Key Points
Reducing Id2 in Gfi-1−/− mice restores radioprotective function of hematopoietic progenitors and partially rescues erythroid development.
Rescue of erythroid development in Gfi-1−/− mice by Id2 reduction directly correlates with an increase of Gata1, Eklf, and EpoR expression.
Abstract
Growth factor independence 1 (Gfi-1) is a part of the transcriptional network that regulates the development of adult hematopoietic stem and progenitor cells. Gfi-1-null (Gfi-1−/−) mice have reduced numbers of hematopoietic stem cells (HSCs), impaired radioprotective function of hematopoietic progenitor cells (HPCs), and myeloid and erythroid hyperplasia. We found that the development of HPCs and erythropoiesis, but not HSC function, was rescued by reducing the expression of inhibitor of DNA-binding protein 2 (Id2) in Gfi-1−/− mice. Analysis of Gfi-1−/−;Id2+/− mice revealed that short-term HSCs, common myeloid progenitors (CMPs), erythroid burst-forming units, colony-forming units in spleen, and more differentiated red cells were partially restored by reducing Id2 levels in Gfi-1−/− mice. Moreover, short-term reconstituting cells, and, to a greater extent, CMP and megakaryocyte-erythroid progenitor development, and red blood cell production (anemia) were rescued in mice transplanted with Gfi-1−/−;Id2+/− bone marrow cells (BMCs) in comparison with Gfi-1−/− BMCs. Reduction of Id2 expression in Gfi-1−/− mice increased the expression of Gata1, Eklf, and EpoR, which are required for proper erythropoiesis. Reducing the levels of other Id family members (Id1 and Id3) in Gfi-1−/− mice did not rescue impaired HPC function or erythropoiesis. These data provide new evidence that Gfi-1 is linked to the erythroid gene regulatory network by repressing Id2 expression.
Introduction
Hematopoiesis is a dynamic cellular process, whereby blood cells are continuously replenished from a pool of hematopoietic stem cells (HSCs).1 This process is dependent upon the correct expression of transcription factors at the appropriate levels and developmental stages, which regulate specific gene expression programs that are required for the proliferation, self-renewal, and differentiation of HSCs.2,3 Deregulation of transcription factor function is frequently associated with the initiation and progression of hematopoietic malignancies. Thus, current efforts are focused on identifying critical transcription factors and transcription factor networks required for hematopoietic development.
Gfi-1 encodes a transcriptional repressor, which is highly expressed in hematopoietic cells and required for the proper maintenance of hematopoietic stem and progenitor cells (HSPCs) and for the development of multiple hematopoietic lineages.4-6 Mice that lack Gfi-1 are viable but have few HSCs, reduced lymphoid cell numbers, myeloid/erythroid hyperplasia, and a maturation arrest in neutrophil development.7-10 Gfi-1−/− bone marrow (BM) cells (BMCs) have a compromised ability to reconstitute lethally irradiated mice.7,10 Mutations in the zinc-finger domains of growth factor independence 1 (Gfi-1) result in hyperplasia and severe congenital neutropenia in humans, characterized by a failure to develop mature neutrophils in patients.11,12 Gfi-1–target genes have been identified, and some have been shown to mediate the hematopoietic defects in Gfi-1−/− mice, which include maturational defect in granulocyte (CSF-1, RasGRP1, Pu.1) and B cell (Pu.1 or Id2) developments and myeloid hyperplasia (Id2 or HoxA9).13-18 However, Gfi-1–target genes that mediate the defects in radioprotection, maintenance of HSCs/hematopoietic progenitor cells (HPCs), and erythroid hyperplasia in Gfi-1−/− mice are unknown.
Id proteins function by inhibiting the DNA-binding ability of basic-helix-loop-helix proteins, which regulate the expression of cell-cycle inhibitors and genes necessary for differentiation.19-21 Because Id2 is upregulated in HSCs/HPCs in Gfi-1−/− mice and it is a direct repressed target of Gfi-1, we hypothesized that the defects in short-term reconstituting cells (STRCs) and HSCs in Gfi-1−/− mice may be rescued by reducing Id2 expression levels.17 In this study, we used 2 mouse models to investigate how the expression levels of Id2 affect hematopoietic development and maintenance of HSCs/HPCs and report that reduction of Id2 in Gfi-1−/− mice rescues the STRC activity of Gfi-1−/− BMCs but does not restore defects in HSC maintenance and self-renewal. Furthermore, rescue of STRCs directly correlated with restoration of erythroid development in Gfi-1−/− mice and with an increase in Gata1, Eklf, and EpoR expression. In contrast, reduction of other Id gene (Id1 or Id3) expression in Gfi-1−/− BMCs did not rescue erythroid development or defects in other hematopoietic lineages. These results provide new evidence that Gfi-1 is linked to the erythroid gene regulatory network by repressing Id2 expression.
Methods
Mice
Gfi-1–, Id1-, Id2-, and Id3-deficient mice have been described in previous manuscripts, and all mice were backcrossed onto C57/Bl6 backgrounds.8,22-24 Animals were treated in accordance with the guidelines of the Frederick National Laboratory for Cancer Research Animal Care and Use Committee in accordance with the Guide for Care and Use of Laboratory Animals. The Frederick National Laboratory for Cancer Research is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals.
BM transplantation (BMT)
BM reconstitution assays were performed by transplanting CD45.2-positive BMCs from femurs and tibias into lethally irradiated (9.5 Gy from a 137Cs source) CD45.1-C57BL/6 recipient mice. For competitive reconstitution assays, 2 × 106 BMCs from Gfi-1+/+, Gfi-1−/−, or Gfi-1−/−;Id2+/− (CD45.2) mice and 2 × 105 BMCs from CD45.1-C57BL/6 mice were transplanted into individual recipients. Survival was monitored for 4 to 6 months, and femurs and tibias were harvested and analyzed for donor reconstitution. To test short-term reconstituting activity, 1 × 106 BMCs from donor mice were transplanted into CD45.1 recipient mice. The survival of recipient mice was monitored for up to 3 months, and then hematopoietic tissues were analyzed for reconstitution by flow cytometry.
BFU-E colony assay and colony-forming unit (CFU) in spleen (CFU-S8) assay
For erythroid burst-forming unit (BFU-E) assays, we plated 3.5 × 104 BMCs or 5.0 × 104 splenocytes in 1 mL of methylcellulose medium (MethoCult M3334; STEMCELL Technologies) supplemented with mSCF, interleukin-3, and Epo in 35-mm Petri dishes, and then cultured cells for 7 to 10 days at 37°C. BFU-E colonies were identified based on shape, hemoglobinization (red), and by staining with 0.03% benzidine (B3383; Sigma-Aldrich, St. Louis, MO), which was dissolved in 0.5 M acetic acid (A6283; Sigma-Aldrich) and 1% hydrogen peroxide.
To determine the number of erythroid progenitors (CFU-S8) within a cell population, we transplanted 1 × 106 donor BMCs into lethally irradiated recipient mice. Eight days after BMT, the spleens were fixed in Tellesniczky’s solution, and spleen colony formation was enumerated.
Retroviral production, transduction, and cell sorting
The pRetro-Id2 short hairpin RNA (shRNA) green fluorescent protein (GFP) or pRetro-nonspecific control shRNA-GFP retroviral plasmids (Cellogenetics, Ijamsville, MD) were transiently transfected into Phoenix “producer” cells with TransIT-293 transfection reagent (Mirus, Madison, WI) to produce retroviral particle containing supernatants, which were used to transduce BMCs according to the manufacturer’s instructions.17
BMCs from Gfi-1+/+ or Gfi-1−/− mice were labeled with anti-CD71-PerCP-Cy5.5 and anti-Ter119-PE (BD Bioscience), sorted using a BD FACS Aria II cell sorter (BD Biosciences), and then cultured in erythroid cell growth medium (described in the supplemental Methods, available at the Blood Web site).25 Ter119−/CD71− sorted BMCs were cultured in erythroid medium for 12 hours, and then the cells were transduced with Id2-shRNA or NS-shRNA retrovirus on nontissue culture-treated 6-well plates, which were previously coated with RetroNectin (50 μg/well; Takara Bio, Shiga, Japan), onto which the viral supernatant had been centrifuged for 2 hours (3600 rpm at 10°C). After centrifugation, residual viral supernatant was removed and cells were added to the plates and incubated for 12 hours. This transduction was repeated 3 times. After the last transduction, the cells were cultured for additional 5 days. Cells were then stained for Ter119 and CD71 and analyzed by flow cytometry as described above.
Results
STRCs are rescued by reducing Id2 levels in Gfi-1−/− BM
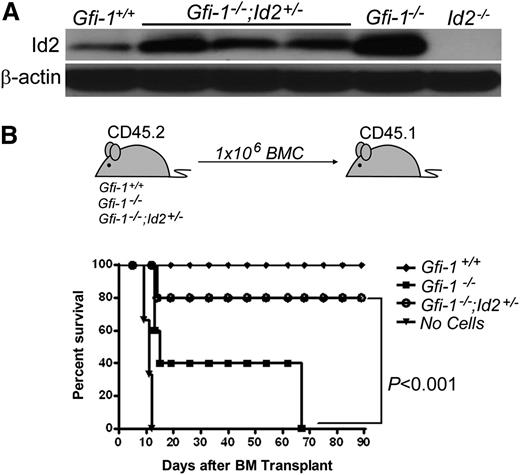
Id2 is a direct repressed target of Gfi-1 and is overexpressed in Gfi-1−/− BMCs compared with Gfi-1+/+ controls.17 Because overexpression of Id genes promotes HSPC proliferation, we hypothesized that lowering Id2 levels might rescue the defects in STRCs and loss of long-term HSCs (LT-HSCs) observed in the Gfi-1−/− mice. To test this, we generated Gfi-1−/−;Id2+/+, Gfi-1−/−;Id2+/−, and control mice in which Id2 protein levels were reduced in Gfi-1−/−;Id2+/− BMCs compared with Gfi-1−/− mice (Figure 1A). Because Gfi-1+/+;Id2+/+, Gfi-1+/+;Id2+/−, and Gfi-1+/−;Id2+/− mice showed the same phenotype, we used Gfi-1+/+;Id2+/− mice as the “control group” for all experiments presented in this manuscript and referred to as “Gfi-1+/+.”
STRC activity failure in Gfi-1−/− BMCs is rescued by Id2 reduction. (A) Western blot analysis showing heterozygosity at the Id2 locus results in reduced expression of Id2 protein levels in Gfi-1−/− mice. β-actin was used to demonstrate equal loading of protein. (B) STRC activity failure of Gfi-1−/− BMCs is rescued by reducing Id2 levels. Schematic representation of transplantation experiment (top). Survival of transplant recipients by 90 days (bottom). Recipient mice were lethally irradiated with 9.5 Gy then transplanted with 1 × 106 BMCs of the indicated genotype (n = 5 for each group except “no cells,” where n = 3).
STRC activity failure in Gfi-1−/− BMCs is rescued by Id2 reduction. (A) Western blot analysis showing heterozygosity at the Id2 locus results in reduced expression of Id2 protein levels in Gfi-1−/− mice. β-actin was used to demonstrate equal loading of protein. (B) STRC activity failure of Gfi-1−/− BMCs is rescued by reducing Id2 levels. Schematic representation of transplantation experiment (top). Survival of transplant recipients by 90 days (bottom). Recipient mice were lethally irradiated with 9.5 Gy then transplanted with 1 × 106 BMCs of the indicated genotype (n = 5 for each group except “no cells,” where n = 3).
To test if reducing Id2 levels rescues the STRC activity of Gfi-1−/− BMCs, we transplanted 1E6 Gfi-1−/−;Id2+/−, Gfi-1−/−, or Gfi-1+/+ BMCs into lethally irradiated recipients (Figure 1B). Gfi-1−/− BMC recipients became moribund within 60 days, whereas only 1 of 5 Gfi-1−/−;Id2+/− BM recipients became moribund, and survived 90 days, demonstrating that lowering Id2 levels in Gfi-1−/− mice rescues STRCs in Gfi-1−/− BMCs. In contrast, transplantation of the same BMC populations in competitive reconstitution assays showed that lowering Id2 levels in Gfi-1−/− mice does not rescue the defect in LT-HSC numbers (supplemental Figure 1A-B).
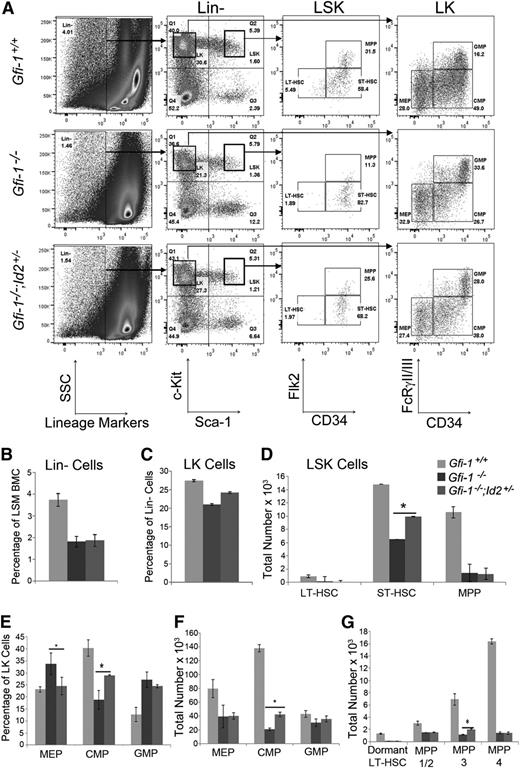
To determine which STRC populations were rescued by lowering Id2 in Gfi-1−/− mice, we analyzed HSPCs by flow cytometry (Figure 2A). We selected lineage-negative (Lin−) Sca-1+c-Kit+ cells (LSK) and measured LT-HSCs (LSK:CD34−Flk2−), short-term HSCs (ST-HSCs) (LSK:CD34+Flk2−), and multipotent progenitors (MPPs) (LSK:CD34+Flk2+). We used the percentages of Lin− (Figure 2B) and LSK cells and total BM cellularity to determine total numbers of LT-HSCs, ST-HSCs, and MPPs (Figure 2D). As expected, the number of LT-HSCs was significantly decreased in Gfi-1−/− BM compared with Gfi-1+/+ BM and was not rescued by reducing Id2 expression in Gfi-1−/− mice (Figure 2A,D). In comparison, ST-HSCs and MPPs were significantly decreased in Gfi-1−/− BM, and ST-HSCs were partially rescued by reducing Id2 expression in vivo (Figure 2D). We confirmed these results by evaluating LSK with CD150 (SLAM markers) in combination with the other surface markers.26 We found that LSK:Flk2−CD34+CD150− (MPP-3), which include ST-HSCs, were rescued by reducing Id2 levels in Gfi-1−/− BM, but not LSK:Flk2−CD34−CD150+ (dormant LT-HSCs) or LSK:Flk2−CD34+CD150+ (MPP-1/2), or LSK:Flk2+CD34+ (MPP-4) (Figure 2G and supplemental Figure 2). Thus, restoration of ST-HSC and MPP-3 cells in Gfi-1−/−;Id2+/− BMCs directly correlated with rescue of STRCs in vivo.
Gfi-1−/− ST-HSCs and CMPs are partially rescued by reducing Id2 levels. (A) Flow cytometric analyses of Lin−/Sca-1+/c-Kit+ (LSK) and Lin−/Sca-1−/c-Kit+ (LK) BMCs for HSPCs. Representative flow cytometry plots indicating LT-HSCs, ST-HSCs, and MPPs in LSK identified by Flk2 and CD34 cell-surface markers and CMPs, MEPs, and GMPs in LK identified by FcRγII/III and CD34. (B) Bar graph showing the percentage of Lin− cells in light-density separation medium (LSM)-purified BMCs harvested from 2 femurs and 2 tibias of each mouse. The data are presented as the mean % ± standard deviation (SD). The percentages of Lin− cells are reduced in Gfi-1−/− mice compared with control mice. (C) Bar graphs showing the percentage of LK cells in Lin− BMC population. The data are presented as the mean % ± SD. (D) Total number of LT-HSCs, ST-HSCs, and MPPs in BMCs, which were obtained by multiplying the percentage Lin− and LSK cells in BMCs, the percentage of each subpopulation identified by Flk2 and CD34, and the total cellularity of 2 femurs and 2 tibias. The data are presented as the mean ± SD. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− ST-HSCs. (E) Bar graph showing the percentage of CMPs, MEPs, and GMPs in LK population. Data are presented as the mean % ± SD. (F) Total number of CMPs, MEPs, and GMPs in BMCs, which were obtained by multiplying the percentage of Lin− and LK cells, the percentage of each subpopulation identified by FcR and CD34, and the total cellularity of 2 femurs and 2 tibias. Data are presented as the mean ± SD. (G) Bar graph showing total numbers of LT-HSC, MPP-1/2, MPP-3, and MPP-4 subpopulations, which were obtained by multiplying the percentages of Lin− and LSK cells in BMCs, the percentage of Flk2-positive or negative cells, the percentage of each subpopulation identified by CD150 and CD34, and the total cellularity of 2 femurs and 2 tibias. The graphs show data combined from 3 individual immunophenotypic analyses. The data are presented as the mean ± SD. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− for MPP-3.
Gfi-1−/− ST-HSCs and CMPs are partially rescued by reducing Id2 levels. (A) Flow cytometric analyses of Lin−/Sca-1+/c-Kit+ (LSK) and Lin−/Sca-1−/c-Kit+ (LK) BMCs for HSPCs. Representative flow cytometry plots indicating LT-HSCs, ST-HSCs, and MPPs in LSK identified by Flk2 and CD34 cell-surface markers and CMPs, MEPs, and GMPs in LK identified by FcRγII/III and CD34. (B) Bar graph showing the percentage of Lin− cells in light-density separation medium (LSM)-purified BMCs harvested from 2 femurs and 2 tibias of each mouse. The data are presented as the mean % ± standard deviation (SD). The percentages of Lin− cells are reduced in Gfi-1−/− mice compared with control mice. (C) Bar graphs showing the percentage of LK cells in Lin− BMC population. The data are presented as the mean % ± SD. (D) Total number of LT-HSCs, ST-HSCs, and MPPs in BMCs, which were obtained by multiplying the percentage Lin− and LSK cells in BMCs, the percentage of each subpopulation identified by Flk2 and CD34, and the total cellularity of 2 femurs and 2 tibias. The data are presented as the mean ± SD. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− ST-HSCs. (E) Bar graph showing the percentage of CMPs, MEPs, and GMPs in LK population. Data are presented as the mean % ± SD. (F) Total number of CMPs, MEPs, and GMPs in BMCs, which were obtained by multiplying the percentage of Lin− and LK cells, the percentage of each subpopulation identified by FcR and CD34, and the total cellularity of 2 femurs and 2 tibias. Data are presented as the mean ± SD. (G) Bar graph showing total numbers of LT-HSC, MPP-1/2, MPP-3, and MPP-4 subpopulations, which were obtained by multiplying the percentages of Lin− and LSK cells in BMCs, the percentage of Flk2-positive or negative cells, the percentage of each subpopulation identified by CD150 and CD34, and the total cellularity of 2 femurs and 2 tibias. The graphs show data combined from 3 individual immunophenotypic analyses. The data are presented as the mean ± SD. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− for MPP-3.
To evaluate if committed progenitors were rescued by reducing Id2 levels in Gfi-1−/− mice, we analyzed Lin−Sca-1−c-Kit+ (LK) cells for common myeloid progenitors (CMPs; LK:CD34+FcγRII/III−), megakaryocyte-erythroid progenitors (MEPs; LK:CD34−FcγRII/III−), and granulocyte-macrophage progenitors (GMPs; LK:CD34+FcγRII/III+) (Figure 2A,E-F). Gfi-1−/− mice showed an increased percentage of GMPs and a decreased percentage of CMPs within the LK population previously demonstrated; however, we observed an increase in the percentage of MEPs in Gfi-1−/− LK cells compared with Gfi-1+/+ LK cells (Figure 2E). Reduction of Id2 in Gfi-1−/− mice restored the percentages of MEPs and partially rescued CMPs, but not GMPs. When these data were normalized using the percent LK (Figure 2C) and BM cellularity, we found that the total numbers of CMPs were partially restored, whereas the total numbers of MEPs and GMPs were not rescued by reducing Id2 expression (Figure 2F). Therefore, ST-HSCs and CMPs were rescued by reducing Id2 expression in Gfi-1−/− mice, whereas the LT-HSCs, MEPs, and GMPs were not. Because purified ST-HSCs, MPPs, CMPs, and MEPs have been shown to radioprotect lethally irradiated recipient mice, the ST-HSC and CMP progenitors are likely to be responsible for the restored STRC activity observed in Gfi-1−/−;Id2+/− BMCs.27
Rescue of STRC activity after BMT correlates with restored numbers of ST-HSCs and MEP-enriched c-Kithi LK cells in Gfi-1−/− BMCs
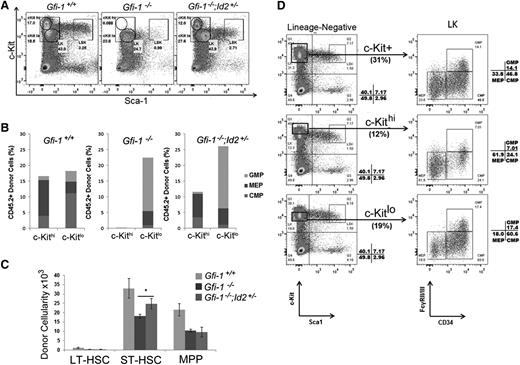
To elucidate which HSCs/HPCs are rescued by reducing Id2 levels in Gfi-1−/− BMCs in vivo, we transplanted BMCs into lethally irradiated recipients and evaluated progenitor cell recovery in 36 days after BMT. LSK and LK numbers were reduced in the BM of Gfi-1−/− BMC recipients compared with mice transplanted with Gfi-1+/+ BMCs (Figure 3A). Interestingly, this experiment showed that LK cells present in Gfi-1−/− BMC-transplanted mice completely lacked a subpopulation that expressed high levels of c-Kit (c-Kithi), but they did have a population that expressed low levels of c-Kit (c-Kitlo) cells (Figure 3A). The c-Kithi LK cells were restored in mice transplanted with Gfi-1−/−;Id2+/− BMCs (Figure 3A). To determine what progenitors were contained within the c-Kithi LK cells, we analyzed normal BMCs and determined that c-Kithi LK cells were enriched for MEPs (62%) compared with c-Kitlo cells (18%) (Figure 3D) and that c-Kitlo LK cells were enriched for CMPs and GMPs (78%). Similar to Gfi-1+/+ mice, the c-Kithi LK cells in Gfi-1−/−;Id2+/− BMC-transplanted mice were also enriched for MEPs (82%), suggesting that MEPs were rescued (Figure 3B). We also determined that donor LSK and ST-HSCs were rescued in mice transplanted with Gfi-1−/−;Id2+/− BMCs (Figure 3C). Thus, the rescue of STRC activity in vivo correlated with restored LSK/ST-HSC numbers and MEP-enriched c-Kithi LK cells, suggesting that lowering Id2 levels in Gfi-1−/− mice may rescue erythropoiesis.
Decreasing Id2 levels in Gfi-1−/− BMCs restores c-Kithi LK subpopulation in short-term BM reconstitution. (A) Flow cytometric analysis of donor LSK and LK c-Kithi/c-Kitlo populations 36 days after BM transplantation. LK cells were divided into c-Kithi and c-Kitlo subpopulations. Lack of donor c-Kithi LK cells in Gfi-1−/− BMCs (middle) was restored in Gfi-1−/−;Id2+/− BMCs (right). (B) Bar graph showing proportion of CMP, MEP, and GMP donor cells in c-Kithi and c-Kitlo cells, as determined by flow cytometry. Results are representative of 3 independent experiments. (C) Bar graph showing total numbers of donor LSK LT-HSCs, ST-HSCs, and MPPs in recipient BMCs. Cellularity was calculated based on total BMCs isolated from 2 femurs and 2 tibias of each recipient. These data are representative of 3 independent experiments. Data are presented as the mean ± SD. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− ST-HSC cellularity. (D) Flow cytometric analyses show the proportion of CMPs, MEPs, and GMPs found in total LK (top), LK c-Kithi (middle), and LK c-Kitlo (bottom) of wild-type mouse BMCs.
Decreasing Id2 levels in Gfi-1−/− BMCs restores c-Kithi LK subpopulation in short-term BM reconstitution. (A) Flow cytometric analysis of donor LSK and LK c-Kithi/c-Kitlo populations 36 days after BM transplantation. LK cells were divided into c-Kithi and c-Kitlo subpopulations. Lack of donor c-Kithi LK cells in Gfi-1−/− BMCs (middle) was restored in Gfi-1−/−;Id2+/− BMCs (right). (B) Bar graph showing proportion of CMP, MEP, and GMP donor cells in c-Kithi and c-Kitlo cells, as determined by flow cytometry. Results are representative of 3 independent experiments. (C) Bar graph showing total numbers of donor LSK LT-HSCs, ST-HSCs, and MPPs in recipient BMCs. Cellularity was calculated based on total BMCs isolated from 2 femurs and 2 tibias of each recipient. These data are representative of 3 independent experiments. Data are presented as the mean ± SD. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− ST-HSC cellularity. (D) Flow cytometric analyses show the proportion of CMPs, MEPs, and GMPs found in total LK (top), LK c-Kithi (middle), and LK c-Kitlo (bottom) of wild-type mouse BMCs.
BFU-E and CFU-S8 progenitors are rescued in Gfi-1−/− mice by decreasing Id2 expression
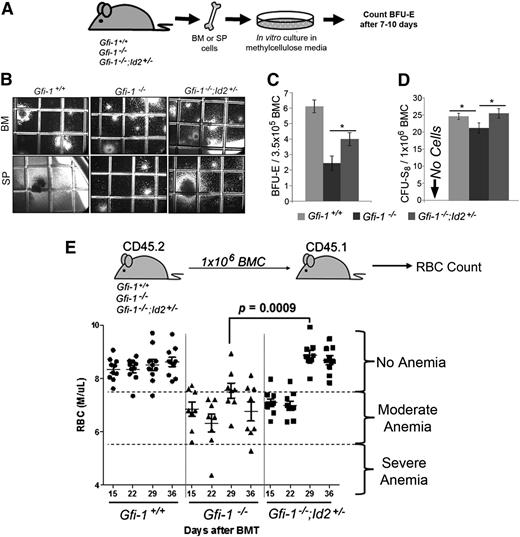
To test if Id2 reduction rescues erythroid development in Gfi-1−/− mice, we quantified erythroid progenitors in mice using BFU-E colony-formation and CFU-S8 assays (Figure 4A-D). The number of BFU-E (Figure 4C) and CFU-S8 (Figure 4D) between groups can be directly compared, because BM cellularity is the same in all animals. Gfi-1−/− BMCs had significantly fewer BFU-E colonies than Gfi-1+/+ BMCs, which were restored by reducing Id2 levels in Gfi-1−/−;Id2+/− mice (Figure 4C). Moreover, the size of BFU-E colony formation in BMCs and large colonies (1.5-2 mm) that contain erythroid and other cell lineages in spleen cells was also restored (Figure 4B). To confirm this observation, we determined the number of CFU-S8 in Gfi-1−/− and Gfi-1−/−;Id2+/− BMCs, which give rise to erythroid-containing colonies in vivo. We found that the number of CFU-S8 are decreased in Gfi-1−/− BMCs and that they are rescued by reducing the levels of Id2 in Gfi-1−/−;Id2+/− mice (Figure 4D). Finally, we have observed increased numbers of BFU-Es in the spleens of Gfi-1−/− mice compared with Gfi-1+/+ mice (supplemental Figure 3C), which is consistent with the previously observed extramedullary hematopoiesis including increased numbers of CFUs in culture and spleen cellularity.17 The number of BFU-E, CFUs in culture, and spleen cellularity17 were reduced in Gfi-1−/− spleens by lowering the levels of Id2 expression. This suggests that red cell development in Gfi-1−/− BMCs is rescued and extramedullary hematopoiesis reduced by lowering Id2 levels in vivo.
Reducing Id2 levels rescues impaired erythroid development of Gfi-1 knockout BM in vitro and in vivo. (A) Schematic representation of the BFU-E colony assay. A total of 5 × 104 BMC or 7.5 × 104 spleen cells were cultured in MethoCult to enumerate BFU-E for 7 to 10 days. Colonies were stained with benzidine as described in “Methods.” (B) Representative photomicrographs (original magnification ×40) of BFU-Es from BMCs and large CFU-mix (1.5-2 mm) from stem progenitor cells. Benzidine-stained colonies were identifiable by the dark brown/black color of the colonies. Grid on culture dish is 2 mm × 2 mm. (C) Bar graph showing total number of the stained BFU-E colonies per plate, which can be directly compared because BM cellularity is equivalent between mice. Large colonies showing typical BFU-E shape that were first identified as red hemoglobinized colonies and then confirmed by benzidine staining were enumerated. These data were obtained by counting triplicate samples and are representative of 3 independent experiments. The data are presented as the mean ± SD. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− BFU-E. (D) Bar graph showing total number of CFU-S8 counted in the spleens of recipient mice. The majority of CFU-S8 are erythroid-committed early progenitors. The data were obtained by counting triplicate spleens and are representative of 2 separate experiments. The data are presented as mean ± SD. *P < .05 for Gfi-1+/+ compared with Gfi-1−/−, and Gfi-1−/− compared with Gfi-1−/−;Id2+/− CFU-S8. (E) Schematic presentation of transplantation experiment (top). RBC counts in the peripheral blood of individual recipients measured 15, 22, 29, and 36 days after BMT (bottom). Column scatterplot showing RBC numbers in recipient mice (n = 10 per group). Data are representative of 2 separate experiments. SP, stem progenitor.
Reducing Id2 levels rescues impaired erythroid development of Gfi-1 knockout BM in vitro and in vivo. (A) Schematic representation of the BFU-E colony assay. A total of 5 × 104 BMC or 7.5 × 104 spleen cells were cultured in MethoCult to enumerate BFU-E for 7 to 10 days. Colonies were stained with benzidine as described in “Methods.” (B) Representative photomicrographs (original magnification ×40) of BFU-Es from BMCs and large CFU-mix (1.5-2 mm) from stem progenitor cells. Benzidine-stained colonies were identifiable by the dark brown/black color of the colonies. Grid on culture dish is 2 mm × 2 mm. (C) Bar graph showing total number of the stained BFU-E colonies per plate, which can be directly compared because BM cellularity is equivalent between mice. Large colonies showing typical BFU-E shape that were first identified as red hemoglobinized colonies and then confirmed by benzidine staining were enumerated. These data were obtained by counting triplicate samples and are representative of 3 independent experiments. The data are presented as the mean ± SD. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− BFU-E. (D) Bar graph showing total number of CFU-S8 counted in the spleens of recipient mice. The majority of CFU-S8 are erythroid-committed early progenitors. The data were obtained by counting triplicate spleens and are representative of 2 separate experiments. The data are presented as mean ± SD. *P < .05 for Gfi-1+/+ compared with Gfi-1−/−, and Gfi-1−/− compared with Gfi-1−/−;Id2+/− CFU-S8. (E) Schematic presentation of transplantation experiment (top). RBC counts in the peripheral blood of individual recipients measured 15, 22, 29, and 36 days after BMT (bottom). Column scatterplot showing RBC numbers in recipient mice (n = 10 per group). Data are representative of 2 separate experiments. SP, stem progenitor.
To determine if the same rescue occurred in vivo, we measured red blood cell (RBC) production after Gfi-1−/−;Id2+/− or Gfi-1−/− BMT (Figure 4E). Because saline-treated recipients were moribund 13 to 14 days after irradiation, we evaluated RBCs 15 days after BMT. We found that Gfi-1−/− BMCs produced significantly lower numbers of RBCs than Gfi-1+/+ BMCs 29 and 36 days after BMTs. In comparison, Gfi-1−/−;Id2+/− recipient RBC levels were similar to those of Gfi-1+/+ recipients, which directly correlated with the survival of transplanted mice. Altogether, these data suggest that reducing Id2 expression restores erythropoiesis of Gfi-1−/− BMCs both in vitro and in vivo.
Rescue of erythroid differentiation in Gfi-1−/− BMCs by lowering Id2 expression
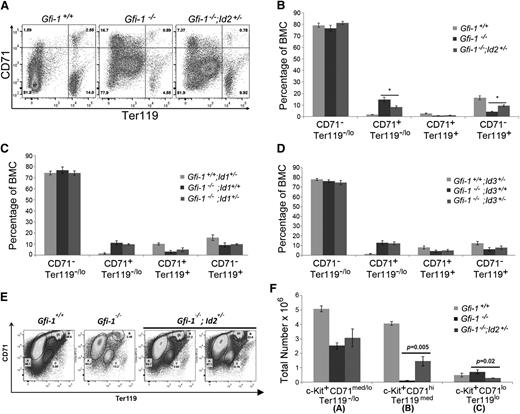
Because reducing Id2 levels in Gfi-1−/− mice rescued anemia and RBC production in mice transplanted with Gfi-1−/−;Id2+/− BMCs, we examined if the levels of Id2 were important for the later stages of erythropoiesis. Therefore, we measured CD71 and Ter119 expression, which are expressed on developing erythroid lineage cells in the BM. Specifically, CD71+Ter119neg/lo cells are enriched for erythroid CFUs (CFU-Es) and proerythroblasts but do not include BFU-Es,28-30 which differentiate into CD71+Ter119+ erythroblasts and into more mature CD71−Ter119+ erythroid cells. The percentage of these erythroid cell populations can be directly compared, since BM cellularity is the same in all animals (supplemental Figure 3D). We found that the CD71+Ter119neg/lo cell population was greatly increased in Gfi-1−/− BMCs compared with Gfi-1+/+ BMCs, which was reduced in Gfi-1−/−;Id2+/− mice, suggesting a potential block in erythroid development due to the high levels of Id2 in Gfi-1−/− mice (Figure 5A-B). In addition, the differentiated CD71−Ter119+ erythroid cells in Gfi-1−/− BMCs were partially rescued by lowering Id2 levels. Similarly, we observed increased numbers of CD71+Ter119neg/lo cells in Gfi-1−/− splenocytes that were also reduced by lowering Id2 levels (supplemental Figure 3A-B). These data indicate that reducing Id2 levels can diminish the accumulation of CD71+Ter119neg/lo erythroid progenitors in BMCs and spleens of Gfi-1−/− mice and normalize erythroid development.
Early and late stages in erythropoiesis are rescued in Gfi-1−/− mice by reducing Id2 levels. (A) Representative flow cytometric analyses of erythroid development in BMCs using CD71 and Ter119 surface markers. Upper left quadrant of contour plot contains CD71+/Ter119−/lo erythroid-committed progenitors, which further develop into CD71+/Ter119+ erythroblasts in the upper right quadrant of the contour plot, which differentiate into more mature CD71−/Ter119+ erythroid contains in the lower right quadrant of the contour plot. (B) Bar graph showing the percentages of erythroid populations identified by CD71 and Ter119 expression on Gfi-1+/+, Gfi-1−/−, and Gfi-1−/−;Id2+/− BMCs. The percentages can be directly compared between mice, because the total BM cellularity of all mice examined was equivalent. The data are presented as mean ± SD and are representative of 3 independent experiments. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− CD71+/Ter119−/lo and CD71−/Ter119+ cell populations. (C-D) Bar graphs showing the percentages of erythroid populations identified by CD71 and Ter119 expression on (C) Gfi-1+/+;Id1+/−, Gfi-1−/−;Id1+/+, and Gfi-1−/−;Id1+/− BMCs and (D) Gfi-1+/+;Id3+/−, Gfi-1−/−;Id3+/+, and Gfi-1−/−;Id3+/− BMCs. The data are presented as the mean ± SD. (E) Analysis of CD71 and Ter119 expression in c-Kit+ BMCs in Gfi-1+/+, Gfi-1−/−, and Gfi-1−/− ; Id2+/− mice. These include the percentages of CD71med/loTer119−/lo (A), CD71hiTer119med (B), and CD71loTer119lo (C) c-Kit+ BMCs. CD71hiTer119med erythroid progenitors in c-Kit+ BMCs were restored by reducing Id2 levels in Gfi-1−/− mice, and the increase in c-Kit+ CD71loTer119lo cell population in Gfi-1−/− BMCs was reduced by decreasing Id2 levels in vivo. (F) Total number of c-Kit+ CD71med/loTer119−/lo, c-Kit+ CD71hiTer119med, and c-Kit+ CD71loTer119lo erythroid progenitor cells in BMCs using total BM cellularity, percentage of c-Kit+ BMCs, and percentage of erythroid progenitors from 2 femurs and 2 tibias of each mouse. Data are presented as the mean ± SD.
Early and late stages in erythropoiesis are rescued in Gfi-1−/− mice by reducing Id2 levels. (A) Representative flow cytometric analyses of erythroid development in BMCs using CD71 and Ter119 surface markers. Upper left quadrant of contour plot contains CD71+/Ter119−/lo erythroid-committed progenitors, which further develop into CD71+/Ter119+ erythroblasts in the upper right quadrant of the contour plot, which differentiate into more mature CD71−/Ter119+ erythroid contains in the lower right quadrant of the contour plot. (B) Bar graph showing the percentages of erythroid populations identified by CD71 and Ter119 expression on Gfi-1+/+, Gfi-1−/−, and Gfi-1−/−;Id2+/− BMCs. The percentages can be directly compared between mice, because the total BM cellularity of all mice examined was equivalent. The data are presented as mean ± SD and are representative of 3 independent experiments. *P < .05 for Gfi-1−/− compared with Gfi-1−/−;Id2+/− CD71+/Ter119−/lo and CD71−/Ter119+ cell populations. (C-D) Bar graphs showing the percentages of erythroid populations identified by CD71 and Ter119 expression on (C) Gfi-1+/+;Id1+/−, Gfi-1−/−;Id1+/+, and Gfi-1−/−;Id1+/− BMCs and (D) Gfi-1+/+;Id3+/−, Gfi-1−/−;Id3+/+, and Gfi-1−/−;Id3+/− BMCs. The data are presented as the mean ± SD. (E) Analysis of CD71 and Ter119 expression in c-Kit+ BMCs in Gfi-1+/+, Gfi-1−/−, and Gfi-1−/− ; Id2+/− mice. These include the percentages of CD71med/loTer119−/lo (A), CD71hiTer119med (B), and CD71loTer119lo (C) c-Kit+ BMCs. CD71hiTer119med erythroid progenitors in c-Kit+ BMCs were restored by reducing Id2 levels in Gfi-1−/− mice, and the increase in c-Kit+ CD71loTer119lo cell population in Gfi-1−/− BMCs was reduced by decreasing Id2 levels in vivo. (F) Total number of c-Kit+ CD71med/loTer119−/lo, c-Kit+ CD71hiTer119med, and c-Kit+ CD71loTer119lo erythroid progenitor cells in BMCs using total BM cellularity, percentage of c-Kit+ BMCs, and percentage of erythroid progenitors from 2 femurs and 2 tibias of each mouse. Data are presented as the mean ± SD.
We have consistently observed a distinct population of CD71loTer119lo cells in Gfi-1−/− BMCs (Figure 5A) and spleen (supplemental Figure 3A) that resembles previously described c-Kit+ stress erythroid progenitor populations.31 Therefore, we gated Gfi-1−/− BMCs expressing c-Kit and reanalyzed the expression of CD71 and Ter119, and we observed a significant increase the in c-Kit+ CD71loTer119lo cell population and a striking reduction in the c-Kit+ CD71hiTer119med population, which are partially restored by reducing Id2 levels in Gfi-1−/− mice (Figure 5E-F). These data provide additional evidence for a developmental block in erythropoiesis in Gfi-1−/− BMCs under steady-state conditions, which is rescued by reducing Id2 levels in vivo.
Because reduction of Id2 in Gfi-1−/− BM facilitated erythroid development and restored STRCs, we asked if this rescue was specific to Id2 among the Id family members. To test this, we generated Gfi-1−/− mice that were heterozygous at the Id1 or Id3 loci and evaluated erythroid development. Similar to Id2, Id1, and Id3, protein levels were increased in Gfi-1−/− BMCs relative to Gfi-1+/+ BMCs (supplemental Figure 4A-B); therefore, it was possible that Id1 and Id3 could contribute to the hematopoietic phenotypes observed in Gfi-1−/− mice. However, we found that reducing expression levels of Id1 or Id3 in Gfi-1−/− mice did not rescue expansion of CD71+Ter119neg/lo erythroid cells in Gfi-1−/− BMCs (Figure 5C-D). Furthermore, the developmental defects in myeloid, B, and T cells in Gfi-1−/− mice were also not rescued by reducing Id1 or Id3 levels (supplemental Figure 4D-L). Taken together, erythroid development in Gfi-1−/− is rescued by reducing Id2 in vivo, but not by reducing Id1 or Id3 levels.
To confirm that Id2 reduction increases normal erythroid development in Gfi-1−/− BM, we isolated CD71−Ter119− cells from total BMCs in Gfi-1−/− or Gfi-1+/+ mice and evaluated their differentiation after reducing Id2 expression in vitro. CD71−Ter119− cells were transduced with a retroviral vector that expresses a shRNA targeting Id2 (Id2-shRNA) or a nonspecific shRNA (NS-shRNA) and GFP, which efficiently reduces Id2 expression.32 After retroviral transduction, the cells were cultured for 5 days in growth conditions that support erythroid differentiation, and then GFP-positive cells were analyzed for CD71 and Ter119 expression. Gfi-1−/− CD71−Ter119− cells transduced with Id2-shRNA gave rise to CD71+Ter119− cells (58%) and CD71+Ter119lo cells (33%), whereas cells transduced with NS-shRNA showed increased CD71+Ter119− cells (68%) and fewer CD71+Ter119lo cells (23%) (supplemental Figure 5A-B). Reducing Id2 expression did not significantly affect erythroid differentiation of Gfi-1+/+ CD71−Ter119− cells. Thus, targeted reduction of Id2 levels in Gfi-1−/− BMCs increases the differentiation of CD71+Ter119lo cells in vitro and supports the hypothesis that Gfi-1−/− mice show a block in erythroid differentiation, which is overcome by reducing Id2 levels in vitro and in vivo.
Id2 expression is dynamically regulated during erythropoiesis
Taken together, these data suggest that erythropoiesis depends upon regulation of Id2 expression for proper development. To examine the expression of Id2 during erythropoiesis, we used a novel “knockin” mouse model in which the Id2 promoter drives an enhanced yellow fluorescent protein (EYFP) reporter (Lei Sun, Ming Ji, W.K., K.D.K., and J.R.K., manuscript in preparation). These mice develop normally and accurately “report” Id2 promoter activity based on EYFP expression. Therefore, we analyzed BMCs of these mice for CD71, Ter119, and EYFP expression to determine if Id2 is expressed in developing erythroid lineage cells (Figure 6A-B). As cells acquired expression of CD71, they expressed comparatively high levels of Id2. However, Id2 expression decreased as cells matured into erythroblasts (CD71+Ter119+) and more differentiated erythroid cells (CD71−/loTer119+). Thus, high levels of Id2 are expressed in CD71+Ter119−/lo progenitors, which include CFU-Es and proerythroblasts, and Id2 levels are decreased during maturation into erythroblasts, supporting the hypothesis that downregulation of Id2 at the proerythroblast stage may be critical for these cells to undergo differentiation.
Expression of Id2 is upregulated in CFU-E and proerythroblast stages. (A) Dynamic changes of EYFP expression levels during the development of erythroid progenitors. BMCs expressing EYFP under control of the Id2 promoter (Id2-EYFP) were evaluated for CD71 and Ter119 expression by flow cytometry (middle). The expression of Id2-EYFP was evaluated in CD71+/Ter119−/lo CFU-E/proerythroblasts (upper left) in CD71+/Ter119+ basophilic erythroblasts (upper right) and in /CD71−/Ter119+ chromatophilic/orthochromatophilic erythroblasts (lower right). Id2-EYFP mice (black line) cells and control mice (gray line) for each histogram are compared. (B) Bar graph represents the percentage of Id2-EYFP–positive cells ± SD found in each stage of erythroid progenitor development from 3 mice. The data are representative of 3 independent experiments.
Expression of Id2 is upregulated in CFU-E and proerythroblast stages. (A) Dynamic changes of EYFP expression levels during the development of erythroid progenitors. BMCs expressing EYFP under control of the Id2 promoter (Id2-EYFP) were evaluated for CD71 and Ter119 expression by flow cytometry (middle). The expression of Id2-EYFP was evaluated in CD71+/Ter119−/lo CFU-E/proerythroblasts (upper left) in CD71+/Ter119+ basophilic erythroblasts (upper right) and in /CD71−/Ter119+ chromatophilic/orthochromatophilic erythroblasts (lower right). Id2-EYFP mice (black line) cells and control mice (gray line) for each histogram are compared. (B) Bar graph represents the percentage of Id2-EYFP–positive cells ± SD found in each stage of erythroid progenitor development from 3 mice. The data are representative of 3 independent experiments.
Lowering Id2 levels increases expression of Gata1, EpoR, and Eklf in Gfi-1−/− mice
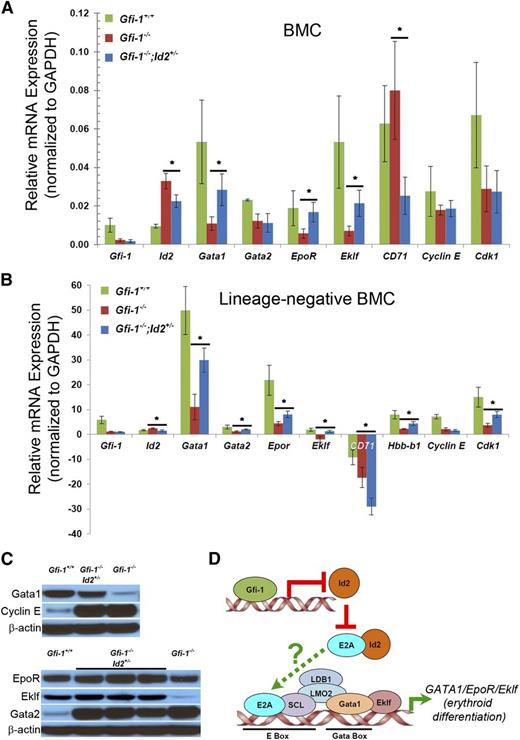
To understand mechanisms by which reducing Id2 rescues impaired erythropoiesis in Gfi-1−/− mice, we compared gene expression in Gfi-1−/−;Id2+/− and Gfi-1−/− BMCs using Affymetrix microarray (Table 1 and supplemental Table 1) (Gene Expression Omnibus accession number GSE59360). We found that genes controlling B-cell and myeloid cell development were differentially expressed, which confirmed our previous findings that B-cell development is rescued by reducing Id2 levels in Gfi-1−/− mice.17 In addition, we found that expression of CD71 (Tfrc) was downregulated and erythropoietin receptor (EpoR) was upregulated by Id2 reduction in Gfi-1−/− BMCs (Table 1). The downregulation of CD71 agrees with our finding that CD71+Ter119−/lo cells were increased by lowering Id2 levels in Gfi-1−/− mice (Figure 5A-B). We validated the expression of EpoR and CD71 and examined the expression of other genes expressed during erythroid differentiation (Gata1, Eklf, Gata2, and Hbb-b1) in whole BMCs (Figure 7A) and in purified HPCs (Lin− BMCs) (bottom graph) (Figure 7B) by quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR). As expected, Gfi-1 was not expressed in Gfi-1−/− BMCs, whereas Id2 expression was increased in Gfi-1−/− BMCs and reduced in Gfi-1−/−;Id2+/− BMCs (Figure 7A). Furthermore, the expression of Gata1, EpoR, and Eklf was significantly decreased, CD71 expression increased in Gfi-1−/− BMCs compared with wild-type mice, and the expression of Gata1, EpoR, and Eklf was rescued in Gfi-1−/−;Id2+/− vs Gfi-1−/− BMCs. We confirmed that the levels of Gata1, Eklf, and, to a lesser extent, EpoR protein expression were reduced in cell lysates from whole Gfi-1−/− BMCs compared with Gfi-1+/+ mice and were restored by reducing Id2 levels in Gfi-1−/− mice (Figure 7C). To evaluate if differences in gene expression between mice were the consequence of changes in BMC populations, we compared gene expression in Lin− BM progenitor cells, which were purified by removing mature BMC populations. We found that Gata-1, Eklf, and EpoR gene expression was similarly rescued in Lin− BMCs by reducing Id2 levels in vivo. In contrast to BMCs, CD71 expression was reduced in the Lin− progenitor cells, though this would be expected, because mature cells are removed by lineage depletion. These results suggest that reduction of Id2 levels in Gfi-1−/− mice increases the expression of transcription factors required for erythroid development (Gata and Eklf) and their target genes EpoR, Hbb-b1(Lin− BMCs), and CD71(BMC), which could account for the rescue of impaired erythroid development in Gfi-1−/−;Id2+/− mice.
Summary of microarray analyses for expression of EpoR and Tfrc (CD71) in BMCs of Gfi-1−/−;Id2+/− mice vs Gfi-1−/− mice
| Gene symbol . | RefSeq . | P . | Fold change (log2) . | Cell/tissue specificity . |
|---|---|---|---|---|
| Tfrc (CD71) | NM_011638 | .01957 | −2.46 | Erythrocyte |
| Id2 | NM_010496 | .00351 | −1.65 | Nonspecific |
| EpoR | NM_010149 | .04279 | 1.60 | Erythrocyte |
| Gene symbol . | RefSeq . | P . | Fold change (log2) . | Cell/tissue specificity . |
|---|---|---|---|---|
| Tfrc (CD71) | NM_011638 | .01957 | −2.46 | Erythrocyte |
| Id2 | NM_010496 | .00351 | −1.65 | Nonspecific |
| EpoR | NM_010149 | .04279 | 1.60 | Erythrocyte |
Id2 reduction increases Gata1, EpoR, Hbb-b1, Eklf, and CD71 expression levels in Gfi-1−/− mice. RNA was purified from (A) whole BMCs and (B) purified Lin− BM progenitors from the indicated mice to determine the expression of genes that regulate erythroid development and cell-cycle genes by real-time qRT-PCR. TaqMan primers were used to determine the expression of each of the indicated genes. Expression levels are reported relative Gapdh expression in mouse BMCs. *P < .05. (C) Western blot analysis of Gata1, Gata2, EpoR, Eklf, and Cyclin E protein levels in Gfi-1+/+, Gfi-1−/−, and Gfi-1−/−;Id2+/− BMCs. (D) Summary of potential mechanism of how Id2 reduction in the Gfi-1−/− background restores impaired erythroid development.
Id2 reduction increases Gata1, EpoR, Hbb-b1, Eklf, and CD71 expression levels in Gfi-1−/− mice. RNA was purified from (A) whole BMCs and (B) purified Lin− BM progenitors from the indicated mice to determine the expression of genes that regulate erythroid development and cell-cycle genes by real-time qRT-PCR. TaqMan primers were used to determine the expression of each of the indicated genes. Expression levels are reported relative Gapdh expression in mouse BMCs. *P < .05. (C) Western blot analysis of Gata1, Gata2, EpoR, Eklf, and Cyclin E protein levels in Gfi-1+/+, Gfi-1−/−, and Gfi-1−/−;Id2+/− BMCs. (D) Summary of potential mechanism of how Id2 reduction in the Gfi-1−/− background restores impaired erythroid development.
Discussion
Mice that lack Gfi-1 have few functioning HSCs and compromised STRC activity, and most HSPCs are in active cell cycle, which results in myeloid and erythroid hyperplasia.7,10 We report a novel erythroid maturation defect in Gfi-1−/− mice during steady-state hematopoiesis and demonstrate that Gfi-1−/− mice have reduced numbers of ST-HSCs, CMPs, and BFU-Es and more committed erythroid lineage cells. In addition, we observed an increase in CD71+Ter119neg/lo cells suggesting a block in proerythroblast development. We show that all of these defects are partially rescued or restored by reducing the levels of Id2 expression in Gfi-1−/− mice during steady-state hematopoiesis. We also found that the compromised ability of Gfi-1−/− BMCs to provide radioprotection in BMT assays during stress hematopoiesis was due, in part, to an impaired ability to give rise to ST-HSC and, to a greater extent, CMP and MEP and subsequent erythroid development resulting in reduced RBC production. Reduction of Id2 levels rescues the compromised STRC activity of Gfi-1−/− BMCs in BMT assays by restoring ST-HSC, CMP, and MEP development in vivo. In summary, our results provide evidence that links Gfi-1 to the erythroid transcription network by repressing Id2 expression levels in HPCs.
We found that the expression of Gata1, Eklf, and EpoR was downregulated in Gfi-1−/− BMCs and purified HPCs and that reduction of Id2 levels in Gfi-1−/− mice resulted in increased expression of these genes in vivo. These proteins are critical transcriptional regulators of erythroid development, and loss of any of these genes leads to profound defects in erythropoiesis.33-36 Scl is a tissue-specific basic-helix-loop-helix protein that dimerizes with E2A and specifically binds to E-box DNA motifs. E2A/Scl and Gata2/Gata1 proteins are part of a multiprotein transcription complex that binds to paired E-box/GATA sites in the promoters of erythroid-specific genes including Gata1, EpoR, Eklf, and β-globin to drive gene expression.37-40 Because it is known that Id2 directly binds to E2A, but not Scl, and inhibits the transcriptional activity of E2A/Scl, we propose that high levels of Id2 in Gfi-1−/− mice could inhibit E2A/Scl activity, resulting in decreased Gata1, Eklf, and EpoR expression and impaired erythropoiesis (Figure 7D).41,42 Interestingly, mice that lack Scl have impaired development of erythroid cells and compromised STRC activity, which is similar to that observed in Gfi-1−/− mice.33,43 Future experiments will be required to determine if Id2 directly inhibits multiprotein complex formation at paired E-box/Gata promoter sites. Thus, our results suggest a model whereby Gfi-1 is directly linked to the erythroid transcription factor networks via Id2, which may inhibit E2A/Scl function to regulate Gata1, Eklf, and EpoR gene expression.
Our data demonstrate that reducing Id2 levels can rescue ST-HSCs, CMPs, and BFU-Es in Gfi-1−/− BMCs during steady-state hematopoiesis as summarized in supplemental Figure 6. In addition, reduction of Id2 in Gfi-1−/− BMCs corrected the expansion of CD71+Ter119neg/lo cells and CFU-Es and restored proerythroblast numbers, indicating that Id2 may also function during the later stages of erythroid development. Thus, we propose that high levels of Id2 in Gfi-1−/− mice leads to expansion of CD71+Ter119neg/lo cells and a block in proerythroblast development. Regulation of the cell cycle is linked to differentiation, and mice that lack Rb have a block in the differentiation of proerythroblasts similar to that observed in Gfi-1−/− mice.44,45 Because Id2 is known to increase cell proliferation by binding and inhibiting Rb function, we speculate that high levels of Id2 may inhibit Rb function in Gfi-1−/− mice, resulting in the expansion of CD71+Ter119neg/lo cells and a developmental block in proerythroblast maturation. However, it has been shown that erythropoiesis is also impaired in Id2−/− mice, suggesting that the presence of Id2 is required at certain stages and that the dynamically regulated levels of expression are the key to functional erythroid development.32
This report shows that Gfi-1 is linked to the erythroid transcription network and functions by repressing Id2 expression in HSCs/HPCs. However, Gfi-1 also regulates the expression of other genes that may contribute to the other hematopoietic phenotypes observed in these mice. For example, Pu.1 is overexpressed in Gfi-1−/− BMCs, and lowering Pu.1 in Gfi-1−/− mice leads to a partial rescue of B cell and neutrophil development. The defect in neutrophil maturation is also rescued by lowering CSF-1 or by increasing RasGRP1 in Gfi-1−/− progenitors.14,15 The levels of Pu.1 are critical for the development of erythroid and myeloid lineages, because high levels of Pu.1 are known to antagonize Gata-1 function and inhibit erythroid development.16,46 Therefore, the high levels of Pu.1 may also contribute to impaired erythroid development in Gfi-1−/− mice. Reducing HoxA9 expression, but not HoxA7, in Gfi-1−/− mice rescues myeloid hyperplasia, but not the developmental block in neutrophil differentiation, similar to that observed by reducing Id2, suggesting that HoxA9 may regulate Id2 expression.18 Although overexpression of Bcl-2 in Gfi-1−/− cells restores the ability of Gfi-1−/− BMCs to reconstitute in lethally irradiated recipients by protecting HSCs from Bax-induced apoptosis, the Bcl-2 overexpression alone does not reverse the hyperproliferative defect in Gfi-1−/− BMCs.47 Thus, it is still unknown how Gfi-1 restricts HSC proliferation/quiescence and maintains normal numbers of HSCs.
Growth factor independence-1b (Gfi-1b), a paralog of Gfi-1, is required for megakaryopoiesis and erythropoiesis, but it is not known how Gfi-1b regulates these processes.48 Gfi-1 and Gfi-1b bind to the same consensus sequence and are known to regulate each other’s expression, so it is possible that Gfi-1b might also directly regulate Id2 expression.49 Expression levels of Gfi-1b and Gfi-1 have been examined during hematopoiesis using Gfi-1b-GFP and Gfi-1-GFP reporter strains. Gfi-1b expression is upregulated in CD71+Ter119− cells and remains at relatively high levels as these cells differentiate to CD71+Ter119+ cells, when Id2 levels begin to decrease. Because Gfi-1 is unlikely to be expressed in these cells, Gfi-1b may be required to downregulate Id2 expression during the later stages of erythroid development. Additional studies will be required to establish if Id2 is directly repressed by Gfi-1b.
In summary, this study provides new insights regarding the necessity of Gfi-1–mediated Id2 repression, which is required for the development of erythroid-committed progenitors and STRC activity during stress-induced hematopoiesis. Specifically, these findings set the foundation for future studies addressing mechanisms of Id2 regulation by Gfi-1 (and/or Gfi-1b) in hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mr Steve Stull and Ms Terri Stull for technical assistance in mouse care, BMT, peripheral blood collection, and tissue harvest; Ms Kathleen Noer, Roberta Matthai, and Guity Mohammadi for technical assistance in fluorescence-activated cell sorting; Drs Xiaolin Wu, Ling Su, and David Sun for technical assistance and data analysis in microarray and real-time qRT-PCR; and Ms Bobby Smith for technical assistance in CBC analysis. We thank Dr Yoshifumi Yokota for the Id2−/− mice and Dr Hanno Hock for the Gfi-1−/− mice.
This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported, in part, by the Intramural Research Program of the National Institutes of Health, Frederick National Laboratory, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Authorship
Contribution: W.K. designed and performed research, analyzed data, and wrote the paper; K.D.K. performed research, analyzed data, and wrote the paper; and J.R.K. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonathan R. Keller, Hematopoiesis and Stem Cell Biology Section, Mouse Cancer Genetics Program, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Basic Science Program, Leidos Biomedical Research, Inc., Building 560, Room 12-03, 1050 Boyles St, PO Box B, Frederick, MD 21701; e-mail: kellerjo@mail.nih.gov.