In this issue of Blood, Simon and colleagues describe the human platelet RNA profile in 154 unrelated individuals and present the transcriptome variability related to their age and gender.1
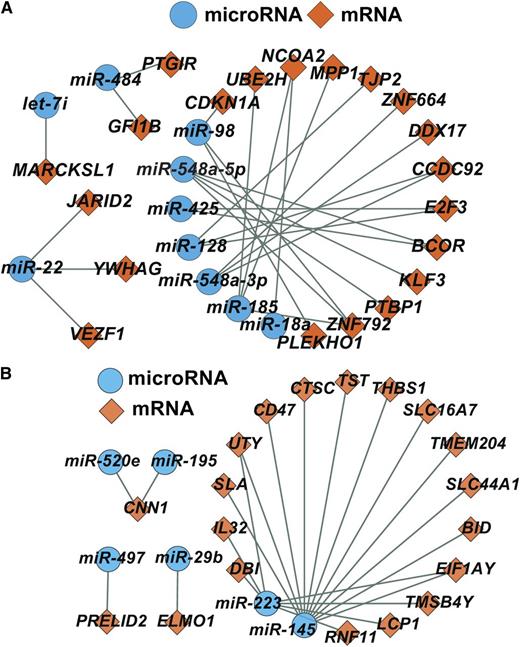
Networks of microRNA and mRNA differentially expressed by age (A) and gender (B). See Figures 3D and 4D in the article by Simon et al that begins on page e37.
Networks of microRNA and mRNA differentially expressed by age (A) and gender (B). See Figures 3D and 4D in the article by Simon et al that begins on page e37.
Although it has been known since the late 1960s that platelets are capable of synthesizing proteins, over the last 30 years, the hematology community has seen the concept of platelet “vestigial” RNA evolved into the description of more complex RNA networks that may regulate megakaryopoiesis and platelet function. Since the experiments by Newman et al in 1988 that showed the presence of messenger RNA (mRNA) in platelets that could be amplified and sequenced, several groups continued to show that platelets synthesize proteins.2,3 More recently, in a seminal publication, Denis et al reported the presence of spliceosomes that regulate synthesis of IL-1β in activated platelets by splicing pre-mRNA, demonstrating that mRNA processing occurs in platelets even without a nucleus.4
At the same time, platelet responsiveness to agonists has been shown to differ in humans of different races, ages, and genders, and these differences might be important not only to determine risk for cardiovascular disease but also to predict adequate response to antiplatelet agents. For example, Knight et al showed age-related differences in platelet fibrinogen binding, αIIbβ3, and P-selectin expression.5 Moreover, variability in the adenosine 5′-diphosphate (ADP) receptor P2Y12 reactivity may lead to clopidogrel hyporesponsiveness.6 The genetic molecular mechanism for these differences is poorly understood; however, it is clear that certain platelet phenotypes are heritable therefore understanding the RNA profile of platelets may lead to important discoveries on how these phenotypes are regulated.
Since 2005, the Bray laboratory has meticulously phenotyped platelet responses to various agonists in healthy individuals.7 This has allowed them not only to understand the degree of variability of the phenotypes but also to obtain quantifiable traits that can be associated with specific differentially expressed RNA transcripts. This group established the Platelet RNA and eXpression-1 (PRAX1) Study that measured platelet aggregation in response to arachidonic acid, ADP, protease activated receptor 1–activating peptide (PAR1-AP), and PAR4-AP, as well as mRNA and microRNA (miRNA) levels in 154 healthy subjects. They recently showed increased PAR4-induced platelet aggregation and calcium mobilization in blacks when compared with white healthy subjects. Among several differentially expressed mRNAs, they established a direct correlation between increased PAR4-induced aggregation and elevated phosphatidylcholine transfer protein (PC-TP) mRNA levels as well as an inverse correlation with levels of miR-376c, an miRNA that targets PCTP, the gene that encodes for the PC-TP. They further showed that miR-376c regulates expression of PC-TP protein in human megakaryocytes, suggesting a direct link between these RNA networks and platelet hyperreactivity in blacks.8
In this issue, Simon et al share additional important information from the PRAX1 study, demonstrating age and gender differences in miRNA and mRNA in 7771 genes expressed in human platelets. Perhaps not surprisingly, they found several mRNAs and miRNAs differentially expressed by age and gender. Several of these mRNAs are known targets of miRNAs, and an inverse relationship between these miRNA and mRNA levels suggests that miRNAs are regulating mRNA expression in platelets. Therefore, the investigators were able to determine specific miRNA-mRNA networks that deserve further exploration with platelet phenotypes. A network of how these miRNAs and mRNAs may interact is shown in the figure.
Although this particular manuscript does not show association with specific platelet activity data, the authors have made all information publically accessible by creating a user-friendly interactive web tool (www.plateletomics.com) that allows investigators to search for specific genes of interest. Once the gene name is entered, several features including relative abundance of the transcript as well as demographic and functional associations are displayed. A multidimensional relationship tab allows for associations between all possible variables.
These data will be invaluable as we learn more about differences in drug response with demographic variability, and differences in mRNA and miRNA expression in disease. It will also serve as a model for evaluation of differential expression of mRNA and miRNA in other human tissues.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal