Key Points
Using state-of-the-art three-dimensional electron microscopy approaches, we show that the onset of the DMS formation is at the megakaryocyte plasma membrane.
A pre-DMS structure is formed in the perinuclear region, through a PM invagination process that resembles cleavage furrow formation.
Abstract
The demarcation membrane system (DMS) in megakaryocytes forms the plasma membrane (PM) of future platelets. Using confocal microscopy, electron tomography, and large volume focused ion beam/scanning electron microscopy (FIB/SEM), we determined the sequential steps of DMS formation. We identified a pre-DMS that initiated at the cell periphery and was precisely located between the nuclear lobes. At all developmental stages, the DMS remained continuous with the cell surface. The number of these connections correlated well with the nuclear lobulation, suggesting a relationship with cleavage furrow formation and abortive cytokinesis. On DMS expansion, Golgi complexes assembled around the pre-DMS, and fusion profiles between trans-golgi network–derived vesicles and the DMS were observed. Brefeldin-A reduced DMS expansion, indicating that the exocytic pathway is essential for DMS biogenesis. Close contacts between the endoplasmic reticulum (ER) and the DMS were detected, suggesting physical interaction between the 2 membrane systems. FIB/SEM revealed that the DMS forms an intertwined tubular membrane network resembling the platelet open canalicular system. We thus propose the following steps in DMS biogenesis: (1) focal membrane assembly at the cell periphery; (2) PM invagination and formation of a perinuclear pre-DMS; (3) expansion through membrane delivery from Golgi complexes; and (4) ER-mediated lipid transfer.
Introduction
The maturation of megakaryocytes (MKs) includes the development of a unique and extensive membrane system known as the demarcation membrane system (DMS), which divides the cytoplasm into small platelet territories. One MK is thought to produce an average of 4000 platelets. Although it has been known for many years that the DMS ultimately forms the cell membrane of the future platelets,1,2 the exact mechanism of the formation of this unique membrane system remains unclear. Early electron microscopy (EM) examination of late stage mature MKs led to the proposal that the DMS demarcates already preformed platelets.3 Observations by De Bruyn of long MK extensions protruding into the sinusoidal lumen modified this idea and suggested that the DMS divides the MK cytoplasm into intertwined and compacted cylindrical regions.4 Such a model had already been proposed earlier by Thiery and Bessis, who observed that MKs from bone marrow (BM) explants extended elongated projections they called proplatelets.5 Studies using cultured MKs led to a more refined model, where the DMS appears to function as a membrane reservoir for the extension of proplatelets, which would then fragment into platelets along their length6 or at their tips.7-9 More recently, using intravital microscopy to visualize platelet generation in mice, Junt et al10 have provided evidence that MKs extend voluminous processes into the lumen of sinusoids, which are sheared off by the flowing blood and thereby produce proplatelets that subsequently fragment into individual platelets.2,11-13
Although the DMS has been well characterized at the end stages of MK maturation, little is known about the biogenesis of this unique membrane system during the initial stages of MK development. Various subcellular origins have been proposed as a source of DMS biogenesis: (1) the MK plasma membrane (PM), (2) specializations of the endoplasmic reticulum (ER) or the Golgi apparatus, and (3) de novo membrane formation.2,14-16 The current view favors a mechanism involving invagination of the MK plasma membrane. This is supported by EM studies using extracellular tracers that demonstrated that the DMS is continuous with the extracellular environment. Using live cell imaging of mature MKs, Mahaut-Smith et al17 showed that the DMS is electrophysiologically contiguous with the peripheral plasma membrane. In this context, it was recently proposed that the term “invaginated membrane system” would be more appropriate than DMS.6,8,18 However, continuity of the DMS with the PM does not provide definitive evidence that PM invagination is the sole source of these membranes. Considering the large amount of membrane required for the biogenesis and expansion of the DMS, it follows that fast de novo membrane synthesis must also be essential. It remains to determine whether the DMS is formed intracellularly from the Golgi and then directed to the cell surface or whether newly formed membrane is first delivered to the PM and then rapidly invaginates. It also remains to define the exact contribution of the ER, the largest intracellular membrane source in eukaryotic cells, where lipids are synthesized and lipid transfer takes place.
In the present study, we used confocal microscopy imaging and high resolution EM and tomography to investigate the biogenesis and expansion of the DMS, both in situ in mouse BM MKs and in vitro in cultured MKs generated from progenitors. In immature MKs, we found the pre-DMS to be a centrally located well-defined membrane complex, which at its earliest detectable stage was continuous with the cell surface. Moreover, we found that it originates from a distinct focal site at the plasma membrane. Our data further show that Golgi-derived membranes and close ER contacts contribute to a continuous membrane supply for DMS growth.
Materials and methods
See supplemental Materials on the Blood Web site for details. All procedures for animal experiments are performed in accordance with the guide for the care and use of laboratory animals as defined by the European laws (Animal Facility Agreement C-67-482-10).
Megakaryocyte culture
The mouse BM progenitor cells were obtained after Lin− selection (Stem Cell Technologies) and prepared as described previously.19 In some experiments, brefeldin-A (BFA: 1 µg/mL) was added to the cultures on day 1, and the cells were fixed after 4 hours.
Confocal microscopy
The cells were fixed at the indicated times using 2% paraformaldehyde and prepared as described.19 The following antibodies were used at 10 µg/mL: Alexa488-conjugated anti-glycoprotein Ib β (GPIbβ), anti–β-tubulin, anti–protein regulator of cytokinesis 1 (PRC-1), anti–γ-tubulin, and anti-giantin. The appropriate secondary antibodies were conjugated with Alexa 546. 4,6 diamidino-2-phenylindole (DAPI) was applied for nucleus staining. Cells were examined under a confocal microscope (TCS SP5; Leica). Confocal analysis of BM tissue was performed according to the method described previously.20
Pulse-chase surface-labeling experiments
Lin− BM cell were first labeled with 10 µg/mL Alexa488-conjugated antibody against GPIbβ at 4°C for 15 minutes. In parallel, nonspecific binding of irrelevant IgG was determined. The unbound antibodies were removed by 3 washes of phophate-buffered saline, and the cells were then cultured in Dulbecco’s modified Eagle medium for various chase times (5 minutes and 1, 2, and 4 hours). In some experiments, cycloheximide (100 µg/mL) was added in the culture medium. The cells were fixed with 2% paraformaldehyde, washed, cytospun, counterstained with DAPI, and prepared for confocal microscopy.
EM
For transmission electron microscopy (TEM), BM were fixed with 2.5% glutaraldehyde and embedded in Epon as described previously.21 In some experiments, the fixed samples were incubated with 1% tannic acid for 1 hour. For immunoelectron microscopy, BM was fixed with 2.5% paraformaldehyde and 0.5% glutaraldehyde, infiltrated with 2.3 M sucrose, and frozen in liquid nitrogen as described.22
For EM tomography, samples were processed as for TEM. Dual-axis tilt series of selected MKs were recorded using a Tecnai 20 (FEI) and processed as previously described.22 The tilt series were aligned and three-dimensionally (3D) reconstructed using IMOD (Boulder Laboratory).23
For focused ion beam/scanning electron microscopy (FIB/SEM), samples were prepared as described previously.24 Briefly, glutaraldehyde-fixed BM cells were incubated with 1.5% potassium ferrocyanide and 1% osmium (contrast-enhancing step), processed as for TEM, and examined under a Helios NanoLab microscope (FEI). The 3D models were computed using Amira software.
For correlative light and electron microscopy (CLEM), cells were pulse-labeled for 1 hour using the anti-GPIb antibody, fixed, allowed to settle on prepatterned Aclar supports, and processed as previously reported.25
Results
Characterization of a DMS precursor: the pre-DMS
DMS formation starts at focal locations at the cell surface.
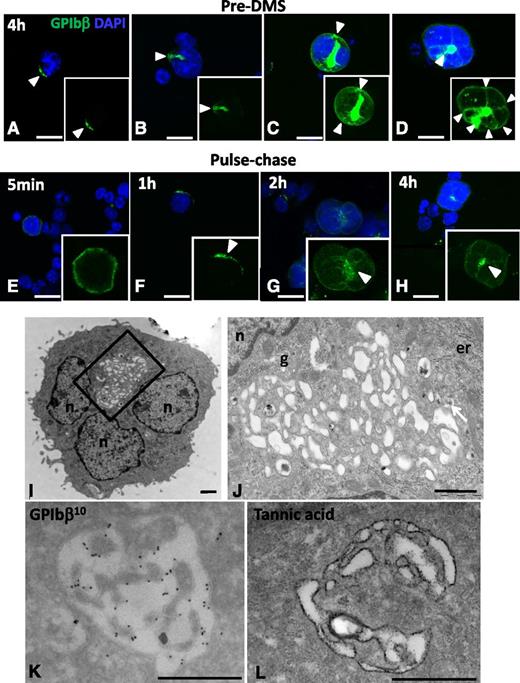
The major difficulty in performing a detailed study of the earliest developmental stages of the DMS is related to the low frequency of immature MKs in fresh BM samples (∼8% of the total MKs).21 Therefore, in our first experiments, we used an in vitro MK culture system that enables the enrichment of immature MKs. Lin− BM cells were fixed after short (4 hours) or long periods (1-4 days), stained for the platelet lineage-specific markers GPIbβ (Figure 1) or αIIbβ3 (supplemental Figure 1), and examined by confocal microscopy. Although the cells appeared heterogeneous in their developmental stage, we could define 4 characteristic stages on the basis of the GPIbβ staining pattern during the first 4 hours of culture (Figure 1A-D). Remarkably, GPIbβ appeared to be clustered on the cell surface (Figure 1A, arrowhead) or localized in a polarized fashion just beneath the cell surface (Figure 1B). More frequently, labeling was found both at the cell surface and in the center of the cell (Figure 1C). Complete Z-stack analysis of whole cells revealed that at these early stages the cell surface pool of GPIbβ is continuous with the centrally located GPIbβ-positive spot (supplemental Movies 1-4). The percentage of MKs showing these 4 characteristic stages was 16 ± 5, 12 ± 3, 13 ± 3, and 8 ± 1, respectively (average of 3 independent experiments; total GPIbβ-positive cells counted, 799). Importantly, no intracellular GPIbβ was detected in cells expressing the distinct cell surface clusters (Figure 1A; supplemental Movie 1). In MKs examined on days 1 and 2, the GPIbβ-positive area increased in size until it covered the entire cytoplasm by day 3 (supplemental Figure 1A). On day 4, fully mature MKs extending proplatelets with GPIbβ surface staining were observed (supplemental Figure 1A). We called these developmental stages, respectively, pre-DMS (Figure 1A-D), intermediate DMS, and late DMS (supplemental Figure 1A). Similar immunofluorescence (IF) images were obtained using an anti-αIIbβ3 antibody (supplemental Figure 1B). To better evaluate the kinetics of the membrane invagination, we performed pulse-chase experiments using Alexa488-conjugated GPIbβ antibodies (supplemental Figure 1B). Lin− BM cells were pulse-labeled for 15 minutes, washed, and fixed after 5 minutes and 1, 2, and 4 hours, respectively. IF in antibody-incubated living cells was lower than in permeabilized cells (Figure 1A-D). However the sequence of events was similar. MKs exhibited an even staining of the cell surface at 5-minute chase. After 1 hour, GPIb-positive focal points appeared at the cell surface. After 2 and 4 hours, staining was observed at the cell center with numerous connections with the surface (Figure 1E-H). These results suggest that plasma membrane invagination represents the first event leading to the formation of the DMS. To determine whether there might be a preexisting intracellular membrane pool formed in parallel to plasma membrane invagination, we performed pulse-chase experiments in presence of cycloheximide, an inhibitor of protein synthesis (supplemental Figure 2). The same sequence of events was observed under these conditions, albeit with lower fluorescence intensities, probably due to inhibition of protein synthesis. CLEM was used to determine whether there exists an intracellular membrane pool at a stage that shows the GPIb-positive cell surface patch (supplemental Figure 3), and we found no internal pre-DMS at the ultrastructural level.
DMS onset and expansion in cultured MKs. (A-D) Confocal images from MK cultures fixed after 4 hours. Cells were stained with anti-GPIbβ (green) and DAPI (blue); full Z-stacks of the representative DMS developmental stages are assembled as maximum intensity projections. (A-B) Distinct GPIbβ-positive territories at and just beneath the cell surface (arrowheads) were observed in MKs. Note the connection with the cell surface (supplemental Movies 1 and 2). (C-D) In larger MKs, GPIbβ-positive areas (pre-DMS) were centrally located between the nuclear lobes and displayed numerous tubular connections with the cell surface (arrowheads in D). Bars, 10 µm. (E-H) Pulse-chase kinetics of anti-GPIbβ in cultured MKs. Lin− BM cells were pulse-labeled for 15 minutes with Alexa488-conjugated anti-GPIb (green), washed, and subsequently fixed after 5 minutes and 1, 2, and 4 hours, respectively. Representative maximal projections of whole cells are shown. Bars, 10 µm (I-L) Ultrastructural characterization of the pre-DMS in cultured MKs. TEM images showing pre-DMS in MKs cultured for 4 hours. (I) At this stage, the pre-DMS is a well-defined circular membrane network located between the lobes of the nucleus. (J) Higher magnification of the network. (K) Immunogold labeling with anti-GPIbβ (10 nm protein A gold particles) showing that the pre-DMS contains GPIbβ. (L) Tannic acid staining reveals that the pre-DMS is connected o the cell surface. Bars, 1 µm. er, endoplasmic reticulum; g, golgi; n, nucleus.
DMS onset and expansion in cultured MKs. (A-D) Confocal images from MK cultures fixed after 4 hours. Cells were stained with anti-GPIbβ (green) and DAPI (blue); full Z-stacks of the representative DMS developmental stages are assembled as maximum intensity projections. (A-B) Distinct GPIbβ-positive territories at and just beneath the cell surface (arrowheads) were observed in MKs. Note the connection with the cell surface (supplemental Movies 1 and 2). (C-D) In larger MKs, GPIbβ-positive areas (pre-DMS) were centrally located between the nuclear lobes and displayed numerous tubular connections with the cell surface (arrowheads in D). Bars, 10 µm. (E-H) Pulse-chase kinetics of anti-GPIbβ in cultured MKs. Lin− BM cells were pulse-labeled for 15 minutes with Alexa488-conjugated anti-GPIb (green), washed, and subsequently fixed after 5 minutes and 1, 2, and 4 hours, respectively. Representative maximal projections of whole cells are shown. Bars, 10 µm (I-L) Ultrastructural characterization of the pre-DMS in cultured MKs. TEM images showing pre-DMS in MKs cultured for 4 hours. (I) At this stage, the pre-DMS is a well-defined circular membrane network located between the lobes of the nucleus. (J) Higher magnification of the network. (K) Immunogold labeling with anti-GPIbβ (10 nm protein A gold particles) showing that the pre-DMS contains GPIbβ. (L) Tannic acid staining reveals that the pre-DMS is connected o the cell surface. Bars, 1 µm. er, endoplasmic reticulum; g, golgi; n, nucleus.
Pre-DMS is continuous with the cell surface.
To better identify the GPIbβ-positive structures in the perinuclear region (Figure 1C), we performed a detailed EM analysis (Figure 1I-L). Similarly as in our IF experiments, well-defined pre-DMS membrane networks were detected between the nuclear lobes (Figure 1I) and were clearly distinguishable from the endoplasmic reticulum and the Golgi apparatus (“er” and “g” in Figure 1J). Immunoelectron microscopy confirmed that these membrane networks contained GPIbβ, which is compatible with the view that they represent the emerging DMS (Figure 1K). In agreement with this hypothesis, incubation of the cells with tannic acid, an electron-dense extracellular tracer, revealed that the centrally located membranes were already at this stage continuous with the cell surface (Figure 1L) and could thus be regarded as early stages of the DMS (pre-DMS as defined above).
Formation of the pre-DMS in situ in BM MKs.
To evaluate the biological relevance of these observations, we also studied the development of the DMS in its natural bone marrow environment (Figure 2A). Confocal microscopic analysis of BM sections revealed the same developmental stages as observed in cultured MKs. An overview of mouse BM showing representative maturation stages of the DMS is presented in Figure 2A and a high magnification of a typical pre-DMS area in situ in Figure 2B. Similarly, as in cultured MKs, the pre-DMS appeared to be connected to the surface, and this was confirmed by Z-stack analysis (supplemental Movie 5). Quantification of the number of cells exhibiting pre-DMS showed that they represented 5 ± 2.7% of the total MKs (n = 103 cells analyzed in 3 independent experiments). We used the same criteria as before to define the different stages of development of the DMS at the EM level. As in cultured MKs, pre-DMS was predominantly found between the nuclear lobes (Figure 2C, arrow) and seemed to be continuous with the cell surface (Figure 2C, arrowhead; supplemental Figure 4). We then adopted a 3D approach using FIB/SEM technology. This method allows a detailed visualization of large cellular volumes, thereby providing a complete 3D representation of the pre-DMS with respect to its intracellular position and connections to the cell surface. A 3D reconstruction of a pre-DMS analyzed by FIB/SEM is shown in Figure 2D. Two cell surface connections are connected to a centrally located pre-DMS structure (Figure 2D, arrowhead; supplemental Movie 6).
Detection of pre-DMS in bone marrow MKs. (A) View of a 100-µm-thick BM section with immunofluorescence staining as indicated on the figure. The different stages of development of the DMS (pre-, intermediate, and late) are identified by the anti-GPIbβ antibody. Bar, 20 µm. (B) Higher-magnification confocal image of a typical stage I MK showing GPIbβ at the cell periphery and the pre-DMS (arrow) connected to and located just beneath the cell surface, between the nuclear lobes (see supplemental Movie 3 for the complete Z-stack analysis). Bar, 10 µm. (C) Representative TEM image of the pre-DMS. Note the well-defined boundary, the precise localization in the perinuclear region (arrow) between the nuclear lobes, and the connection with the cell surface (arrowhead). Bar, 2 µm (D) 3D reconstruction from an FIB/SEM analysis of the pre-DMS (orange) in a bilobulated immature MK. Note that the pre-DMS is precisely positioned between the 2 lobes of the nucleus and is already branched at this early stage of development. The arrowhead indicates a connection with the cell surface.
Detection of pre-DMS in bone marrow MKs. (A) View of a 100-µm-thick BM section with immunofluorescence staining as indicated on the figure. The different stages of development of the DMS (pre-, intermediate, and late) are identified by the anti-GPIbβ antibody. Bar, 20 µm. (B) Higher-magnification confocal image of a typical stage I MK showing GPIbβ at the cell periphery and the pre-DMS (arrow) connected to and located just beneath the cell surface, between the nuclear lobes (see supplemental Movie 3 for the complete Z-stack analysis). Bar, 10 µm. (C) Representative TEM image of the pre-DMS. Note the well-defined boundary, the precise localization in the perinuclear region (arrow) between the nuclear lobes, and the connection with the cell surface (arrowhead). Bar, 2 µm (D) 3D reconstruction from an FIB/SEM analysis of the pre-DMS (orange) in a bilobulated immature MK. Note that the pre-DMS is precisely positioned between the 2 lobes of the nucleus and is already branched at this early stage of development. The arrowhead indicates a connection with the cell surface.
Relationship with the endomitotic process.
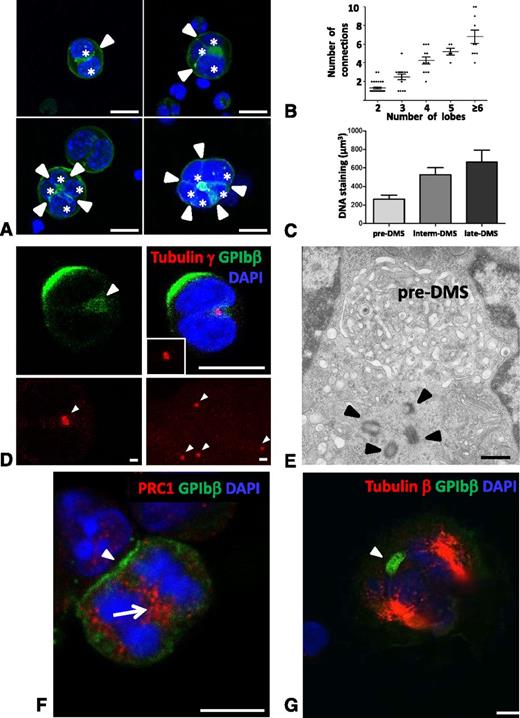
As these structures were systematically found between the lobes of the MK nucleus, we wanted to investigate the possible relationship of these pre-DMS invaginations with the MK endomitotic cell cycle. From our IF observations using DAPI and GPIbβ, it became apparent that the inward growth of the PM was not random but rather seemed to coincide with the nuclear lobulation (Figure 3A). To further characterize this, we performed a detailed Z-stacks analysis in cultured MKs and quantified the number of cell surface connections and the total number of nuclear lobes. Interestingly, we found that the number of connections exactly correlated (r = 0.994) with the number of nuclear lobes (Figure 3B). We also observed a proportional increase in DNA content when comparing pre-, intermediate, and late DMS-containing MKs (Figure 3C). We next used an anti-γ-tubulin antibody to further correlate the number of pre-DMS connections with γ-tubulin fluorescent spots, representing the centrosomes. At the early stages, it appeared difficult to quantify the exact ploidy level of the cell based on γ-tubulin staining, because the centrosomes remain detected as a single fluorescent spot (Figure 3D, upper panel). However, the size of γ-tubulin–positive spots were larger in the early stages compared with the late stage MKs (Figure 3D, lower panel), suggesting that multiple centrosomes assemble at these stages of pre-DMS formation. Indeed using TEM analysis, we regularly found centrosome doublets closely positioned at pre-DMS sites (compare Figure 3E with upper panel of Figure 3D). Of note, the position of the centrosomes near the nucleus indicates that the MKs are in a nonmitotic stage. To further assess the correlation between the pre-DMS and the endomitotic process, we analyzed the pre-DMS in anaphase cells transitioning from 2N to 4N (Figure 3F-G). MKs in this anaphase were identified using the PRC-1 staining, a microtubule-binding protein that controls the midzone organization, and the β1-tubulin staining of the mitotic spindle. Cells in anaphase are rare, but on occasion we could observe MKs where PRC-1 labeling was centrally localized between the nuclei, and clustered GPIb was at a focal area on the cell surface (Figure 3F). On another image (Figure 3G), β-tubulin staining identified 2 opposite mitotic spindles, and the GPIb-positive pre-DMS was located at 1 extremity of the midzone. Altogether, these observations support the concept that pre-DMS formation is somehow linked to the endomitotic process.
Relationship with the endomitotic process. (A-C) The number of invaginating pre-DMS correlates with the number of nuclear lobes. (A) Confocal images show that the tubular membrane connections (green) are situated between the nuclear lobes (blue). Note that the number of cell surface connections (arrowheads) increased with the number of nuclear lobes (stars). Bars, 10 µm. (B) Quantification of the number of invaginating pre-DMS and nuclear lobes using Z-stack analysis on whole cells. Note that the number of nuclear lobes are not equal to the number of chromosome copies. The Z-step size was 0.25 µm. Overall, a total of 80 MKs (39 × 2 lobes; 14 × 3 lobes; 12 × 4 lobes, and 10 × >6 lobes) were included in our analysis, and the means ± standard error of the mean are presented. (C) Quantification of the DNA staining as a function of the different developmental stages of the DMS (pre-, intermediate, and late DMS). The Z-step size was 0.25 µm, and the Amira Software was used to calculate the DNA content in whole cells. (D) Immunostaining for the centrosome marker γ tubulin reveals the presence of a single fluorescent spot with an increased fluorescent size (arrowhead in upper left panel) compared with γ tubulin staining in late stage MK cells (arrowheads in lower right panel), suggesting multiple centrosomes assembly. The images are maximum projections of whole cells. Bars: upper panels, 10 µm; lower panels, 1 µm. (E) TEM image showing the presence of four centrioles (arrowheads) in proximity to the pre-DMS. Bar, 500 nm. (F) Image of an MK undergoing its first anaphase. Immunostaining for PRC-1, a marker of the midzone. PRC-1 is localized between the condensed chromosomes (arrow). Note the distinct GPIbβ-positive cell surface pool (arrowhead). Bar, 10 µm. (G) Identification of a pre-DMS (arrowhead) in MKs undergoing the first anaphase. Cells were permeabilized with 0.5% Triton X-100, stained with β-tubulin and DAPI to characterize the mitotic spindle and the condensed chromosomes, respectively. Bar, 10 µm.
Relationship with the endomitotic process. (A-C) The number of invaginating pre-DMS correlates with the number of nuclear lobes. (A) Confocal images show that the tubular membrane connections (green) are situated between the nuclear lobes (blue). Note that the number of cell surface connections (arrowheads) increased with the number of nuclear lobes (stars). Bars, 10 µm. (B) Quantification of the number of invaginating pre-DMS and nuclear lobes using Z-stack analysis on whole cells. Note that the number of nuclear lobes are not equal to the number of chromosome copies. The Z-step size was 0.25 µm. Overall, a total of 80 MKs (39 × 2 lobes; 14 × 3 lobes; 12 × 4 lobes, and 10 × >6 lobes) were included in our analysis, and the means ± standard error of the mean are presented. (C) Quantification of the DNA staining as a function of the different developmental stages of the DMS (pre-, intermediate, and late DMS). The Z-step size was 0.25 µm, and the Amira Software was used to calculate the DNA content in whole cells. (D) Immunostaining for the centrosome marker γ tubulin reveals the presence of a single fluorescent spot with an increased fluorescent size (arrowhead in upper left panel) compared with γ tubulin staining in late stage MK cells (arrowheads in lower right panel), suggesting multiple centrosomes assembly. The images are maximum projections of whole cells. Bars: upper panels, 10 µm; lower panels, 1 µm. (E) TEM image showing the presence of four centrioles (arrowheads) in proximity to the pre-DMS. Bar, 500 nm. (F) Image of an MK undergoing its first anaphase. Immunostaining for PRC-1, a marker of the midzone. PRC-1 is localized between the condensed chromosomes (arrow). Note the distinct GPIbβ-positive cell surface pool (arrowhead). Bar, 10 µm. (G) Identification of a pre-DMS (arrowhead) in MKs undergoing the first anaphase. Cells were permeabilized with 0.5% Triton X-100, stained with β-tubulin and DAPI to characterize the mitotic spindle and the condensed chromosomes, respectively. Bar, 10 µm.
Golgi apparatus contributes to DMS biogenesis and its expansion
Multiple Golgi complexes assemble at pre- and intermediate DMS.
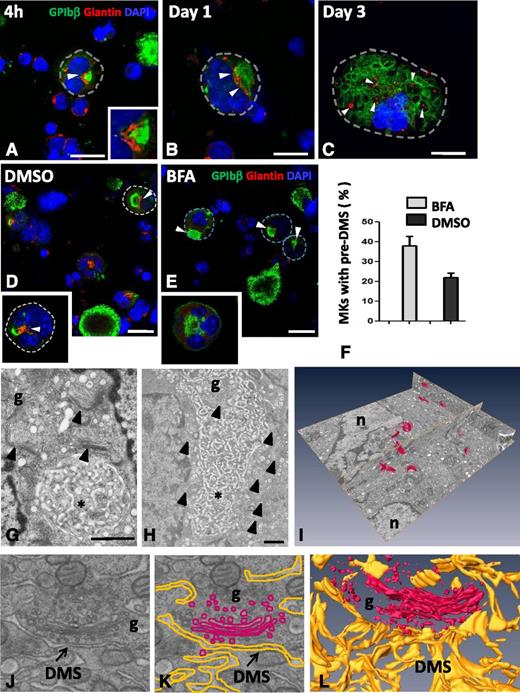
As the pre-DMS was located in the perinuclear region, which is the exit site of the trans-Golgi network (TGN) in many mammalian cells, we next investigated the possible contribution of this secretory pathway. Immunofluorescence was used to correlate the distribution pattern of known Golgi markers (giantin and GM130) with GPIbβ at the different stages of MK development. Although pre-DMS (green) was often found in close proximity to giantin-positive structures (red), it never colocalized with them (arrowheads and inset in Figure 4A). At stages where intermediate DMS were formed (1-2 days of culture), the giantin-positive areas became larger, suggesting the assembly of multiple Golgi complexes. Golgi complexes remained in close proximity, but distinct from the intermediate DMS (arrowhead in Figure 4B). At stages when late DMS became apparent, numerous distinct but smaller giantin-positive spots appeared dispersed throughout the cytoplasm (Figure 4C). The same results were obtained using the cis-Golgi marker GM130. To further evaluate the contribution of Golgi-derived vesicular transport, cells were exposed to the fungal toxin BFA, a potent inhibitor of anterograde Golgi trafficking (Figure 4E). BFA treatment did not modify the MK proliferation and the ploidy levels as measured by cell counting and FACS, respectively (data not shown). The Golgi staining was completely lost (Figure 4E, inset), and the number of MKs displaying pre-DMS was significantly increased compared with the control (arrows in Figure 4D-E and F) (38 ± 4.6% vs 22 ± 2.2%, respectively). These findings suggest that inhibition of anterograde Golgi transport slows down the progression of DMS formation.
Golgi complexes are in close position to the pre-DMS and contribute to DMS growth. MKs were cultured for 4 hours or for 1 or 3 days. The cells were permeabilized and immunostained for the Golgi marker giantin (red) and GPIbβ (green). (A-C) Representative confocal images are shown at different developmental stages of the DMS. (A) Giantin staining is confined to the perinuclear region and lies close to the pre-DMS (inset of boxed area). (B) Close association but no colocalization of giantin with the intermediate DMS. Note that the size of the giantin spots increases at these later stages (white arrowheads). (C) Giantin spots become randomly distributed in MKs at the latest stages of maturation (white arrowheads). Bars, 10 µm. (D-E) Confocal images of MKs cultured for 20 hours and treated as indicated. Note the absence of Golgi staining after BFA treatment. Dotted lines indicate cell perimeters. Bars, 10 µm. (F) Quantification of the number of MKs displaying typical pre-DMS areas. (G-H) Electron micrographs showing the presence of Golgi complexes (arrowheads) in proximity to the pre-DMS between (G) 2 nuclear lobes or (H) the intermediate DMS. Bars, 500 nm. (I) A typical FIB/SEM analysis illustrating the 3D visualization of all the Golgi complexes (red) detected in a large volume (∼34 µm3) of a stage II MK. (J) Higher magnifications of the FIB/SEM analysis showing close apposition of Golgi-derived vesicles at the boundary of the DMS. (K) Segmentation of the DMS (yellow) and the Golgi (red). (L) 3D reconstruction. *DMS. Bars, 1 µm. g, Golgi stacks; n, nucleus.
Golgi complexes are in close position to the pre-DMS and contribute to DMS growth. MKs were cultured for 4 hours or for 1 or 3 days. The cells were permeabilized and immunostained for the Golgi marker giantin (red) and GPIbβ (green). (A-C) Representative confocal images are shown at different developmental stages of the DMS. (A) Giantin staining is confined to the perinuclear region and lies close to the pre-DMS (inset of boxed area). (B) Close association but no colocalization of giantin with the intermediate DMS. Note that the size of the giantin spots increases at these later stages (white arrowheads). (C) Giantin spots become randomly distributed in MKs at the latest stages of maturation (white arrowheads). Bars, 10 µm. (D-E) Confocal images of MKs cultured for 20 hours and treated as indicated. Note the absence of Golgi staining after BFA treatment. Dotted lines indicate cell perimeters. Bars, 10 µm. (F) Quantification of the number of MKs displaying typical pre-DMS areas. (G-H) Electron micrographs showing the presence of Golgi complexes (arrowheads) in proximity to the pre-DMS between (G) 2 nuclear lobes or (H) the intermediate DMS. Bars, 500 nm. (I) A typical FIB/SEM analysis illustrating the 3D visualization of all the Golgi complexes (red) detected in a large volume (∼34 µm3) of a stage II MK. (J) Higher magnifications of the FIB/SEM analysis showing close apposition of Golgi-derived vesicles at the boundary of the DMS. (K) Segmentation of the DMS (yellow) and the Golgi (red). (L) 3D reconstruction. *DMS. Bars, 1 µm. g, Golgi stacks; n, nucleus.
To better examine the Golgi contribution, we performed a detailed 3D analysis. As expected from the IF results, Golgi complexes were frequently located in close position to the pre-DMS (on average, 2-4) (Figure 4G and compare with Figure 4A). With respect to the intermediate DMS, the number of stacks increased up to as many as 10 (Figure 4H and compare with Figure 4B). Using FIB/SEM large volume analysis (ie, 34 µm3 corresponding to ∼2% of a whole MK), we were able to detect 12 individual Golgi stacks surrounded by numerous vesicles in close proximity to the DMS (Figure 4I-L; supplemental Movie 7). Thus, the FIB/SEM data confirmed our IF results showing that Golgi complexes were randomly distributed through the cytoplasm but remained in close proximity to the DMS.
Membrane delivery from TGN to the pre-DMS.
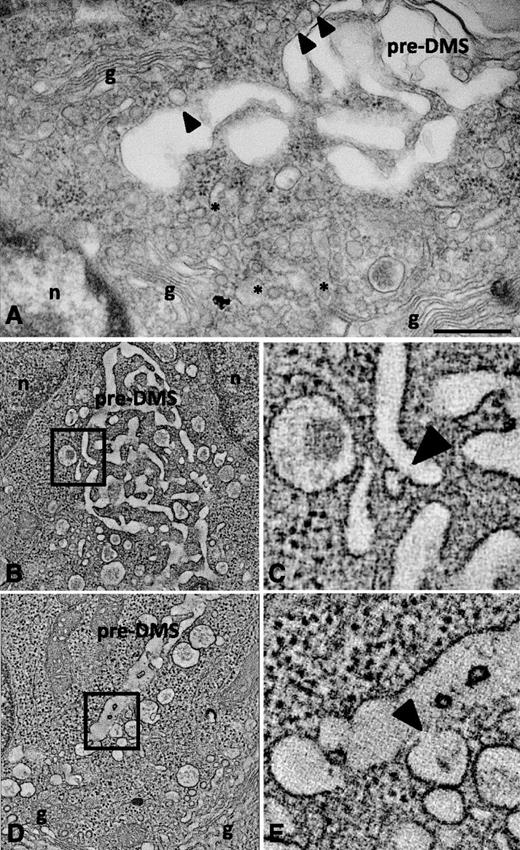
Golgi complexes always appeared to be oriented with the trans-sites (TGN) facing the pre- and intermediate DMS. Numerous clathrin-coated and uncoated vesicles were located between the Golgi stacks and the DMS, some of them in close apposition (arrowhead in Figure 5A), which suggests active trafficking. Immunoelectron microscopy analysis revealed that these vesicles were positive for the TGN marker TGN 38 (supplemental Figure 5). In search of fusion profiles of TGN-derived vesicles, we analyzed the cells by dual axis electron tomography, focusing on TGN exit sites and pre-DMS. Besides small vesicles fusing with the DMS (arrowheads in Figure 5B-C; supplemental Movie 8), we found numerous large electron-lucent vesicles aligned along the pre-DMS. These vesicles have the characteristics of immature secretory granules. Some of them were captured in our tomograms in clear continuity with the pre-DMS (Figure 5D-E; supplemental Movie 9). Overall, these observations suggest that membrane delivery from the TGN exit site occurs at all developmental stages of DMS formation and involves multiple vesicle populations.
Dual axis electron tomography of the TGN and DMS reveals fusion of vesicles with the pre-DMS. (A) TEM image of the Golgi complexes oriented so that the trans-sites (TGN) lie facing the pre-DMS. Some vesicles are in close apposition to the pre-DMS (arrowheads). Bar, 500 nm. (B-C) Selected tomographic sections showing fusion of TGN-derived vesicles with the pre-DMS. A higher magnification of the boxed area in B is shown in C, and the arrowhead points to the zone of continuity between the membranes of the vesicle and the pre-DMS. (D-E) Alignment of larger vesicles along the pre-DMS. Fusion of a vesicle with the pre-DMS (arrowhead in D). g, Golgi stacks; n, nucleus.
Dual axis electron tomography of the TGN and DMS reveals fusion of vesicles with the pre-DMS. (A) TEM image of the Golgi complexes oriented so that the trans-sites (TGN) lie facing the pre-DMS. Some vesicles are in close apposition to the pre-DMS (arrowheads). Bar, 500 nm. (B-C) Selected tomographic sections showing fusion of TGN-derived vesicles with the pre-DMS. A higher magnification of the boxed area in B is shown in C, and the arrowhead points to the zone of continuity between the membranes of the vesicle and the pre-DMS. (D-E) Alignment of larger vesicles along the pre-DMS. Fusion of a vesicle with the pre-DMS (arrowhead in D). g, Golgi stacks; n, nucleus.
DMS forms close contacts with specific domains of the ER
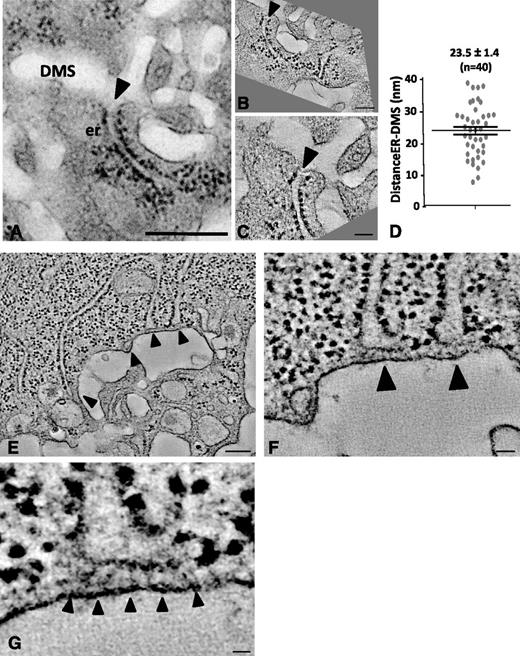
It is well established that the ER contributes to the delivery of lipids for membrane building.26 In the later stages of DMS development, we frequently found the ER and DMS to be in close proximity, as illustrated in the TEM image of Figure 6A (arrowhead). High-resolution dual axis electron tomography enabled us to define the nature of these contacts. The 2 tomographic sections in Figure 6B-C, which represent the boxed area of Figure 6A in different orientations, revealed tight contact but no apparent membrane continuity. In 6 separate tomograms, we measured an average distance of 23.5 ± 1.4 nm (n = 40 contact sites; Figure 6D). This is further illustrated in Figure 6E-H, where several ER cisternae are seen in close proximity (arrowheads) to the DMS. Importantly, these ER-DMS contact sites were free of ribosomes and contained filamentous electron-dense material possibly reflecting cross-linking/anchoring molecules (supplemental Movie 10).
Dual axis electron tomography of ER-DMS contacts. (A) TEM image of a stage II MK showing a tubular ER cisterna extending toward the DMS. Bar, 500 nm. (B-C) Tomographic sections from a dual axis tomogram of the area in A. Note that the membranes of the DMS and ER are closely apposed but not in continuity (arrowheads). Bars, 100 nm. (D) Minimal distance in nm between the ER and DMS, as quantified from 40 contact sites in 6 tomograms. (E-G) Selected tomographic sections (∼5 nm thick) of tubular ER cisternae (arrowheads) extending toward the DMS. Bar, 100 nm. (F-G) Selected Z-orientations from the tomogram revealing filamentous structures connecting ribosome-free ER domains with the DMS. Bars, 10 nm. er, endoplasmic reticulum.
Dual axis electron tomography of ER-DMS contacts. (A) TEM image of a stage II MK showing a tubular ER cisterna extending toward the DMS. Bar, 500 nm. (B-C) Tomographic sections from a dual axis tomogram of the area in A. Note that the membranes of the DMS and ER are closely apposed but not in continuity (arrowheads). Bars, 100 nm. (D) Minimal distance in nm between the ER and DMS, as quantified from 40 contact sites in 6 tomograms. (E-G) Selected tomographic sections (∼5 nm thick) of tubular ER cisternae (arrowheads) extending toward the DMS. Bar, 100 nm. (F-G) Selected Z-orientations from the tomogram revealing filamentous structures connecting ribosome-free ER domains with the DMS. Bars, 10 nm. er, endoplasmic reticulum.
Discussion
A combination of high-resolution microscopy approaches (IF, immunogold electron microscopy [IEM], EM tomography, and FIB/SEM) was used to perform a detailed characterization of the DMS at various stages of MK development, both in cultured cells and in in situ BM MKs. Use of GPIb as a platelet lineage-specific plasma membrane marker revealed distinct peripheral GPIb-positive loci at the cell surface, which became centrally located in the perinuclear region at later stages, thereby maintaining their plasma membrane connections. We called this earliest recognizable membrane network the pre-DMS and established that it originated at the plasma membrane. The pre-DMS displayed several characteristic features: (1) its territory was well defined with clear boundaries, (2) it was located between the lobes of the nucleus in the central region of the cell, (3) it was linked to the cell surface through tubular membrane connections, and (4) Golgi complexes were selectively positioned at the boundary of the pre-DMS.
With respect to the origin of the DMS, we here favor the option that DMS biogenesis starts at the cell surface as a GPIb-positive cluster. Our full confocal Z-stack analysis revealed that the intracellular GPIbβ-positive membrane pools are all in connection with the cell surface and never present as isolated structures within the cytoplasm. Pulse-chase experiments using cell surface labeling also demonstrated the presence of an intracellular pre-DMS following a 2-hour chase. These experiments suggest that this intracellular pool derives from the cell surface. However, we cannot exclude the possibility of an independent preexisting intracellular pool. When we performed IEM analysis using an anti-GPIb antibody, we consistently found a small membrane pool but did not present the features of the pre-DMS and corresponded to Golgi stacks and TGN-related vesicles (supplemental Figure 6). These vesicles likely represent the anterograde transport pathway and as such also contribute to membrane delivery. Using 4 different approaches (confocal Z-stack analysis, electron tomography, CLEM, and tannic acid tracer), we were unable to identify an isolated intracellular pre-DMS–like structure. However, it is still possible that other intracellular membrane populations (ie, GPIb negative) contribute to DMS biogenesis. We found that numerous Golgi complexes were clustered around the pre-DMS. The Golgi complexes were positioned in a fashion whereby the trans-Golgi sites (TGN) were facing the DMS. Tomography analysis revealed numerous TGN-derived membrane carriers of different sizes in close proximity to the DMS. Some vesicles were found to be continuous with the centrally located DMS structures, suggesting direct vesicular membrane insertion. The fact that BFA impaired DMS expansion suggests an important role of TGN-mediated delivery in DMS biogenesis. Because BFA may also have impact on endosomal trafficking, there may still be a role of the endocytic pathway in DMS biogenesis.This will require further investigation. Our study thus provides evidence that the growing DMS requires, besides invagination of the plasma membrane, the direct insertion of vesicular membrane from intracellular stores.
An interesting aspect of our study was that in early developmental stages, the tubular membrane connections were always positioned between the nuclear lobes, a characteristic that resembles cytokinesis. Using time-lapse confocal microscopy, it was previously shown that a cleavage furrow is induced normally from the 2N to 4N transition, which fails to close completely, resulting in cells with bilobulated nucleus.27-30 This is essentially what we observed when the pre-DMS was formed. TEM analysis and IF staining of γ-tubulin at the stages where the pre-DMS is apparent revealed that multiple centrosomes have assembled, suggesting that the MK at this stage have undergone at least one endomitotic cycle. Our Z-stack analysis also revealed that the number of tubular connections was proportional to the number of nuclear lobes. Together, these results recall several elements characteristic of the membrane growth observed during cellularization in Drosophila melanogaster.31 During this special kind of cytokinesis, the plasma membrane invaginates and separates each nuclear lobe, thereby forming a pseudo-cleavage furrow. Similarly as in our study, numerous Golgi stacks are targeted to the sites of furrow formation, thereby contributing to active membrane secretion.32 Little information is available regarding the fate of the Golgi during subsequent endomitotic cycles. During normal mitosis, the Golgi complexes disassemble and subsequently reform during telophase.33,34 Whether such a mechanism is also operational during endomitosis is not known. Occasionally we found MKs in anaphase cycle with typically polarized centrosomes35 and mitotic spindles. During this typical event, the pre-DMS was located where the cleavage furrow will form and where proteins essential for division should be located. Indeed PRC1 was located essentially at this site. On the basis of the developmental stages of the DMS presented here, we hypothesize that formation of the pre-DMS is coupled to the process of abortive cytokinesis whereby the tubular invaginations represent an MK specific pseudo-cleavage furrow.36,37
At later stages of MK development, ribosome-free ER domains were frequently observed in close contact with the DMS. Although we were unable to detect DMS-ER fusions, the average distance between the 2 membrane leaflets (30 nm) lay within the range recognized as a true association and would point to a functional role of ER-DMS communication.38 Our tomograms indeed frequently revealed protein linkages between the 2 types of membrane, suggesting selective ER-DMS membrane tethering, possibly for lipid transfer. Nonvesicular lipid transport between the ER and other membranes has been suggested previously. Indeed, several studies have shown that physical membrane contacts exist between the ER and the PM (the so-called “plasma membrane-associated membranes” or “PAM fraction”)38 and the mitochondria (“MAM” fraction),39 and this has been reported to be a prerequisite for lipid translocation between the 2 compartments. Vance et al40 demonstrated that in mammalian cells a specific subfraction of the ER, highly enriched in enzymes responsible for phospholipid biosynthesis, is associated with the PM. Close ER contacts were also described recently in association with de novo membrane formation in the biogenesis and expansion of the autophagosomal membrane.41 Nevertheless, at this stage it must remain speculative as to what extent the observed tethering of the ER to the DMS may be required for lipid transfer and DMS expansion. In addition, the presence of ER subdomains in close proximity to the DMS could also support local calcium sequestration, which is required for membrane fusion events and directioning during expansion of the DMS. Such a mechanism has been proposed to occur during formation of the T-tubules in skeletal muscle.42
In previous studies of guinea pig MKs,43 it was suggested that membrane sheets were formed from tubular DMS structures through multiple fusion events. In our 3D analysis, we found tubules rather than sheets. It is well established that the DMS functions as a membrane reservoir for future platelets.8 On the basis of previous studies of DMS architecture, it has been proposed that the DMS has a “ball of yarn” configuration and that additional membrane for de novo platelets is obtained by sequestering membrane from the edge and continuously replenishing it from the interior. This is not consistent with large volume 3D analysis of the mature MKs (supplemental Figure 7), showing that the DMS is not a single membrane invagination but a highly intertwined membrane network with numerous side branches and multiple connections with the cell surface, which is similar to descriptions of the open canalicular system in platelets.22 This might suggest that the open canalicular system derives from the DMS.
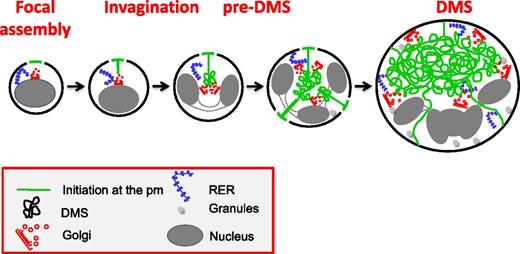
In conclusion, we propose the following model for the biogenesis of the DMS (Figure 7): (1) DMS formation starts at focal points on the cell surface and (2) is followed by an invagination process resembling cleavage furrow formation; and (3) formation of the pre-DMS and its further expansion require vesicular membrane delivery from numerous Golgi complexes targeted to the DMS and possibly direct lipid transfer through ER-DMS tethering. Finally, abnormal development of the DMS has been implicated in several hereditary diseases with thrombocytopenia including Bernard-Soulier syndrome and Myosin Heavy chain 9-related disorders.44,45 Thus, an understanding of the molecular mechanisms of DMS biogenesis and development is not only fundamentally important, but also highly relevant to understand how structural defects in the DMS are related to known and unknown diseases such as (macro/micro) thrombocytopenia.
Proposed model of DMS formation and expansion. DMS formation initiates at focal areas on the cell surface. During subsequent MK maturation, a distinct pre-DMS structure is formed in the perinuclear region, through a PM invagination process that resembles cleavage furrow formation. Further expansion requires vesicular membrane delivery from multiple Golgi complexes that are targeted to the developing DMS and possibly via direct lipid transfer through ER-DMS tethering.
Proposed model of DMS formation and expansion. DMS formation initiates at focal areas on the cell surface. During subsequent MK maturation, a distinct pre-DMS structure is formed in the perinuclear region, through a PM invagination process that resembles cleavage furrow formation. Further expansion requires vesicular membrane delivery from multiple Golgi complexes that are targeted to the developing DMS and possibly via direct lipid transfer through ER-DMS tethering.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank S. Moog for expert technical assistance.
This work was supported by Association de Recherche et Développement en Médecine et Santé Publique and the European Union through the European Regional Development Fund.
Authorship
Contribution: A.E. and H.H. conceived and designed the research, acquired the data, analyzed and interpreted the data, and wrote the manuscript; F. Pertuy and C.L. acquired the confocal images on bone marrow sections; J.-Y.R. and F. Proamer acquired the TEM, IEM, and FIB/SEM data; W.G. participated in the acquisition of the dual axis tomography data; and F.L. and C.G. conceived and designed the research, wrote the manuscript, and handled funding and supervision.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: C. Gachet, UMR S949, INSERM-Université de Strasbourg, Etablissement Français du Sang-Alsace (EFS-Alsace), 10 rue Spielmann, B.P. N°36, 67065 Strasbourg Cedex, France; e-mail: christian.gachet@efs-alsace.fr.