Key Points
Latent membrane protein 2A augments MYC oncogene in driving the cell cycle by increasing protein instability of a tumor suppressor p27kip1.
Latent membrane protein 2A potentiates MYC expression to overcome a cell cycle checkpoint without disrupting p53 tumor suppressor pathway.
Abstract
Elevated expression of MYC is a shared property of many human cancers. Epstein-Barr virus (EBV) has been associated with lymphoid malignancies, yet collaborative roles between MYC and EBV in lymphomagenesis are unclear. EBV latent membrane protein 2A (LMP2A) functions as a B-cell receptor (BCR) mimic known to provide survival signals to infected B cells. Co-expression of human MYC and LMP2A in mice (LMP2A/λ-MYC) accelerates B lymphoma onset compared with mice expressing human MYC alone (λ-MYC mice). Here we show a novel role of LMP2A in potentiating MYC to promote G1-S transition and hyperproliferation by downregulating cyclin-dependent kinase inhibitor p27kip1 in a proteasome-dependent manner. Expressing a gain-of-function S10A mutant of p27kip1 has minor effect on tumor latency. However, pretumor B cells from λ-MYC mice expressing homozygous S10A mutant show a significant decrease in the percentage of S-phase cells. Interestingly, LMP2A is able to counteract the antiproliferative effect of the S10A mutant to promote S-phase entry. Finally, we show that LMP2A expression correlates with higher levels of MYC expression and suppression of p27kip1 before lymphoma onset. Our study demonstrates a novel function of EBV LMP2A in maximizing MYC expression, resulting in hyperproliferation and cellular transformation into cancer cells in vivo.
Introduction
The cell cycle is a tightly regulated process governed by cyclins, cyclin-dependent kinases (CDKs), and CDK inhibitors. Cell cycle checkpoints are important cellular mechanisms that prevent uncontrolled proliferation caused by oncogenic stimuli. Retinoblastoma (Rb) and p53 pathways are critical pathways preventing premature cell cycle progression by inducing cell cycle arrest or apoptosis. Although pathways and mutations leading to malignancies are different depending on cancer types, many routes converge to increase MYC expression through chromosomal translocation, amplification of c-MYC transcription, and protein stabilization. MYC is a basic helix-loop-helix (bHLH) transcription factor regulating many target genes in most, if not all, cell types. MYC heterodimerizes with a binding partner, Max, and binds to CACGTG-containing DNA sequences.1,2 Recent studies suggest that the main function of MYC is to upregulate transcription of its target genes which in turn indirectly lead to repression of certain genes,3,4 including those encoding CDK inhibitors. MYC is important for B-lymphocyte activation and proliferation.5 Constitutive expression of MYC in murine B cells leads to increases in the percentage of cells in S and G2/M phases of the cell cycle.6,7 MYC activation increases activities of cyclin-CDK complexes, resulting in the hyperphosphorylation of Rb8 and release of E2F transcription factors to upregulate S-phase genes. MYC promotes the expression of D-type cyclins9,10 and E2F,11,12 and it contributes to the transcription repression of genes encoding CDK inhibitors p27Kip1, p21Cip1, and p15Ink4b, leading to the progression into S-phase.13 MYC also induces apoptosis via the upregulation of p19ARF and induction of the p53 pathway.14 Mice lacking p53, Cdkn2a, and Cdkn1b show rapid onset of MYC-induced lymphomagenesis,15,16 suggesting importance of cell cycle regulators in preventing excessive proliferation caused by MYC.
Latent infection of Epstein-Barr virus (EBV), a member of gammaherpesviruses, is associated with cellular hyperproliferation observed in posttransplant lymphoproliferative disorders (PTLD), as well as malignancies including Hodgkin disease, and non-Hodgkin lymphoma.17 EBV is linked to a role in suppressing apoptosis induced by MYC in infected B cells and thus aids MYC in cell growth and proliferation.18 Latent membrane protein 2A (LMP2A) is encoded by EBV and found in most latency programs. LMP2A contains an ITAM (immunoreceptor tyrosine activation motif) similar to that of the host BCR19 and provides BCR-like survival signals to B cells.20-22 Furthermore, a recent study suggests that LMP2A can contribute to proliferation of B cells during an early phase of EBV-induced B-cell proliferation,23 suggesting a pro-proliferative role of LMP2A in the cellular transformation process.
In a mouse model of EBV latent infection, mice expressing LMP2A and human MYC transgene (LMP2A/λ-MYC) demonstrate accelerated lymphomagenesis compared with mice expressing human MYC alone (λ-MYC mice),24,25 which is similar to another study showing that constitutive activation of the BCR leads to a rapid onset of MYC-induced lymphoma.26 The p19ARF-p53 pathway is an important mechanism controlling aberrant proliferation induced by pathologic expression of MYC. Therefore, mutations and inactivation of the p53 pathway are frequently found in MYC-induced tumorigenesis.27 However, p53 pathway inactivation is absent in LMP2A/λ-MYC tumors, suggesting that LMP2A uses a different mechanism to promote MYC-driven lymphomagenesis.
In this study, we investigate a role of LMP2A in altering the cell cycle regulation independent of the p53 pathway. We demonstrate that LMP2A cooperates with MYC in disrupting the G1 checkpoint through the downregulation of a CDK inhibitor p27kip1, leading to premature S-phase entry. We also show that LMP2A expression leads to high MYC levels, indicating a novel relationship between MYC and LMP2A. Our study sheds new light onto an intricate collaboration of EBV and host oncoproteins in malignant transformation.
Materials and methods
Mice
The Tg6 lines of EµLMP2A transgenic mice20,21 and λ-MYC mice28 have been described and are in the C57BL/6 background. Mice expressing an S10A mutant of p27kip1 (Cdkn1btm2Jro mice) were constructed previously29 and obtained from The Jackson Laboratory and are in the 129 background. Tumor mice were sacrificed when lymph node tumors could be observed externally or mice were moribund. Animals were maintained at Northwestern University’s Center for Comparative Medicine, in accordance with the university’s animal welfare guidelines.
Tumor and spleen cell isolation
Pretumor spleens were isolated from 21- to 25-day-old mice. Magnetic activated cell sorting using CD19 positive selection (Miltenyi Biotec) were used to purify splenic B cells. Tumor-bearing lymph nodes were dissociated and treated with 155 mM ammonium chloride to lyse red blood cells. Approximately 90% of tumor cells in tumor-bearing lymph nodes were of B-cell lineage and thus did not required further cell sorting.
Flow cytometry and cell cycle analysis
Five million purified pretumor B cells were fixed in ice-cold 70% ethanol. Cells were stained with Ki-67-FITC antibody and DNA was stained with PI/RNase staining buffer according to the manufacturer’s instructions (BD Biosciences). For the viability assay, primary tumor cells were stained with 7-AAD (Invitrogen) and anti–CD19-APC antibody (eBioscience) after Nutlin-3 (CalBiochem) treatments. All samples were analyzed using the FACS-CantoII flow cytometer (BD Biosciences) and FlowJo v 9.2 software (Tree Star, Inc.).
Histology
Tumor-bearing lymph nodes were fixed in 10% buffered formalin phosphate (Fisher Scientific) followed by paraffin embedding. Pretumor spleens were frozen in tissue-embedding medium (Tissue-Tek). Tumors were sectioned and stained with hematoxylin and eosin (tumor lymph nodes) or with anti-B220 antibody (pretumor spleens) at the Mouse Histology and Phenotyping Laboratory at Northwestern University. Stained tissue sections were imaged using an EVOS XL Core digital inverted microscope (Advanced Microscopy Group) using a 20× magnification lens.
Real-time polymerase chain reaction
Total RNA was isolated from purified pretumor B cells using the RNeasy RNA extraction kit with DNase I digestion from QIAGEN. To generate cDNA, a High-Capacity cDNA Reverse Transcription kit from Applied Biosystems was used. Real-time polymerase chain reaction (RT-PCR) was carried out on an Applied Biosystems Step One Plus machine using primers specific for the indicated genes. Fast SYBR green master mix from Applied Biosystems was used in all reactions. The ΔΔCt method was used to normalize gene expression to Gapdh.
Immunoblots
Pretumor B cells or tumor cells were lysed in lysis buffer with protease and phosphatase inhibitor cocktails (Roche). Lysates were electrophoretically separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Bio-Rad). Protein was transferred to a nitrocellulose membrane (Whatmann) and incubated with primary antibodies. Membranes were probed with IRDye secondary antibodies (Li-Cor Biosciences). The protein bands were visualized with an Odyssey Fc Western Blotting Imager and analyzed with Image Studio version 2.0 (Li-Cor Biosciences).
Statistical analysis
A 2-tailed Student t test, 1-way and 2-way analysis of variance (ANOVA), and survival analysis were analyzed using GraphPad (GraphPad Software, Inc.). P < .05 was considered statistically significant.
Results
LMP2A couples with MYC to promote G1-S transition
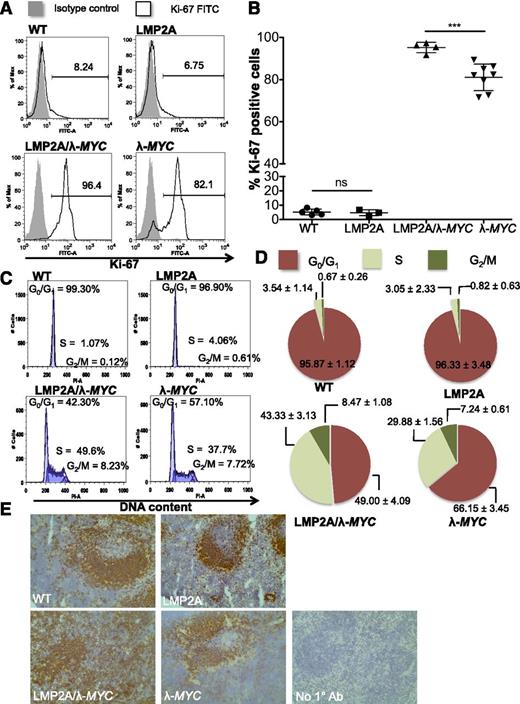
Because previous studies suggested that LMP2A promotes cell cycle progression in the context of deregulated MYC,24,25 we chose to investigate mechanisms by which LMP2A disrupts the cell cycle regulation. Because p53 pathway inactivation is frequent in MYC-driven lymphomagenesis, the status of the p53 pathway may interfere with analysis of the cell cycle when comparing tumor cells from LMP2A/λ-MYC and λ-MYC mice. Thus we chose to examine cell cycle profiles of pretumor B cells because the p53 pathway is intact at this stage in both LMP2A/λ-MYC and λ-MYC mice. Purified B cells from 3-week-old pretumor LMP2A/λ-MYC mice demonstrated high proliferative characteristics, with a significant increase in the percentage of Ki-67–positive cells (Figure 1A-B) and percentage of S-phase cells (Figure 1C-D) compared with those isolated from λ-MYC mice of the same age. The hyperproliferative phenotype of LMP2A/λ-MYC pretumor B cells was also evident histologically. Although wild-type (WT) C57BL/6 and LMP2A and λ-MYC transgenic mice formed B-cell follicles in secondary lymphoid organs such as the spleen, LMP2A/λ-MYC mice showed disorganized B-cell follicles (Figure 1E) and an increase in B220+ population. WT, LMP2A, and λ-MYC mice had comparable percentages of immature and mature B cells. LMP2A/λ-MYC B220+ cells were IgM–IgD–, likely as a result of LMP2A replacing the requirement for the BCR20,30,31 (see supplemental Figures 1 and 2 available on the Blood Web site for other detailed analyses). These data indicate that LMP2A promotes S-phase entry and hyperproliferation of B lymphocytes before malignant transformation.
Pretumor B cells from LMP2A/λ-MYC mice demonstrate highly proliferative phenotypes. (A) Histograms of Ki-67+ purified B cells from 3-week-old representative mice. The number in each histogram indicates the percentage of Ki-67+ cells in purified B cells. (B) Combined Ki-67 data from 3 experiments shown as mean ± SD. The differences in the percentage of Ki-67+ cells were analyzed by 1-way ANOVA. ***P < .001; ns, not significant. (C) Representative histograms of propidium iodine staining for DNA content in purified pretumor B cells. (D) Pie charts show mean ± SD of cells in different phases of the cell cycle as analyzed in (C). WT: n = 3, LMP2A: n = 3, LMP2A/λ-MYC: n = 4, λ-MYC: n = 4. (E) Immunohistochemistry of B220+ cells in representative 3-week-old mouse spleens (original magnification ×20). No 1° Ab, negative control with secondary antibody only.
Pretumor B cells from LMP2A/λ-MYC mice demonstrate highly proliferative phenotypes. (A) Histograms of Ki-67+ purified B cells from 3-week-old representative mice. The number in each histogram indicates the percentage of Ki-67+ cells in purified B cells. (B) Combined Ki-67 data from 3 experiments shown as mean ± SD. The differences in the percentage of Ki-67+ cells were analyzed by 1-way ANOVA. ***P < .001; ns, not significant. (C) Representative histograms of propidium iodine staining for DNA content in purified pretumor B cells. (D) Pie charts show mean ± SD of cells in different phases of the cell cycle as analyzed in (C). WT: n = 3, LMP2A: n = 3, LMP2A/λ-MYC: n = 4, λ-MYC: n = 4. (E) Immunohistochemistry of B220+ cells in representative 3-week-old mouse spleens (original magnification ×20). No 1° Ab, negative control with secondary antibody only.
LMP2A expression disrupts a cell cycle checkpoint during G1
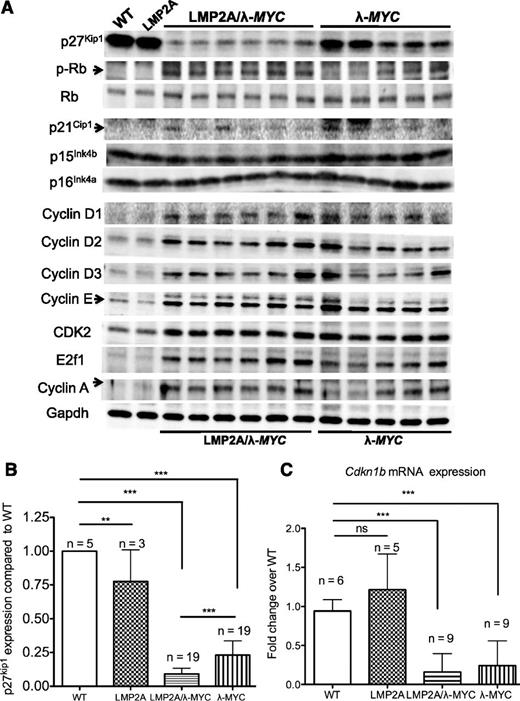
We further delineated changes in cell cycle regulators of the G1 checkpoint in LMP2A– and LMP2A+ cells by western blot analyses. Interestingly, of all CDK inhibitors important for preventing premature progression from G1 to S phase, p27kip1 was the only CDK inhibitor significantly downregulated in pretumor LMP2A/λ-MYC B cells compared with λ-MYC B cells (Figure 2A-B). There was also a moderate increase in phosphorylated Rb and cyclin A (a marker for S phase) in LMP2A/λ-MYC B cells, suggesting that low levels of p27kip1 result in the disruption of the Rb pathway in G1 checkpoint and lead to premature S-phase entry. However, quantitative RT-PCR (qRT-PCR) revealed that Cdkn1b mRNA levels were similar between LMP2A/λ-MYC and λ-MYC pretumor B cells (Figure 2C). These data agreed with previous studies demonstrating that MYC can promote Cdk1b transcriptional repression.32,33 Taken together, LMP2A does not contribute to Cdkn1b transcriptional repression mediated by MYC, but rather downregulates p27kip1 at the protein level to promote cell cycle progression.
P27Kip1is downregulated at the protein level, but not at the mRNA level, in pretumor B cells from LMP2A/λ-MYC mice compared with those in λ-MYC mice. (A) Representative western blots of cell cycle regulators in purified B cells from 3-week-old mice of indicated genotypes (C57BL/6 background). Each lane represents a protein sample from one mouse. (B) P27kip1 protein expression in B cells from 3-week-old mice shown as a ratio to WT expression from 5 independent experiments. (C) qRT-PCR analyses of Cdkn1b expression from 2 independent experiments using a WT mouse as a control for each experiment. All differences in mRNA and protein expression were analyzed by 1-way ANOVA. **P < .01, ***P < .001; ns, not significant. Data represent the mean ± SD.
P27Kip1is downregulated at the protein level, but not at the mRNA level, in pretumor B cells from LMP2A/λ-MYC mice compared with those in λ-MYC mice. (A) Representative western blots of cell cycle regulators in purified B cells from 3-week-old mice of indicated genotypes (C57BL/6 background). Each lane represents a protein sample from one mouse. (B) P27kip1 protein expression in B cells from 3-week-old mice shown as a ratio to WT expression from 5 independent experiments. (C) qRT-PCR analyses of Cdkn1b expression from 2 independent experiments using a WT mouse as a control for each experiment. All differences in mRNA and protein expression were analyzed by 1-way ANOVA. **P < .01, ***P < .001; ns, not significant. Data represent the mean ± SD.
Instability of p27kip1 in LMP2A/λ-MYC and λ-MYC pretumor B cells
Because p27kip1 is largely controlled by posttranslational modification and proteolysis,34-36 we further explored the possibility that LMP2A could increase the instability of p27kip1 to promote cell cycle progression. Pretumor B cells of LMP2A/λ-MYC and λ-MYC littermates were treated with the protein synthesis inhibitor cycloheximide (CHX) for 1, 2, and 3 hours. LMP2A/λ-MYC B cells showed a slightly shorter half-life compared with λ-MYC B cells after CHX treatment. However, the reduction of p27kip1 was significantly diminished when LMP2A/λ-MYC B cells were co-treated with both CHX and MG-132, a proteasome inhibitor (Figure 3A-B). Furthermore, when treated with MG-132 alone, LMP2A/λ-MYC B cells had twice the level of p27kip1 compared with vehicle-treated cells. In contrast, MG-132 had minimal effect on p27kip1 expression in λ-MYC B cells (Figure 3C). These data indicate that LMP2A promotes p27kip1 degradation through a proteasome-dependent process.
LMP2A facilitates p27kip1degradation in a proteasome-dependent manner. (A) Representative western blot analyses of p27kip1 expression after treatments with 25 μg/mL cycloheximide (CHX) with or without 40 μM MG-132 at indicated time points. The levels of p27kip1 were normalized to calnexin (a loading control), and the fold reductions of proteins were calculated using cells at 0 hour of each genotype as a control. (B) Combined data from 3 experiments represent the mean ± SD of the p27kip1 level. Differences in p27kip1 protein expression were analyzed by 2-way ANOVA. *P < .05, **P < .01, ***P < .001. (C) Representative western blot analyses of pretumor B-cell treatment with vehicle control (dimethyl sulfoxide [DMSO]) 40 μM, or 60 μM MG-132 for 2 hours. Gapdh and calnexin were used as loading controls. The left and right parts of the images shown are from the same membrane, and the middle, irrelevant, lanes were removed. Experiments were repeated at least 3 times.
LMP2A facilitates p27kip1degradation in a proteasome-dependent manner. (A) Representative western blot analyses of p27kip1 expression after treatments with 25 μg/mL cycloheximide (CHX) with or without 40 μM MG-132 at indicated time points. The levels of p27kip1 were normalized to calnexin (a loading control), and the fold reductions of proteins were calculated using cells at 0 hour of each genotype as a control. (B) Combined data from 3 experiments represent the mean ± SD of the p27kip1 level. Differences in p27kip1 protein expression were analyzed by 2-way ANOVA. *P < .05, **P < .01, ***P < .001. (C) Representative western blot analyses of pretumor B-cell treatment with vehicle control (dimethyl sulfoxide [DMSO]) 40 μM, or 60 μM MG-132 for 2 hours. Gapdh and calnexin were used as loading controls. The left and right parts of the images shown are from the same membrane, and the middle, irrelevant, lanes were removed. Experiments were repeated at least 3 times.
Expression of S10A mutant form of p27kip1 (p27S10A) in MYC-driven lymphomagenesis
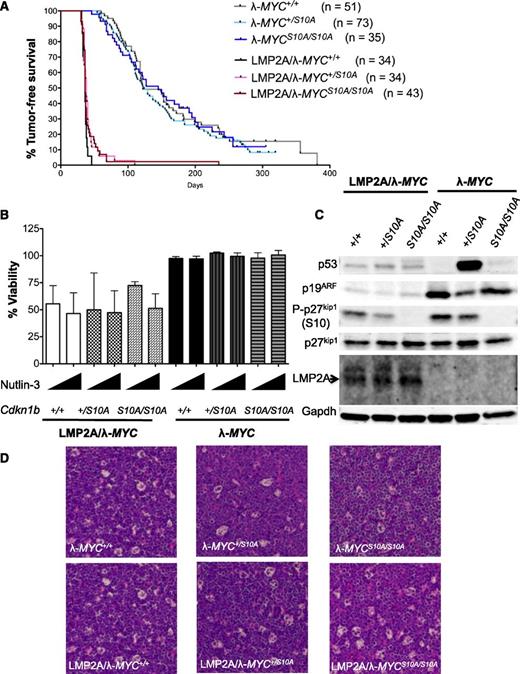
To examine the importance of p27kip1 in LMP2A-mediated lymphomagenesis, we crossed our transgenic mice to knock-in mice in which serine 10 of p27kip1 was replaced with alanine (p27S10A). We chose the p27S10A knock-in mutant mice because previous studies demonstrated a tumor-suppressing phenotype compared with WT p27kip1 in a mouse model of lung cancer,29 a lymphoproliferative disorder,37 and SV40-induced tumorigenesis.38 In addition, murine embryonic fibroblasts (MEFs) from p27S10A/S10A mice have a nuclear localization phenotype29 and are more refractory to degradation mediated by SV40-induced transformation compared with MEFs from WT mice.38 Because the role of S10 phosphorylation of p27kip1 had not been tested in the context of deregulated MYC in vivo, the S10A mutant could act as a gain-of-function form of p27kip1 and would allow us to test whether the expression of this mutant would influence lymphoma development in our model. We hypothesized that expression of p27S10A would prevent LMP2A-mediated p27kip1 suppression and would delay lymphomagenesis to be similar to that of λ-MYC mice. However, we found the median survival time of all LMP2A/λ-MYC mice was very similar (36-38 days). We noted that a small subset of LMP2A/λ-MYC+/S10A and LMP2A/λ-MYCS10A/S10A mice were tumor-free at day 50 when all LMP2A/λ-MYC+/+ developed tumors. One LMP2A/λ-MYCS10A/S10A mouse lived tumor-free beyond the median survival time of λ-MYC+/+ (122 days). All λ-MYC mice regardless of p27kip1 genotypes had similar time-to-lymphoma onset. This observation suggests that p27S10A has a minimal effect on the latency time of lymphoma development in our model (Figure 4A).
Expression of S10A mutant confers minor effect on time-to-tumor onset of LMP2A/λ-MYC mice and has no effect on the p53 pathway. (A) Kaplan-Meier curves indicating the percent survival of indicated genotypes. (B) Viability of primary tumor cells (7-AAD–, CD19+) at 3 hours after 5 μM or 10 μM Nutlin-3 treatments from 3 independent experiments. Percentage of viability in vehicle control in each genotype was set at 100%. (C) Western blot analyses of representative primary tumor cells showed aberrant stabilization of p53 and/or p19ARF in tumors from λ-MYC mice. (D) Hematoxylin and eosin (H&E) staining of tumor-bearing lymph nodes demonstrating a “starry sky” appearance in all tumor genotypes (original magnification ×20).
Expression of S10A mutant confers minor effect on time-to-tumor onset of LMP2A/λ-MYC mice and has no effect on the p53 pathway. (A) Kaplan-Meier curves indicating the percent survival of indicated genotypes. (B) Viability of primary tumor cells (7-AAD–, CD19+) at 3 hours after 5 μM or 10 μM Nutlin-3 treatments from 3 independent experiments. Percentage of viability in vehicle control in each genotype was set at 100%. (C) Western blot analyses of representative primary tumor cells showed aberrant stabilization of p53 and/or p19ARF in tumors from λ-MYC mice. (D) Hematoxylin and eosin (H&E) staining of tumor-bearing lymph nodes demonstrating a “starry sky” appearance in all tumor genotypes (original magnification ×20).
P53-mediated apoptosis is an important tumor-suppressive mechanism counteracting MYC oncogenic function. To examine whether p27S10A expression would change the status of the p53 pathway, primary tumor cells were treated with the MDM2 inhibitor Nutlin-3, which leads to the release of p53 from its negative regulator. All LMP2A/λ-MYC tumor cells were sensitive to Nutlin-3 treatments, resulting in a sharp decrease in viability compared with control at 3 hours post treatment, suggesting that these cells have an intact p53 pathway after activation of p53 upon Nutlin-3 treatment. All λ-MYC tumor cells were resistant to Nutlin-3 treatments (Figure 4B), indicating that p53 is mutated or the pathway is inactivated, similar to previous reports.24,27 Previous studies reported that tumors with a defect in the p53-19ARF-MDM2 pathway show aberrant accumulation or stabilization of p19ARF and p53 caused by the disruption of a p53 negative-feedback loop.24,27,39-41 Excessive accumulation of p53 and p19ARF proteins in λ-MYC tumor cells was evident but was absent in all LMP2A/λ-MYC tumors, regardless of genotypes of p27kip1 (Figure 4C). These data indicate that LMP2A effectively promotes lymphoma development without perturbing the p53 pathway, and tumor promotion by LMP2A is unaffected by p27S10A expression. Despite the strong differences in tumor onset time and p53 status, all tumors from LMP2A/λ-MYC and λ-MYC mice were histologically similar, with the “starry sky” appearance resembling Burkitt lymphoma (Figure 4D and supplemental Figure 3). Taken together, these data indicate that p27S10A expression does not prevent p53 pathway inactivation in λ-MYC tumors and that the p53 pathway is still functionally intact in all LMP2A/λ-MYC tumors examined.
LMP2A antagonizes p27S10A function
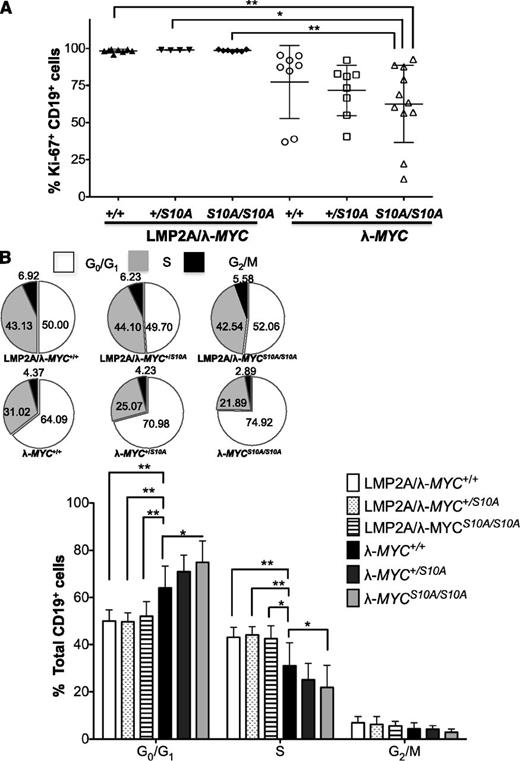
To investigate whether the p27S10A mutant could alter the cell cycle profile at earlier steps of lymphomagenesis, spleen mass was analyzed in mice at 3 weeks of age. P27S10A expression did not significantly change the relative spleen mass or the ratios of B:T cells, although there was a small decrease in relative spleen weight of mice with homozygous p27S10A compared with those with WT p27kip1 (supplemental Figure 4). We found that all LMP2A/λ-MYC pretumor B cells were >90% Ki-67+ (Figure 5A). Interestingly, there was a dose-dependent decrease in the percentage of Ki-67+ B cells with the expression of heterozygous and homozygous p27S10A in λ-MYC pretumor B cells (Figure 5A). Furthermore, cell cycle analyses showed a significant dose-dependent increase in G1 cells and a significant decrease in S-phase cells in λ-MYC pretumor B cells with the expression of 1 or 2 alleles of p27S10A. These data indicate that p27S10A impedes S-phase entry in early events of lymphoma development. However, the effect of p27S10A in blocking G1-S transition was lost in all LMP2A/λ-MYC pretumor B cells (Figure 5B), indicating that LMP2A can counteract the antiproliferative function of p27S10A to promote MYC-driven cell cycle progression.
Homozygous p27S10Aexpression significantly decreases S-phase pretumor B cells from λ-MYC mice but has no effect in LMP2A/λ-MYC pretumor B cells. (A) Percentage of Ki-67+ pretumor B cells of each genotype. Data were combined from 5 experiments and represent the mean ± SD. (B) Pie charts and bar graphs demonstrating the cell cycle profile of pretumor B cells. Data were analyzed with 1-way ANOVA and represent the mean ± SD. *P < .05, **P < .01. The inset shows pie charts representing mean percentages of pretumor B cells in each phase of the cell cycle.
Homozygous p27S10Aexpression significantly decreases S-phase pretumor B cells from λ-MYC mice but has no effect in LMP2A/λ-MYC pretumor B cells. (A) Percentage of Ki-67+ pretumor B cells of each genotype. Data were combined from 5 experiments and represent the mean ± SD. (B) Pie charts and bar graphs demonstrating the cell cycle profile of pretumor B cells. Data were analyzed with 1-way ANOVA and represent the mean ± SD. *P < .05, **P < .01. The inset shows pie charts representing mean percentages of pretumor B cells in each phase of the cell cycle.
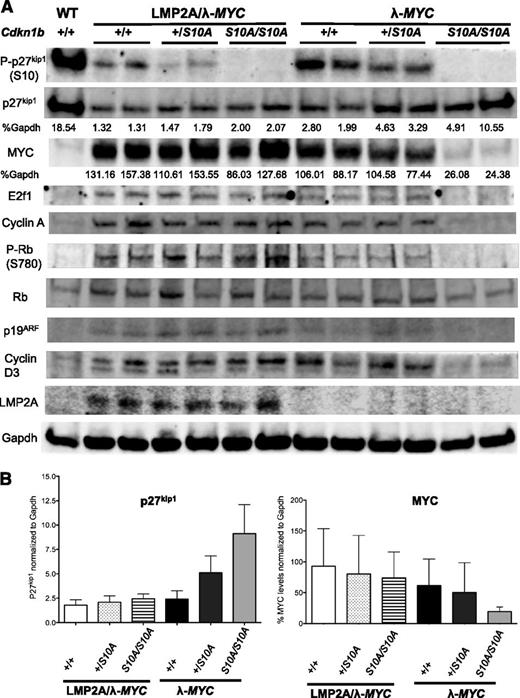
To delineate the molecular mechanisms responsible for the failure of p27S10A in blocking G1-S transition in LMP2A/λ-MYC pretumor B cells but successfully preventing S-phase entry in λ-MYC pretumor B cells, pretumor B cells were subjected to western blot analyses. We found an opposite trend of increasing p27kip1 and decreasing MYC levels in representative λ-MYC+/S10A and λ-MYCS10A/S10A compared with λ-MYC+/+. In addition, we found a decrease in cyclin A in λ-MYCS10A/S10A pretumor B cells, corresponding to a lower percentage of B cells in S-phase shown in Figure 5B. LMP2A/λ-MYCS10A/S10A pretumor B cells did not show an increase in p27kip1 level compared with LMP2A/λ-MYC+/+ cells. Furthermore, MYC was expressed at high levels in all LMP2A/λ-MYC pretumor B cells, regardless of p27S10A expression (Figure 6A-B).
LMP2A expression associates with high MYC level, which inversely correlates with low p27kip1levels. (A) Western blot analyses of cell cycle regulators in purified B cells from 3-week-old WT, LMP2A/λ-MYC+/+, LMP2A/λ-MYC+/S10A, LMP2A/λ-MYCS10A/S10A, λ-MYC+/+, λ-MYC+/S10A, and λ-MYCS10A/S10A mice. Cyclin A was used as an S-phase marker. Normalized levels of p27kip1 and MYC are shown as percentages to Gapdh (loading control). (B) Bar graphs demonstrating MYC and p27kip1 protein expression in B cells from 3-week-old mice from 2 independent experiments. Each bar represents combined data from 4 experiments, with data represented as the mean ± SD.
LMP2A expression associates with high MYC level, which inversely correlates with low p27kip1levels. (A) Western blot analyses of cell cycle regulators in purified B cells from 3-week-old WT, LMP2A/λ-MYC+/+, LMP2A/λ-MYC+/S10A, LMP2A/λ-MYCS10A/S10A, λ-MYC+/+, λ-MYC+/S10A, and λ-MYCS10A/S10A mice. Cyclin A was used as an S-phase marker. Normalized levels of p27kip1 and MYC are shown as percentages to Gapdh (loading control). (B) Bar graphs demonstrating MYC and p27kip1 protein expression in B cells from 3-week-old mice from 2 independent experiments. Each bar represents combined data from 4 experiments, with data represented as the mean ± SD.
Because p27S10A/S10A showed a nuclear translocation phenotype,29,37 the low level of total p27kip1 in LMP2A/λ-MYC pretumor B cells suggests that another degradation process plays a role in LMP2A-mediated p27kip1 degradation. Previous studies showed that Cks1 and Skp2, both are constituents of SCFSkp2 complex regulating nuclear p27kip1 degradation, were increased in precancerous B cells from Eμ–Myc mice compared with WT B cells.42 We found similar Skp2 levels in both LMP2A/λ-MYC and λ-MYC pretumor B cells, whereas there was an increase in Cks1 protein level in LMP2A/λ-MYC B cells. There was also an elevated Cks1 mRNA in LMP2A/λ-MYC compared with λ-MYC B cells, but the difference was not statistically significant (supplemental Figure 5). These data suggest that high MYC levels in LMP2A/λ-MYC B cells promotes further p27kip1 degradation, at least in part by upregulating Cks1 in the SCFSkp2 complex, which is active in the late G1 and S phases. Overall, these results indicate that LMP2A expression can antagonize the function of p27S10A by maintaining MYC at a high level, leading to continued downregulation of p27kip1 levels. In the absence of LMP2A, the gain-of-function p27S10A is able to increase total p27kip1 level, which in turn inversely correlates with MYC expression.
Discussion
This study reports the collaborative efforts of MYC and EBV LMP2A in disrupting the cell cycle before progression to malignancy. The complex relationship of a tumor virus and a human oncogene is exemplified in our model, which demonstrates that EBV provides an optimal intracellular environment conducive to malignant transformation. Our studies confirm previous studies on how 2 important tumor suppressor pathways counteract MYC and reveal how EBV changes the equation to favor the pro-proliferative function of MYC.
Low levels of p27kip1 are observed in several human cancers,43 and murine models of cancers indicate that p27kip1 is a dose-dependent tumor suppressor.44-47 Mice hemizygous for p27kip1 (p27−/+) display an intermediate phenotype in body weight and cell proliferation between p27−/− and p27+/+ mice.48 The loss of 1 or 2 copies of p27kip1 demonstrates a dose-dependent decrease in tumor latency in γ-irradiation and carcinogen-induced tumor models.49,50 Our studies are compatible with these studies because expression of 1 or 2 copies of the gain-of-function S10A mutant in λ-MYC pretumor B cells results in a dose-dependent decrease in the percentage of cells in S-phase and that are Ki-67+.
Germline deletion or mutation of the p27kip1 gene, CDKN1B, in human cancer is rare, suggesting that other mechanisms play a major role in regulating p27kip1 in cancer. MYC promotes transcriptional repression of the p27kip1 gene,32 but p27kip1 is largely controlled at the protein level. MYC promotes p27kip1 protein degradation by upregulating transcription of negative regulators of p27Kip1,42,51-53 including Cks1 and Skp2. Furthermore, phosphorylation of p27kip1 at different sites controls degradation and subcellular localization of p27kip1 at different time points in the cell cycle. There are at least 2 pathways regulating p27kip1 degradation. In G0 and early G1, p27kip1 is phosphorylated at serine 10 and is exported from the nucleus. Cytoplasmic p27kip1 is targeted for proteasomal degradation mediated by the Kip1 ubiquitination-promoting complex (KPC).54 In the late G1 and S phases, p27kip1 is phosphorylated at threonine 187 and recognized by the SCFSkp2 complex, which ubiquitinates p27kip1 and targets it for the degradation by the proteasome in the nucleus.55,56 Mice lacking Csk1 or Skp2 showed an increase in p27kip1 level and delayed MYC-induced tumorigenesis.42,52 We found that p27S10A fails to increase total p27kip1 levels in LMP2A/λ-MYC+/S10A and LMP2A/λ-MYCS10A/S10A pretumor B cells, indicating that LMP2A still efficiently promotes nuclear degradation of p27kip1 mediated by SCFSkp2 complex in late G1 and S-phase and may do so via Cks1 upregulation. Another possibility is that high MYC levels in pretumor B cells from LMP2A/λ-MYC mice make the degradation of p27kip1 very rapid (Figure 6). However, both cytoplasmic and nuclear degradation pathways may be required to act in concert. Future studies will further examine the role of the SCFSkp2 complex in LMP2A-mediated lymphoma development.
It is important to note that regulatory processes of p27kip1 in cancers can be different in different cell types and oncogenic stimuli. Immortalized MEFs from p27S10A/S10A mice are more resistant to in vitro transformation by K-Ras but show similar transformation ability induced by MYC compared with WT MEFs.50 These data suggest that K-Ras and MYC suppress p27kip1 through different mechanisms during cellular transformation and could explain the difference in the tumor-suppressing function of p27S10A in our MYC model and K-Ras–induced tumorigenesis in previous studies.29,38 Furthermore, the localization of p27kip1 in quiescent or activated B cells is different from that of T cells and MEFs. P27kip1 is mainly localized in the cytoplasm of resting B cells and is translocated to the nucleus upon BCR activation. Defects in the suppression of serine 10 phosphorylation on p27kip1 inhibit the nuclear translocation process and thus contribute to autoimmunity and a lymphoproliferative disorder, and expression of p27S10A corrects this anomaly.37 Although homozygous p27S10A expression significantly reduced the percentage of S-phase pretumor B cells from λ-MYC mice (Figure 6B), it had only a small effect in tumor latency (Figure 4A). Therefore, p27S10A may be a mild gain-of-function mutant in an MYC-driven lymphoma model.
Our study is the first to show an underlying mechanism of EBV LMP2A in promoting cell cycle progression via p27kip1 downregulation. A recent study showed that LMP2A is important for cell proliferation and efficient long-term growth after the transformation of primary B cells.23 LMP2A may be dominant in promoting MYC-induced p27kip1 downregulation in latency program I, where EBNA1 and LMP2A appear to be uniquely expressed.57,58 Furthermore, in vitro BCR stimulation in primary normal B cells results in the downregulation of p27kip1, G1-S transition, and proliferation of B cells.37 Given that LMP2A is a BCR mimic, pathways leading to p27kip1 degradation in this study may be similar to a stimulated BCR, which involves the Erk and/or Akt pathway.37 Inhibition of Akt induces apoptosis in B cells from LMP2A transgenic mice.59 Furthermore, the inhibition of the Lyn kinase or the mammalian target of rapamycin (mTOR) pathway leads to decreases in spleen and tumor sizes in LMP2A/λ-MYC mice compared with λ-MYC mice,60,61 suggesting that LMP2A is able to mimic the BCR signal through the Akt/mTOR pathway to promote lymphomagenesis.
The dramatic downregulation of p27kip1 in pretumor B cells and the accelerated lymphomagenesis in LMP2A/λ-MYC mice correspond with a previous study showing that Eμ–Myc mice lacking p27kip1 have rapid tumor onset compared with those with WT p27kip1,16 suggesting the importance of p27kip1 in suppressing MYC-driven lymphomagenesis. However, p53 status in tumors of Eμ–Myc; p27−/− was not assessed. The relationship of how MYC represses p27kip1 is better documented than how p27kip1 can negatively regulate MYC. It is possible that p27kip1 is a part of a negative-feedback network controlling the MYC level. Because LMP2A expression associates with the continued p27kip1 repression, even when homozygous p27S10A is present, the importance of p27kip1 in LMP2A-mediated lymphomagenesis is still unclear and future studies should address this issue.
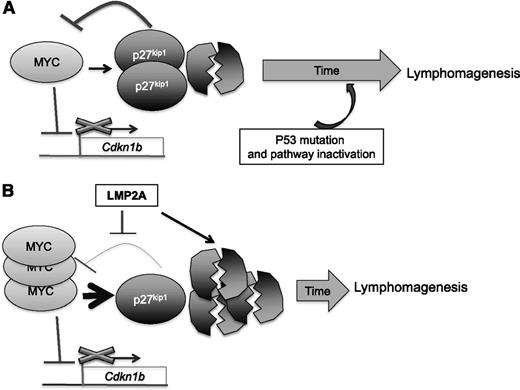
In λ-MYC B cells, our data suggest that the downregulation of the p27kip1 and p53 pathway inactivation occurs at different time points—p27kip1 downregulation precedes p53 inactivation. We propose that MYC downregulates p27kip1 both at the transcription and protein levels, leading to cell cycle progression. The remaining pool of p27kip1 then functions as part of an MYC repression network to control MYC. MYC can also induce apoptosis through the induction of the p53 pathway. Both p53 and p27kip1 act in concert or independently to prevent uncontrolled proliferation. However, selection pressure promotes the survival of only cells that contain a disruption in one or more components of the p53 pathway. P27kip1 may act dominantly at an early stage of lymphomagenesis, and the downregulation of p27kip1 may be less important after p53 pathway inactivation. In the presence of LMP2A, such as in EBV-infected B cells, p27kip1 is downregulated and MYC may be able to drive proliferation to a greater extent, thus outpacing apoptosis. Therefore, B cells expressing both deregulated MYC and LMP2A expand more readily without the need to mutate or inactivate the p53 pathway (Figure 7).
The proposed relationship of MYC, LMP2A, and p27kip1in lymphoma development. (Top) In an λ-MYC mouse model, overexpression of MYC results in repression of Cdkn1b transcription and increases p27kip1 degradation, leading to an increase in proliferation. The remaining pool of p27kip1 partially impedes MYC-induced cell cycle progression and downregulates MYC. Abundant expression of MYC also induces p53 pathway activation. Both p27kip1 and p53 are able to prevent uncontrolled proliferation for a certain length of time. At a later step, selection pressure forces deregulated MYC cells to overcome the apoptosis mechanism mediated by p53, and a lymphoma develops. (Bottom) In the presence of LMP2A, p27kip1 turnover is more rapid and weakens the MYC repression network, resulting in a high level of MYC. Although the p53 pathway is induced as a result of overexpression of MYC, cell cycle progression and proliferation outpaces the rate of apoptosis. A severely low level of p27kip1 may relieve the selection pressure, and a lymphoma develops without the need to inactivate the p53 pathway.
The proposed relationship of MYC, LMP2A, and p27kip1in lymphoma development. (Top) In an λ-MYC mouse model, overexpression of MYC results in repression of Cdkn1b transcription and increases p27kip1 degradation, leading to an increase in proliferation. The remaining pool of p27kip1 partially impedes MYC-induced cell cycle progression and downregulates MYC. Abundant expression of MYC also induces p53 pathway activation. Both p27kip1 and p53 are able to prevent uncontrolled proliferation for a certain length of time. At a later step, selection pressure forces deregulated MYC cells to overcome the apoptosis mechanism mediated by p53, and a lymphoma develops. (Bottom) In the presence of LMP2A, p27kip1 turnover is more rapid and weakens the MYC repression network, resulting in a high level of MYC. Although the p53 pathway is induced as a result of overexpression of MYC, cell cycle progression and proliferation outpaces the rate of apoptosis. A severely low level of p27kip1 may relieve the selection pressure, and a lymphoma develops without the need to inactivate the p53 pathway.
In summary, we demonstrated for the first time the cooperative role of LMP2A and MYC in disrupting the cell cycle through promoting p27kip1 degradation at the early stage of lymphomagenesis. LMP2A perturbs the MYC negative feedback loop, partially mediated by p27kip1, to accentuate MYC expression and accelerate lymphomagenesis without altering the p53 pathway. Our data shed a new light on how EBV infection aids MYC in counteracting with antiproliferative cellular mechanisms, and our results also emphasize critical roles of EBV infection in creating an intracellular state favorable for malignant transformation.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
The authors thank members of the Longnecker laboratory for help in completion of this study. J.C. is a fellow of the American Heart Association. R.L. is Dan and Bertha Spear Research Professor.
This work is supported by the Northwestern University Mouse Histology and Phenotyping Laboratory and a Cancer Center Support Grant, National Institutes of Health (NCI CA060553); the National Cancer Institute, National Institutes of Health (CA073507, CA133063) (R.L.); and the Training Program in Viral Replication (T32AI060523-09) (K.F.).
Authorship
Contribution: K.F. and R.L. designed research, analyzed data, and wrote the manuscript; and K.F. and J.C. performed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard Longnecker, Northwestern University, 303 East Chicago Ave, Chicago, IL 60611; e-mail: r-longnecker@northwestern.edu.

![Figure 3. LMP2A facilitates p27kip1 degradation in a proteasome-dependent manner. (A) Representative western blot analyses of p27kip1 expression after treatments with 25 μg/mL cycloheximide (CHX) with or without 40 μM MG-132 at indicated time points. The levels of p27kip1 were normalized to calnexin (a loading control), and the fold reductions of proteins were calculated using cells at 0 hour of each genotype as a control. (B) Combined data from 3 experiments represent the mean ± SD of the p27kip1 level. Differences in p27kip1 protein expression were analyzed by 2-way ANOVA. *P < .05, **P < .01, ***P < .001. (C) Representative western blot analyses of pretumor B-cell treatment with vehicle control (dimethyl sulfoxide [DMSO]) 40 μM, or 60 μM MG-132 for 2 hours. Gapdh and calnexin were used as loading controls. The left and right parts of the images shown are from the same membrane, and the middle, irrelevant, lanes were removed. Experiments were repeated at least 3 times.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/4/10.1182_blood-2013-07-517649/4/m_530f3.jpeg?Expires=1769192527&Signature=P1CGHCK1QAW~N8B9lPrZ5lAXE0S3QPcrZSvjUgJjuA1E2kLQ0YuUu31loBLbpTfHxCS5JVBgW5RRlO9bf~gV7-iFc--49gT8cNDCOf-u0w4T7tl7wJ4ofom7pROrGSbVWT7~XYNR32coomrb0yjof47VwOeYa3xskv6UhufEa9omIHcxQ48XOgo74TDMWnIJOnpKoTk3kunPTCRghnHuxgwoKbJLzvUEg~DLnuaKEBpzIbqRpXPdxjydvGoGrw-sv2VWwXmGS50g8ZkxYsUDhBtx2RvXQxPI~tQH3OW-cgUMYS2IwDSnfv07nBuTOB8GEWkXj6oQQH9aDWeK8TfblQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal