To the editor:
On December 5, 2013, chikungunya virus (CHIKV) was introduced into the Western Hemisphere. The first cases of autochthonous chikungunya fever were reported in Saint Martin, French West Indies (FWI), demonstrating local transmission.1 As of March 30, 2014, autochthonous cases have been recorded on 9 Caribbean islands, including more than 15 000 suspected cases in FWI.2
Chikungunya fever is an arboviral disease transmitted to humans by Aedes mosquitoes. It is characterized by febrile arthralgia and is responsible for massive epidemics.3
For years, concerns were raised about transfusion-transmitted infections (TTIs) during CHIKV outbreaks,4,5 and attempts were made to estimate the risk.6,7 During the La Reunion outbreak in 2006,6 modeling of TTI risk was based on (1) a proportion of asymptomatic infections of 15%, (2) a 6-day mean duration of viremia after clinical onset in symptomatic patients, (3) a 1.5-day mean duration between the beginning of viremia and clinical onset in symptomatic patients, and (4) a 7.5-day total duration of viremia in asymptomatic patients. Items (1) and (2) were obtained from observational studies,3,6 (3) and (4) being assumed. By using the same model, we estimated that the risk of CHIKV contamination from asymptomatic and presymptomatic viremic blood donors would be roughly equivalent.
In early 2014, the French Blood Agency implemented a strategy to prevent CHIKV TTIs in the FWI that relied on specific CHIKV nucleic acid testing (NAT) combined with postdonation self-reporting of febrile symptoms, accompanied by a 72-hour postdonation quarantine of nonpathogen reduced blood products.
CHIKV NAT screening was performed by using a Biomek NXP Workstation (Beckman Coulter), a NucleoSpin 96 Virus Extraction Kit (MACHEREY-NAGEL), and the RealStar Chikungunya RT-PCR Kit 1.1 (Altona Diagnostics).
Among the first 2149 plasma samples collected from February 24 to April 9, 4 were positive (104 to 2.0 × 108 CHIKV genome-equivalents/mL or 101 to 2.0 × 105 plaque forming units (PFU)/mL); all were males living in Martinique who had not recently traveled abroad. Two remained asymptomatic (41 and 43 years old), and two reported febrile syndrome 12 to 24 hours postdonation (61 and 66 years old).
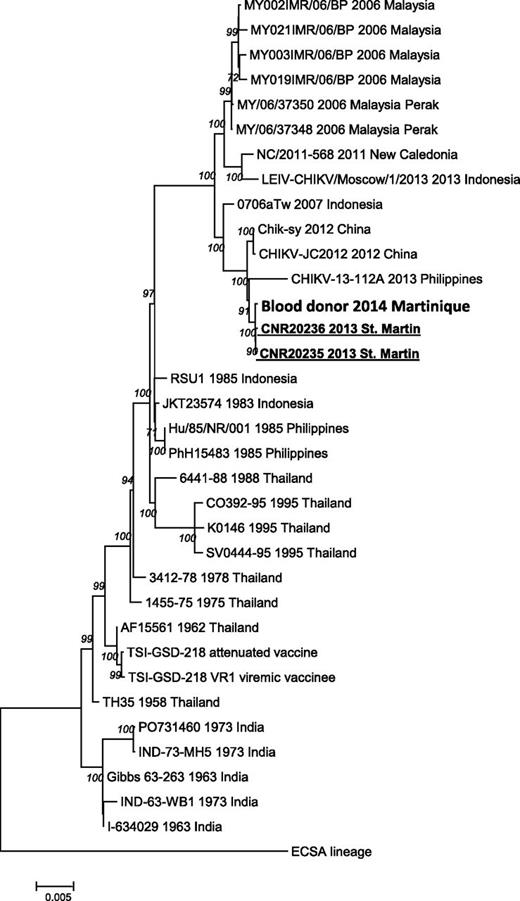
The full-length viral RNA genome, which was characterized directly from the plasma of the first patient by using next-generation sequencing, indicated Asian lineage and close similarity to Caribbean sequences described earlier (Figure 1).1,8
Phylogeny of CHIKVs causing the outbreak in the French Caribbean Islands, 2013-2014. Complete concatenated nucleotide coding sequences of CHIKV belonging to the Asian genotype were obtained by using Ion Torrent next-generation sequencing (Life Technologies) and aligned according to the amino acid sequence. Mega6 software was used to select the best-fit model for the analysis (generalized time reversible model of nucleotide substitution with a gamma distributed rate variation among sites) and to infer a phylogenetic tree using the maximum likelihood method followed by a bootstrap analysis (1000 replicates; values above 70% indicated).10 Sequence information for each virus corresponds to virus strain name (according to GenBank nomenclature) followed by date and country of isolation. The tree was rooted by using a sequence belonging to the East Central South African lineage (LR2006_OPY1 2006 Reunion).
Phylogeny of CHIKVs causing the outbreak in the French Caribbean Islands, 2013-2014. Complete concatenated nucleotide coding sequences of CHIKV belonging to the Asian genotype were obtained by using Ion Torrent next-generation sequencing (Life Technologies) and aligned according to the amino acid sequence. Mega6 software was used to select the best-fit model for the analysis (generalized time reversible model of nucleotide substitution with a gamma distributed rate variation among sites) and to infer a phylogenetic tree using the maximum likelihood method followed by a bootstrap analysis (1000 replicates; values above 70% indicated).10 Sequence information for each virus corresponds to virus strain name (according to GenBank nomenclature) followed by date and country of isolation. The tree was rooted by using a sequence belonging to the East Central South African lineage (LR2006_OPY1 2006 Reunion).
These cases are exemplary because they showed that the duration of presymptomatic viremia can be ≥12 hours and, at early stage, the CHIKV viral load can be the same order of magnitude as during the symptomatic stage. This conclusion supports previous suggestions that CHIKV viral loads peak at the time of clinical onset.9
Appassakij et al10 reported that, upon detection, the median viral load of symptomatic patients was 5.6 × 105 PFU/mL (1.3 × 105 in our patient) and that of asymptomatic patients was 3.4 × 103 PFU/mL (3.2 × 103 in our patient). Our test proved capable of detecting both, in agreement with the expected detection limit of our NAT (0.5 to 1 PFU/mL).
Proportions of symptomatic and asymptomatic patients were in agreement with our aforementioned modeling estimates and with previous observations.6 Therefore, in our study, postdonation reporting of febrile syndromes only would have accounted for detecting 2 of these 4 patients.
At the outset of CHIKV spread into the Americas, this study suggests that NAT together with postdonation self-reporting of fever and quarantine of blood products can play a key role for maintaining safe blood donation during an epidemic situation.
Authorship
Contribution: P.G., P.R., and L.P. collected the samples and performed the detection; G.P. and X.d.L. performed the complete genome sequencing and did the phylogenetic analysis; P.G., N.S., I.L.-G., and P.T. established the real-time polymerase chain reaction platform; and P.G., X.d.L., R.N.C., R.D., and J.C. designed the study, interpreted the data, and wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Remi N. Charrel, Unité Mixte de Recherche190 “Emergence des Pathologies Virales”, Faculte de Medecine, 27, Blvd Jean Moulin, 13005 Marseille, France; e-mail: remi.charrel@univ-amu.fr.