In this issue of Blood, Komeno et al characterize a murine Runx1 splicing isoform, Runx1bEx6e, as a positive regulator for the pool size of hematopoietic stem cells (HSCs).1
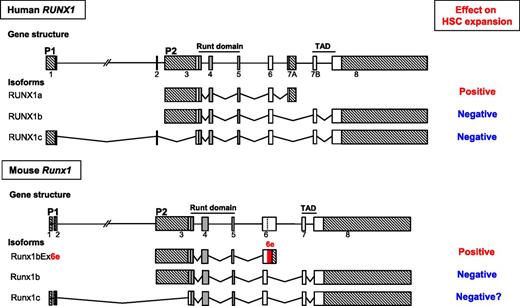
Schematic representation of the gene structures, major splicing isoforms of human and murine Runx1 genes, and their effects on expansion of hematopoietic stem cells. Exons are indicated by boxes. The vertical dashed line within exon 6 indicates an internal splice site. Exons constituting the runt domain are indicated in gray. The two alternative promoters are denoted as P1 and P2. Crosshatched boxes indicate 5′- and 3′-untranslated regions. The coding region in the extended exon 6 is highlighted in red.
Schematic representation of the gene structures, major splicing isoforms of human and murine Runx1 genes, and their effects on expansion of hematopoietic stem cells. Exons are indicated by boxes. The vertical dashed line within exon 6 indicates an internal splice site. Exons constituting the runt domain are indicated in gray. The two alternative promoters are denoted as P1 and P2. Crosshatched boxes indicate 5′- and 3′-untranslated regions. The coding region in the extended exon 6 is highlighted in red.
The RUNX1 gene, also known as AML1, CBFA2, or PEBP2αB, was cloned as the gene located at a chromosomal breakpoint in one of the most common leukemia-associated chromosomal translocations, t(8;21), in 1991,2 roughly 20 years after the first description of t(8;21) by Janet D. Rowley in 1973.3 In the cloning report, its short truncated isoform, later termed RUNX1a, but not the full-length (FL) transcript, was described.2 Soon after, two distinct FL forms, driven by two different promoters (P1 and P2), were identified and named RUNX1b and RUNX1c, respectively (see figure). RUNX1 encodes a transcription factor consisting of two functionally important moieties, the runt domain (RD) responsible for DNA binding and heterodimerization with CBFβ and the transcriptional activation domain (TAD). Since RUNX1a lacks the TAD but retains the RD, it was expected to and in fact shown to behave as a dominant negative form toward the FL-mediated transcriptional regulation.4 Biological functional consequences of this RUNX1a-dependent inhibitory effect have since been extensively investigated in the field, particularly in the areas of HSCs and leukemia.5,6 Through such endeavors over the past two decades, a significant body of knowledge has been accumulated, and we now know that the two FL forms, RUNX1b and RUNX1c, act as negative regulators for HSC pool size, and RUNX1a functions as a positive regulator (see figure). Fine-tuning of the balance between these positive and negative regulators transcribed from a single hematopoietic gene, RUNX1, is clearly shown to be one of the critical mechanisms in determining HSC population size.
Besides the above-mentioned 3 isoforms of RUNX1, many other alternative splicing forms have also been reported in multiple species, including primates (human, chimpanzee, and rhesus monkey), mouse, chicken, zebrafish, elephant shark, fugu, Xenopus, Drosophila, C elegans, Ciona, and sea urchin. However, very strangely, an ortholog of human RUNX1a has never been captured in mice, despite the extensive studies on the mouse as a model organism not only in hematology but also in many other biological fields. Considering its unique and seemingly indispensable role for HSC expansion, the murine equivalent of human RUNX1a was expected to be present, but no successful attempt has been made so far. In the study by Komeno et al, the authors revisit this old research question.
The authors first adopted a comparative genomics approach and found that human RUNX1a orthologs are found only in primates. They therefore expanded their search by using a similar short truncated isoform and noticed a previously reported but uncharacterized Runx1bEx6e isoform, which retains the RD but lacks TAD, like RUNX1a.7 Human RUNX1a is generated by the use of exon 7A, whereas human RUNX1b and RUNX1c are formed by the use of exon 7B. In mice, there are no nucleotide or amino acid (aa) sequences homologous to those of exon 7A in human RUNX1a. The truncated isoform Runx1bEx6e is generated by the use of an extended exon 6 which includes an additional stretch of intron 6 sequence spanning immediately after the end of known exon 6. This extra stretch of sequence encodes 63 aa (189-bp) followed by 171-bp 3′-untranslated region. The Runx1bEx6e ortholog is detected in Runx1 and two other Runx family genes, Runx2 and Runx3, of fugu,8 suggesting that all vertebrates except for primates might use the extended exon 6 to generate a key positive regulator for HSC expansion.
The Runx1bEx6e, together with two other isoforms, FL (Runx1bEx6+) and exon 6–skipping isoform (Runx1bEx6−), were subsequently cloned and subjected to molecular and hematologic analyses. As expected, Runx1bEx6e exhibited loss of transcriptional activity in a reporter assay but enhanced colony-forming capability compared with the FL isoform. Bone marrow transplantation assay using cells that have been retrovirally transduced with these isoforms also clearly demonstrated that the mice that were recipients of Runx1bEx6e-transduced cells revealed continuous increase in engraftment, but the recipients of Runx1bEx6+-transduced cells showed a decrease of transplanted cells. Runx1bEx6− also showed low engraftments, although this isoform exhibited behaviors similar to Runx1bEx6e in some assays.
The strongest evidence for the Runx1bEx6e-mediated HSC expansion came from the data of Runx1-IRES-GFP knockin (KI) mice, originally generated for other purposes.9 Fortunately for the authors, these KI mice lack the exon 6–related alternative splicing isoforms that include Runx1bEx6e, hence serving as the best platform to prove the in vivo functionality of the isoforms of interest. The KI mice had significantly fewer c-Kit+Sca1+ lineage marker− cells, which include HSCs and multipotent progenitors, than control mice. In vivo competitive repopulation assays demonstrated a sevenfold difference of functional HSCs between wild-type and KI mice. On the basis of these results, the authors concluded that the Runx1bEx6e isoform functions as a counter player against FL isoforms: Runx1bEx6e regulates HSC pool size in a positive manner, whereas Runx1bEx6+ does so negatively. The long-awaited positive regulator for HSC expansion in mice, equivalent to human RUNX1a, is now clearly shown to be Runx1bEx6e.
Fine-tuning of the balance between such positively and negatively regulating Runx1 isoforms is reported to work in hematopoietic commitment of embryonic stem cells10 and many other biological processes. Notably, imbalance of RUNX1 isoforms has been suspected to be an underlying mechanism for leukemogenesis. Recent advances in sequencing technology have unraveled a plethora of mutations in splicing factors (SFs) in human hematologic malignancies. The precise mechanisms by which these SF mutations cause diseases remain unknown. Deregulation of RUNX1 splicing machinery could be the key driving force of leukemia stem cells carrying SF mutation. Further investigations on splicing mechanism would provide us with deeper insights into both normal and pathological stem cell behaviors.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal