Key Points
Liver-targeted gene therapy for hemophilia can be improved by using computational promoter design in conjunction with hyperfunctional FIX.
Low and safe vector doses allow for stable supraphysiologic FIX that result in the induction of immune tolerance.
Abstract
The development of the next-generation gene therapy vectors for hemophilia requires using lower and thus potentially safer vector doses and augmenting their therapeutic efficacy. We have identified hepatocyte-specific transcriptional cis-regulatory modules (CRMs) by using a computational strategy that increased factor IX (FIX) levels 11- to 15-fold. Vector efficacy could be enhanced by combining these hepatocyte-specific CRMs with a synthetic codon-optimized hyperfunctional FIX-R338LPadua transgene. This Padua mutation boosted FIX activity up to sevenfold, with no apparent increase in thrombotic risk. We then validated this combination approach using self-complementary adenoassociated virus serotype 9 (scAAV9) vectors in hemophilia B mice. This resulted in sustained supraphysiologic FIX activity (400%), correction of the bleeding diathesis at clinically relevant, low vector doses (5 × 1010 vector genomes [vg]/kg) that are considered safe in patients undergoing gene therapy. Moreover, immune tolerance could be induced that precluded induction of inhibitory antibodies to FIX upon immunization with recombinant FIX protein.
Introduction
Significant progress has recently been made toward the development of gene therapy for hemophilia B. Adenoassociated virus (AAV) vectors are among the most promising vectors for liver-directed gene therapy that are capable of achieving therapeutic factor IX (FIX) expression levels in patients suffering from severe hemophilia B.1,2 Nevertheless, there are still some issues related to the induction of AAV capsid-specific T-cell–mediated immune response against the AAV-transduced cells that need to be addressed.1-4 These inadvertent immune reactions curtailed long-term gene expression by eliminating the gene-modified cells and accounted for liver toxicity. Furthermore, the performance of these AAV vectors must be improved to achieve a bona fide cure.2 Consequently, there is a need to create the next-generation AAV vectors for liver-directed gene therapy that express higher FIX levels at lower vector doses, to the extent that stable physiologic levels of FIX can be attained, while preventing inadvertent AAV capsid-specific T-cell responses and liver toxicity. The availability of more potent vectors would also ease manufacturing needs. To increase the potency of AAV-FIX vectors, we explored the use of a bioinformatics algorithm that resulted in the identification of transcriptional cis-regulatory modules (CRMs) associated with robust hepatocyte-specific expression (M.K.C., M.Y.R., I. Petrus, P.D.B., S.S., D. Danso-Abeam, C. Le Guiner, G. Gernoux, O. Adjali, N.N., J. Willems, H.E., J. Matrai, M. Di Matteo, E.S.-K., B. Yan, A. Acosta-Sanchez, A. Meliani, G. Cherel, V. Blouin, O. Christophe, P. Moullier, F. Mingozzi, and T.V., manuscript submitted March 2013).5 These CRMs contained evolutionary conserved clusters of transcription factor binding-site motifs that confer high tissue-specific gene expression. We then combined these hepatocyte-specific CRMs (HS-CRMs) with a synthetic codon-optimized hyperfunctional FIX transgene (ie, Padua R338L) that conferred 15-fold higher expression and activity levels than its wild-type counterpart.6,7 This novel combination approach substantially reduced the dose requirement for reaching therapeutic efficacy and thus facilitates future scale-up and clinical translation.
Study design
Additional information can be found in supplemental Methods, available at the Blood Web site. Identification of the HS-CRM relied on computational design based on a modified distance difference matrix multidimensional scaling approach, as described elsewhere.5 Generation and initial characterization of the codon-optimized FIX (coFIX) with the hyperactivating Padua mutation (ie, coFIX-R338L) were described previously.7 The most robust HS-CRM (designated as HS-CRM8) was cloned upstream of a minimal transthyretin promoter (TTR) that drives the coFIX-R338L and was incorporated into a self-complementary AAV (scAAV) vector (kindly provided by Dr Srivastava, University of Florida College of Medicine).8 Alternatively, a green fluorescent protein (GFP) reporter gene was used to verify the tissue-specific expression patterns. The effect of the HS-CRM and the hyperactivating Padua R338L mutation was first tested by hydrodynamic transfection in normal mice. The scAAV serotype 9 (scAAV9)-HS-CRM8-TTR-co-human factor IX (hFIX)-R338L and scAAV9-HS-CRM8-TTR-co-hFIX vectors were subsequently produced and characterized, as described.9 Adult hemophilia B mice were injected intravenously at the indicated AAV vector doses (mice were kindly provided by Dr I. Verma, Dr L. Wang, and Dr M. Kay).10 D-dimer and FIX antigen levels were determined by enzyme-linked immunosorbent assay, and FIX activity was determined with a chromogenic assay (HYPHEN BioMed, Andresy, France). Phenotypic correction was assessed by tail clipping. Animal experiments were approved by the university’s ethics committee.
Results and discussion
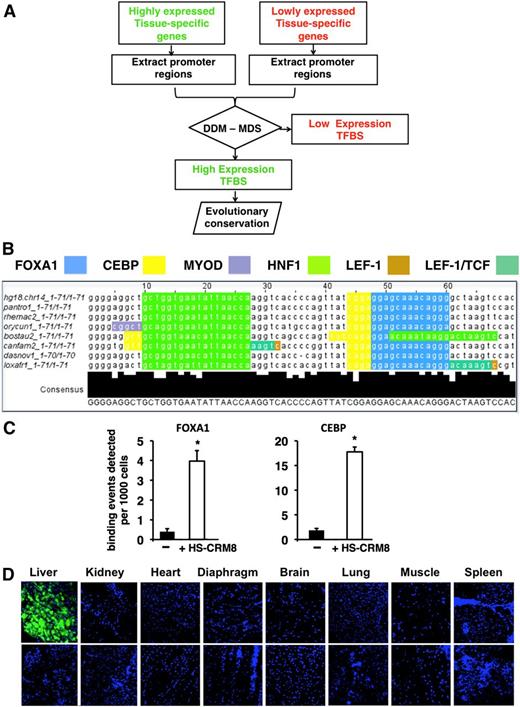
To improve the performance of AAV for liver-directed gene therapy, we explored a computational approach (Figure 1A) that led to the identification of an HS-CRM (designated as HS-CRM8)5 (Figure 1B) that was then cloned upstream of a strong liver-specific TTR promoter. This 72-bp HS-CRM8 element is derived from the human SERPINA1 gene and contains several putative transcription factor binding sites, including FOXA1, CEBP, HNF1, MyoD, LEF-1, LEF-1/TCF, that are strongly associated with robust liver-specific expression in vivo. By using a chromatin immunoprecipitation assay, we subsequently confirmed robust binding of FOXA1 and CEBP on the HSCRM8 element in livers from mice that were injected with AAV vectors containing HS-CRM8 (Figure 1C). By using GFP as reporter, we then assessed the tissue-specific expression pattern. We demonstrated by confocal microscopy that transgene expression was restricted to hepatocytes, whereas there was no detectable GFP expression in nonparenchymal cells (ie, Kupffer cells, sinusoidal endothelial cells) or in any other tissue (Figure 1D). This is consistent with the liver-specific GFP messenger RNA expression (supplemental Figure 1A-B), despite AAV9 transduction in nonhepatic tissues (supplemental Figure 1C).
Computational approach to identifying tissue-specific CRM. (A) The algorithm is based on the following steps: (1) identification of tissue-specific genes that are highly or lowly expressed based on statistical analysis of microarray expression data of normal human tissues; (2) extraction of the corresponding promoter sequences from publicly available databases; (3) mapping transcription factor binding sites (TFBSs) to these promoters by using the TRANSFAC database and identification of the tissue-specific CRM using a differential distance matrix (DDM)/multidimensional scaling (MDS) approach5 ; and (4) searching the genomic context of the highly expressed genes for evolutionary conserved CRM. (B) Evolutionary conservation, nucleotide sequence, and TFBSs located within the 71-bp HS-CRM8 element from human SERPINA1 identified by the aforementioned algorithm. The TFBSs include binding sites for FOXA1 (blue), CEBP (yellow), HNF1 (light green), MyoD (purple), LEF-1 (dark green), and LEF-1/TCF (brown). Some of these TFBSs are partially overlapping. (C) Chromatin immunoprecipitation assay confirming the binding of FOXA1 and CEBP on HS-CRM8. Antibodies specific to FOXA1 and CEBP and polymerase chain reaction (PCR) primers specific for the corresponding TFBS were used. In particular, PCR primers were designed to amplify a region within the vector corresponding to HS-CRM8 (that binds FOXA1 and CEBP), an untranscribed region on chromosome 6 was used as negative control (–). Binding events per 103 cells (mean + standard deviation) were determined for each of the corresponding primer pairs. Significant differences compared with the negative control were indicated (Student t test, *P ≤ .05). (D) Confocal microscopy of different organs of mice injected with AAV9-HS-CRM8-TTR-GFP (5 × 1011 vg/mouse; n = 4) with 4′,6 diamidino-2-phenylindole nuclear staining (top panels). A representative confocal scan is shown. Noninjected mice were used as controls (bottom panels). Pictures were taken at ×20 magnification.
Computational approach to identifying tissue-specific CRM. (A) The algorithm is based on the following steps: (1) identification of tissue-specific genes that are highly or lowly expressed based on statistical analysis of microarray expression data of normal human tissues; (2) extraction of the corresponding promoter sequences from publicly available databases; (3) mapping transcription factor binding sites (TFBSs) to these promoters by using the TRANSFAC database and identification of the tissue-specific CRM using a differential distance matrix (DDM)/multidimensional scaling (MDS) approach5 ; and (4) searching the genomic context of the highly expressed genes for evolutionary conserved CRM. (B) Evolutionary conservation, nucleotide sequence, and TFBSs located within the 71-bp HS-CRM8 element from human SERPINA1 identified by the aforementioned algorithm. The TFBSs include binding sites for FOXA1 (blue), CEBP (yellow), HNF1 (light green), MyoD (purple), LEF-1 (dark green), and LEF-1/TCF (brown). Some of these TFBSs are partially overlapping. (C) Chromatin immunoprecipitation assay confirming the binding of FOXA1 and CEBP on HS-CRM8. Antibodies specific to FOXA1 and CEBP and polymerase chain reaction (PCR) primers specific for the corresponding TFBS were used. In particular, PCR primers were designed to amplify a region within the vector corresponding to HS-CRM8 (that binds FOXA1 and CEBP), an untranscribed region on chromosome 6 was used as negative control (–). Binding events per 103 cells (mean + standard deviation) were determined for each of the corresponding primer pairs. Significant differences compared with the negative control were indicated (Student t test, *P ≤ .05). (D) Confocal microscopy of different organs of mice injected with AAV9-HS-CRM8-TTR-GFP (5 × 1011 vg/mouse; n = 4) with 4′,6 diamidino-2-phenylindole nuclear staining (top panels). A representative confocal scan is shown. Noninjected mice were used as controls (bottom panels). Pictures were taken at ×20 magnification.
We subsequently determined the impact of the HS-CRM8 element on the expression of a synthetic hyperfunctional codon-optimized coFIX-R338L in an scAAV backbone (Figure 2B). Hydrodynamic transfection of the pAAV-HS-CRM8-TTR-co-hFIX-R338L plasmid (all plasmids are abbreviated as p) (Figure 2B) resulted in a significant (2 μg, P < .01; 5 μg, P < .001) 11- to 15-fold dose-dependent increase in circulating FIX expression levels (Figure 2D-E) compared with the pAAV-TTR-co-hFIX-R338L (Figure 2A) control plasmid devoid of HS-CRM8. We then showed by hydrodynamic transfection of 2 microgram pAAV-HS-CRM8-TTR-co-hFIXR338L (Figure 2B) or pAAV-HS-CRM8-TTR-co-hFIX (Figure 2C) plasmids in FIX-deficient hemophilia B mice that the Padua R338L mutation resulted in a significant fivefold enhancement of FIX activity on both day 1 (P < .01) and day 2 (P < .001) (Figure 2F).
Vector design and functional validation.Schematic representation of pAAV-TTR-co-hFIX-R338L (A), pAAV-HS-CRM8-TTR-co-hFIX-R338L (B) and pAAV-HS-CRM8-TTR-co-hFIX (C) plasmids used in this study. The liver-specific minimal transthyretin (TTR) promoter drives the codon-optimized human FIX (co-hFIX) complementary DNA (cDNA) with or without the Padua R338L mutation (co-hFIX-R338L). The HS-CRM8 is located upstream of the TTR promoter. The minute virus of mouse (MVM) mini-intron and bovine growth hormone polyadenylation site (pA) are also indicated. The expression cassettes were cloned in an scAAV backbone, flanked by the 5′ and 3′ AAV inverted terminal repeats (ITRs), as indicated, and were used to generate the cognate scAAV9-HS-CRM8-TTR-co-hFIX-R338L and scAAV9-HS-CRM8-TTR-co-hFIX vectors. The effect of the HSCRM8 element was assessed by hydrodynamic transfection of the pAAV-HS-CRM8-TTR-co-hFIX-R338L (indicated as +HS-CRM8) (B) and control pAAV-TTR-cohFIX-R338L (indicated as –HS-CRM8) (A) plasmids in C57BL/6 mice at doses of 2 µg/mouse and 5 µg/mouse (D-E). FIX expression was measured by using a validated hFIX-specific enzyme-linked immunosorbent assay (ELISA) (n = 4) on plasma samples collected at day 1 or 2 posttransfection. Similarly, to assess the impact of the Padua R338L mutation, hemophilic mice were hydrodynamically transfected with pAAV-HS-CRM8-TTR-cohFIX-R338L (indicated as co-hFIX-R338L) compared with the pAAV-HS-CRM8-TTR-co-hFIX control (indicated as co-hFIX) (F). The clotting factor activity was measured by using a functional chromogenic FIX assay. Subsequently, we injected the cognate scAAV9-HS-CRM8-TTR-co-hFIX-R338L (designated as co-hFIX-R338L) (G-I) and scAAV9-HS-CRM8-TTR-co-hFIX (designated as co-hFIX) (J-L) in FIX-deficient hemophilic mice at a dose of 1 × 109 vg/mouse (5 × 1010 vg/kg), 5 × 109 vg/mouse (2.5 × 1011 vg/kg), and 2 × 1010 vg per mouse (1012 vg/kg) (n = 3 per group) (G-L). FIX activity and antigen levels were determined at the indicated times after AAV administration by using a chromogenic FIX activity assay and hFIX-specific ELISA, respectively. Results are presented as mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001 (Student t test); N.S., not significant (P > .1).
Vector design and functional validation.Schematic representation of pAAV-TTR-co-hFIX-R338L (A), pAAV-HS-CRM8-TTR-co-hFIX-R338L (B) and pAAV-HS-CRM8-TTR-co-hFIX (C) plasmids used in this study. The liver-specific minimal transthyretin (TTR) promoter drives the codon-optimized human FIX (co-hFIX) complementary DNA (cDNA) with or without the Padua R338L mutation (co-hFIX-R338L). The HS-CRM8 is located upstream of the TTR promoter. The minute virus of mouse (MVM) mini-intron and bovine growth hormone polyadenylation site (pA) are also indicated. The expression cassettes were cloned in an scAAV backbone, flanked by the 5′ and 3′ AAV inverted terminal repeats (ITRs), as indicated, and were used to generate the cognate scAAV9-HS-CRM8-TTR-co-hFIX-R338L and scAAV9-HS-CRM8-TTR-co-hFIX vectors. The effect of the HSCRM8 element was assessed by hydrodynamic transfection of the pAAV-HS-CRM8-TTR-co-hFIX-R338L (indicated as +HS-CRM8) (B) and control pAAV-TTR-cohFIX-R338L (indicated as –HS-CRM8) (A) plasmids in C57BL/6 mice at doses of 2 µg/mouse and 5 µg/mouse (D-E). FIX expression was measured by using a validated hFIX-specific enzyme-linked immunosorbent assay (ELISA) (n = 4) on plasma samples collected at day 1 or 2 posttransfection. Similarly, to assess the impact of the Padua R338L mutation, hemophilic mice were hydrodynamically transfected with pAAV-HS-CRM8-TTR-cohFIX-R338L (indicated as co-hFIX-R338L) compared with the pAAV-HS-CRM8-TTR-co-hFIX control (indicated as co-hFIX) (F). The clotting factor activity was measured by using a functional chromogenic FIX assay. Subsequently, we injected the cognate scAAV9-HS-CRM8-TTR-co-hFIX-R338L (designated as co-hFIX-R338L) (G-I) and scAAV9-HS-CRM8-TTR-co-hFIX (designated as co-hFIX) (J-L) in FIX-deficient hemophilic mice at a dose of 1 × 109 vg/mouse (5 × 1010 vg/kg), 5 × 109 vg/mouse (2.5 × 1011 vg/kg), and 2 × 1010 vg per mouse (1012 vg/kg) (n = 3 per group) (G-L). FIX activity and antigen levels were determined at the indicated times after AAV administration by using a chromogenic FIX activity assay and hFIX-specific ELISA, respectively. Results are presented as mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001 (Student t test); N.S., not significant (P > .1).
Subsequently, we produced the corresponding scAAV9-HS-CRM8-TTR-coFIX-R338L and assessed its performance in hemophilic FIX-deficient mice compared with the scAAV9-HS-CRM8-TTR-coFIX vectors at different vector doses corresponding to 1 × 109 vg/mouse (5 × 1010 vg/kg), 5 × 109 vg/mouse (2.5 × 1011 vg/kg), and 2 × 1010 vg/mouse (1012 vg/kg) (Figure 2G-L). AAV9 was chosen because of its hepatotropic properties and reduced sero-prevalence compared with AAV2 in human patients.8,11 Although AAV9 can cross the blood-brain barrier, much higher vector doses are required.12 This is consistent with the low transduction in the brain after AAV9-HS-CRM8-TTR-GFP transduction (supplemental Figure 2C). Sustained supraphysiologic FIX levels can be attained in a dose-dependent manner. Although the FIX antigen levels were comparable between both groups of mice, a significant increase in FIX activity was apparent in those mice treated with the co-hFIX-R338L transgene compared with the co-hFIX control (Figure 2G-L). Similarly, the hyperfunctional co-hFIX-R338L resulted in a significant five- to sevenfold increased FIX activity over protein levels at all vector doses in those mice injected with scAAV9-HS-CRM8-TTR-co-hFIX-R338L (Figure 2G-I). In contrast, an activity:antigen ratio equal to 1 was observed for the co-hFIX control at all time points and vector doses (Figure 2J-L).
This is consistent with our previous results using integration-competent and integration-defective lentiviral vectors7 and with the increased FIX activity after muscle-directed AAV transduction in mouse and dog models.13 Most importantly, even at the lowest vector dose tested (5 × 1010 vg/kg), it was possible to obtain supraphysiologic FIX levels that corrected the bleeding diathesis (supplemental Figure 2A). These FIX activity levels represent a robust improvement compared with the state-of-the-art AAV-FIX vectors, including those used in the most recent scAAV8-based clinical trial.2,14 hFIX messenger RNA expression was confined to the liver but not in other organs and tissues in mice injected with either scAAV9-HS-CRM8-TTR-co-hFIX-R338L or scAAV9-HS-CRM8-TTR-co-hFIX (supplemental Figure 2B), consistent with the GFP expression pattern (Figure 1). Transduction efficiency by quantitative polymerase chain reaction confirmed the predominant hepatotropic properties of AAV9 at the vector doses tested (109 vg scAAV9-HS-CRM8-TTR-co-hFIX-R338L: 9.2 × 103 copies per 100 ng genomic liver DNA; scAAV9-HS-CRM8-TTR-co-hFIX: 4.7 × 103 copies per 100 ng genomic liver DNA; P > .1; not significant).
To assess the immune consequences of expressing the hyperfunctional FIX Padua protein at high levels, we analyzed the anti-FIX antibody response before and after active immunization with wild-type FIX protein and Freund's incomplete adjuvant. The results show that immune tolerance could be achieved because none of the mice treated with the scAAV9-HS-CRM8-TTR-coFIX-R338L vectors developed anti-FIX antibodies in contrast to the controls that were not treated with this vector (supplemental Figure 2C). This implies that hepatocyte-specific expression of the hyperfunctional FIX Padua does not increase the risk of antibody development but instead enables induction of FIX-specific immune tolerance. Previous studies have already shown that the success rates of immune tolerance induction to FIX antigen in hemophilia B mice correlates with the FIX transgene expression levels and that these higher expression levels may favor induction of regulatory T cells.15 Consequently, since higher FIX levels could be attained with the improved scAAV9-HS-CRM8-TTR-coFIX-R338L vector compared with the vectors containing the unmodified coFIX or the construct without CRM8, it can thus be inferred that the new gene construct is better suited for the induction of immune tolerance. To estimate the possible thrombotic risks associated with expression of hyperfunctional FIX, we determined d-dimer levels as a measure of fibrin degradation16 but detected no significant increase, even in those mice that expressed 3000% hyperfunctional FIX (supplemental Figure 3D), at least in the short term.
Collectively, our data indicate that the combination of computational vector design and the use of synthetic hyperactive coFIX-R338L represents a promising strategy for improving the efficacy of hemophilia B gene therapy by using AAV vectors, which will allow the use of lower and thus safer vector doses in patients suffering from hemophilia. A phase 1/2 clinical trial is currently underway to begin safety, efficacy, and optimal dose testing of an scAAV8 vector containing the Padua FIX gene, but preclinical and clinical data are not yet available.17
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Srivastava for the self-complementary AAV backbone and Dr Verma, Dr Wang, and Dr Kay for the hemophilic mice.
This work was supported by grants from European Union 7th Framework Programme (222878, PERSIST), Bayer-Schering, Fonds Wetenschappelijk Onderzoek, European Hematology Association, Association française contre les myopathies, Geconcerteerde Onderzoeks Actie (GOA: EPIGEN), Willy Gepts Fonds, Strategic Research Programme-Grower Grant (SRP: GeneFIX), Industrieel Onderzoeksfonds-Groups of Expertise in Applied Research (IOF GEAR: GeneCure) and Industrieel Onderzoeksfonds-Proof of Concept grant (IOF PoC) (T.V. and M.C.). We thank L. Batota for technical assistance.
Authorship
Contribution: N.N. designed and performed experiments, analyzed data, and wrote the paper; M.Y.R., H.E., S.S., S.D., E.S.K., and O.G. performed experiments and analyzed data; P.D.B. conducted the bioinformatics analysis; B.T. and H.M.V. performed confirmatory experiments and analyzed data; and M.K.C. and T.V. designed experiments, coordinated the work, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thierry VandenDriessche, Free University of Brussels, Faculty of Medicine and Pharmacy, Department of Gene Therapy and Regenerative Medicine, Laarbeeklaan 103, 1090 Brussels, Belgium; e-mail: thierry.vandendriessche@vub.ac.be; Marinee K. L. Chuah, Free University of Brussels, Faculty of Medicine and Pharmacy, Department of Gene Therapy and Regenerative Medicine, Laarbeeklaan 103, 1090 Brussels, Belgium; e-mail: marinee.chuah@vub.ac.be.
References
Author notes
N.N. and M.Y.R. contributed equally to this study.
T.V. and M.K.C. were senior authors.