. This is consistent with the widely held viewpoint that HSC pool size is regulated by establishing equilibrium between commitment/differentiation vs HSC self-renewal fates. The LSK cell pool contains seven functionally distinct subpopulations of HSCs and multipotent progenitor cells with myeloid-biased or lymphoid-biased or myeloid-lymphoid balanced reconstitution capacity in transplanted mice.41 Therefore, these changes in expression of lineage specific genes could reflect a direct effect of HOXB4 in generating early B-cell and myeloid progenitors through balanced induction of both asymmetric and symmetric HSC replication.
Prdm16 was one of the most markedly downregulated genes in HOXB4-transduced LSK cells. At first, this appears paradoxical given that Prdm16 plays an essential role in HSC maintenance.22,23 Activation of sPRDM16 has also been associated with a variety of hematopoietic malignancies including t(1;3)(p36;q21)-positive myelodysplastic syndromes, AML, and T-cell leukemia.37,42-44 Zhang et al described cases of HOXB4-dependent myeloid leukemia in dogs and a macaque after receiving HOXB4-overexpressing CD34+ BM cells.38 In 2 of 3 cases, the HOXB4 vector had integrated into the PRDM16 locus and activated PRDM16 expression. Consistent with these findings, 2 of 54 HOXB4 transplanted mice in our study also developed AML posttransplantation, and 1 case was associated with an activating vector integration site within Prdm16 locus. In addition, cotransduction experiments demonstrated cooperation between HOXB4 and sPrdm16 for inducing self-renewal in myeloid progenitors and for inducing a preleukemic syndrome in 2 transplanted mice. These effects were unlikely due to Prdm16 activation alone because it was not seen in a large number of mice transplanted using control vector that lacked HOXB4.
In our study, we found that overexpression of HOXA9 or HOXA10 in HSCs resulted in markedly increased levels of Prdm16 in vivo relative to that seen with HOXB4, potentially explaining why overexpression of HOXA9 or HOXA10, but not HOXB4, causes acute leukemia. The mechanisms by which sPrdm16 cooperates with HOXB4 to induce leukemia are yet undefined. Expression of sPRDM16 blocks granulocyte-colony stimulating factor–induced myeloid differentiation,37,44 and overexpression of sPRDM16 in mice causes a myeloproliferative disorder and leukemia when enforced in p53-null BM cells.45 Therefore, downregulation of Prdm16 by HOXB4 may function by inducing differentiation in the face of increased HSC self-renewal.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Richard Ashmun and the Flow Cytometry Core for assistance in flow cytometry experiments, the Hartwell Center for Bioinformatics and Biotechnology for expression array analysis, Laura Janke for the pathology diagnosis of mice, Svetlana Rogulina for skilled technical assistance, and Animal Resource Center for animal care.
This work was supported by the National Institutes of Health, National Heart, Lung, and Blood Institute (grant P01 HL 53749), the National Cancer Center (support grant P30 CA 21765), the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities.
Authorship
Contribution: H.Y., H.Z., and H.M.L designed and performed the research, analyzed data, and wrote the paper; Z.M. performed the research; G.N. and S.Z. analyzed data; and B.G.F and B.P.S designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Brian P. Sorrentino, Experimental Hematology Division, Hematology Department, MS 341, Room D-3007D, Thomas Tower, St. Jude Children’s Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105-3678; e-mail: brian.sorrentino@stjude.org.