Key Points
Targeted mutation of a zebrafish fibrinogen gene leads to a bleeding phenotype, analogous to human congenital afibrinogenemia.
This first heritable coagulopathy model validates the use of zebrafish for thrombosis and hemostasis research.
Abstract
Mutations in the human fibrinogen genes can lead to the absence of circulating fibrinogen and cause congenital afibrinogenemia. This rare bleeding disorder is associated with a variable phenotype, which may be influenced by environment and genotype. Here, we present a zebrafish model of afibrinogenemia. We introduced targeted mutations into the zebrafish fga gene using zinc finger nuclease technology. Animals carrying 3 distinct frameshift mutations in fga were raised and bred to produce homozygous mutants. Using a panel of anti-zebrafish fibrinogen antibodies, fibrinogen was undetectable in plasma preparations from homozygous mutant fish. We observed hemorrhaging in fga mutants and reduced survival compared with control animals. This model will now serve in the search for afibrinogenemia modifying genes or agents and, to our knowledge, is the first transmissible zebrafish model of a defined human bleeding disorder.
Introduction
Afibrinogenemia is defined by undetectable circulatory fibrinogen that can lead to a variety of bleeding and thrombotic complications for affected individuals.1 The soluble precursor to fibrin, fibrinogen is a conserved hexameric protein with its 3 polypeptide chains, Aα, Bβ, and γ, encoded by 3 genes: FGA, FGB, and FGG in humans. Minor isoforms of Aα (Aα-E) and γ (γ′) arise by differential messenger RNA (mRNA) splicing. Mutations in one of the fibrinogen genes can cause afibrinogenemia, but causal mutations have been found predominantly in FGA.2 Afibrinogenemia is rare, affecting about 1 in 106 individuals, and can be treated by replacement therapy. However, as with other heritable bleeding disorders, the variable phenotype of afibrinogenemia is not fully understood, and an experimental system where genetic and environmental influences on phenotype could be assessed would be a useful research tool.
While targeted disruption of Fga in mice clearly leads to an afibrinogenemia-like phenotype,3 we sought to produce a zebrafish model of afibrinogenemia. The zebrafish (Danio rerio) is used as a model vertebrate in developmental biology and for modeling human disease.4 Coagulation factors have been described in zebrafish,5 their genes are identifiable in the zebrafish genome,6 and hemostasis assays have been adapted to the model.7 With large numbers of offspring and rapid external early development, the zebrafish is compatible with genetic or chemical screening approaches, such as searching for phenotypic modifier genes or therapeutic agents.8,9 In addition, we anticipated that female zebrafish lacking circulating fibrinogen may not incur the lethality seen in gestating afibrinogenemic mice.3
We previously characterized the zebrafish fibrinogen genes and their expression during development.10 Furthermore, a transitory model of low fibrinogen levels in zebrafish larvae was recently reported using antisense morpholino oligonucleotides targeting fibrinogen mRNAs.11 However, no evidence for lowered fibrinogen protein levels was provided. A zebrafish model of a congenital human bleeding disorder, with a stable genetic change linked to a relevant phenotype, has not been described. Here, we present a zebrafish model of congenital afibrinogenemia. We engineered mutations into the zebrafish fga gene that lead to afibrinogenemic fish with a bleeding phenotype.
Study design
Zebrafish
AB strain zebrafish were maintained at 28°C, pH 7.5, and 500 µS conductivity. Experimentation was authorized by the local veterinary authority.
fga mutants
Plasmids encoding zinc finger nucleases (ZFNs) designed to target the fga gene were purchased from Sigma-Aldrich. In vitro transcribed mRNAs encoding each half of the dimeric ZFN were microinjected into early zebrafish embryos. Surviving fish were raised to reproductive age and screened for transmission of mutated fga by polymerase chain reaction (PCR)-genotyping embryos from natural crosses. The ZFN used to generate the mutations described, targets a sequence that overlaps a BceAI restriction site in fga exon 2 (Figure 1A). This enabled identification of mutated fga by BceAI digestion of PCR products. Oligonucleotides used were forward: 5′-AATGGCCTATGTTGGCAGAC-3′ and reverse: 5′-CAGTGGTTATCAGCTGACAG-3′. BceAI-resistant PCR products were sequenced, and 2 fish transmitting predicted null alleles were crossed with wild-type fish and progeny raised and screened for heterozygosity. Heterozygotes were in-crossed. We identified heterozygous animals from this subsequent generation, for each mutation, because 1 line had transmitted 2 mutations, presumably from separate ZFN-directed editing events in the founder animal. Heterozygotes for each of the 3 mutations were confirmed by sequencing cloned genotyping PCR products (pCRII TOPO; Life technologies). Subsequent in-crosses gave wild-type, heterozygous, and homozygous fish. Zebrafish experimentation was authorized by the local veterinary authority.
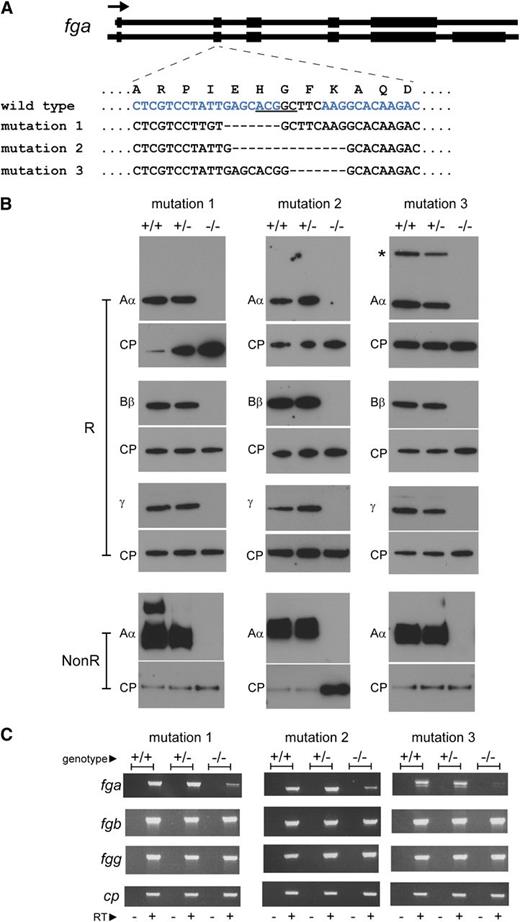
Generation of afibrinogenemic zebrafish. Mutations were introduced in exon 2 of the zebrafish fga gene by ZFN-mediated editing. (A) The mutations in zebrafish lines described are represented. The exon-intron composition of the 2 zebrafish fga transcripts, encoding fibrinogen Aα (upper) and Aα-E (lower), are sketched at the top of the panel. Coding regions are broad, noncoding parts of the transcripts and introns are narrow. The arrow represents the transcriptional orientation. The mutated region of exon 2 is zoomed, showing the local single-letter amino acid sequence in the wild-type Aα or Aα-E above the nucleotide sequence within exon 2 for wild-type and mutant zebrafish described. The underlined nucleotides in the wild-type sequence are a BceAI recognition sequence, disrupted in the mutants described and used for genotyping. The blue letters represent the position of the designed interaction site for the ZFN used; only one DNA strand is shown for clarity. Hyphens represent deleted nucleotides. Mutation 1 is a 10-nucleotide deletion and 3-nucleotide insertion, mutation 2 a 14-nucleotide deletion, and mutation 3 a 7-nucleotide deletion. (B) Reduced (R) plasma preparations from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mutant fish were subjected to western blotting using antibodies to zebrafish fibrinogen Aα, Bβ, and γ chains and ceruloplasmin (CP) as a loading control for each blot. A second Aα blot was made with nonreduced samples to detect the fibrinogen hexamer (NonR). The additional high-molecular weight band in the mutation 1 wild-type (+/+) nonreduced Aα blot is thought to be a small amount of fibrin polymer that may have rapidly formed upon tail sectioning prior to immersion in heparin solution. Fibrinogen was not detected in any of the homozygous mutant fish plasma samples (−/−). The additional higher molecular weight anti-Aα reactive band in the mutation 3 +/+ and +/− samples, marked with an asterisk, is thought to be the Aα-E isoform. Its absence in mutation 1 and 2 +/+ and +/− samples is due to a polymorphism seen on the nonmutated fga allele in these lines that is predicted to truncate the Aα-E isoform (supplemental Data). (C) RT-PCR products run on agarose gels are shown for fga, fgb, fgg, and ceruloplasmin (cp) cDNA in wild-type (+/+), heterozygous (+/−), and homozygous mutant fish (−/−) liver samples for each zebrafish line. The − or + under each lane denotes PCR reactions made on cDNA synthesis reactions without (−) or with (+) reverse transcriptase.
Generation of afibrinogenemic zebrafish. Mutations were introduced in exon 2 of the zebrafish fga gene by ZFN-mediated editing. (A) The mutations in zebrafish lines described are represented. The exon-intron composition of the 2 zebrafish fga transcripts, encoding fibrinogen Aα (upper) and Aα-E (lower), are sketched at the top of the panel. Coding regions are broad, noncoding parts of the transcripts and introns are narrow. The arrow represents the transcriptional orientation. The mutated region of exon 2 is zoomed, showing the local single-letter amino acid sequence in the wild-type Aα or Aα-E above the nucleotide sequence within exon 2 for wild-type and mutant zebrafish described. The underlined nucleotides in the wild-type sequence are a BceAI recognition sequence, disrupted in the mutants described and used for genotyping. The blue letters represent the position of the designed interaction site for the ZFN used; only one DNA strand is shown for clarity. Hyphens represent deleted nucleotides. Mutation 1 is a 10-nucleotide deletion and 3-nucleotide insertion, mutation 2 a 14-nucleotide deletion, and mutation 3 a 7-nucleotide deletion. (B) Reduced (R) plasma preparations from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mutant fish were subjected to western blotting using antibodies to zebrafish fibrinogen Aα, Bβ, and γ chains and ceruloplasmin (CP) as a loading control for each blot. A second Aα blot was made with nonreduced samples to detect the fibrinogen hexamer (NonR). The additional high-molecular weight band in the mutation 1 wild-type (+/+) nonreduced Aα blot is thought to be a small amount of fibrin polymer that may have rapidly formed upon tail sectioning prior to immersion in heparin solution. Fibrinogen was not detected in any of the homozygous mutant fish plasma samples (−/−). The additional higher molecular weight anti-Aα reactive band in the mutation 3 +/+ and +/− samples, marked with an asterisk, is thought to be the Aα-E isoform. Its absence in mutation 1 and 2 +/+ and +/− samples is due to a polymorphism seen on the nonmutated fga allele in these lines that is predicted to truncate the Aα-E isoform (supplemental Data). (C) RT-PCR products run on agarose gels are shown for fga, fgb, fgg, and ceruloplasmin (cp) cDNA in wild-type (+/+), heterozygous (+/−), and homozygous mutant fish (−/−) liver samples for each zebrafish line. The − or + under each lane denotes PCR reactions made on cDNA synthesis reactions without (−) or with (+) reverse transcriptase.
Detection of larval bleeding
Plasma collection and western blotting
Plasma was prepared from adult fish using a described protocol as a guide.14 Blood was collected into 300 µL of heparin solution (420 IU/mL) after tail sectioning of anesthetized fish. Samples of dilute plasma were subjected to western blotting. Antibodies were prepared by immunization of rabbits with peptides found in each of the zebrafish fibrinogen chains (Covalab, France). Peptide sequences, their positions in each fibrinogen chain and associated UniProtKB accession numbers were as follows: Aα IEHGFKAQDTCQTKEW (amino acids 33-48) E9QCD1, Bβ GKAVQEKEEQPESGGC (amino acids 75-90) Q6NYE1, γ (amino acids 50-65): DYLQRYKPDMDKKLDD (50-65) Q7ZVG7. Anti-zebrafish ceruloplasmin antibodies were purchased from Eurogentec.
RNA isolation and RT-PCR
RNA was isolated from adult liver samples using TRIzol reagent (Life Technologies) and reverse transcription-PCR (RT-PCR) performed as previously described using unchanged oligonucleotides.10
Results and discussion
Generating fga mutant zebrafish
The fibrinogen hexamer is assembled from 3 polypeptide chains; severe truncation of one chain is predicted to cause the absence of circulating fibrinogen. Exon 2 of the zebrafish fga gene, encoding fibrinogen Aα (and Aα-E), was targeted for mutation using ZFN technology15 (Figure 1A). Fish subjected to the ZFN were raised, and animals transmitting mutations predicted to give null alleles were out-crossed. Three mutation-transmitting lines were maintained (Figure 1A). The mRNA resulting from each modification is predicted to have an open reading frame shift and encode a nonfunctional truncated fibrinogen Aα (supplemental Data available at the Blood Web site). A generation from mating heterozygotes of each mutant line was genotyped aged 4-5 months; in total, 236 fish were analyzed (Figure 2A).
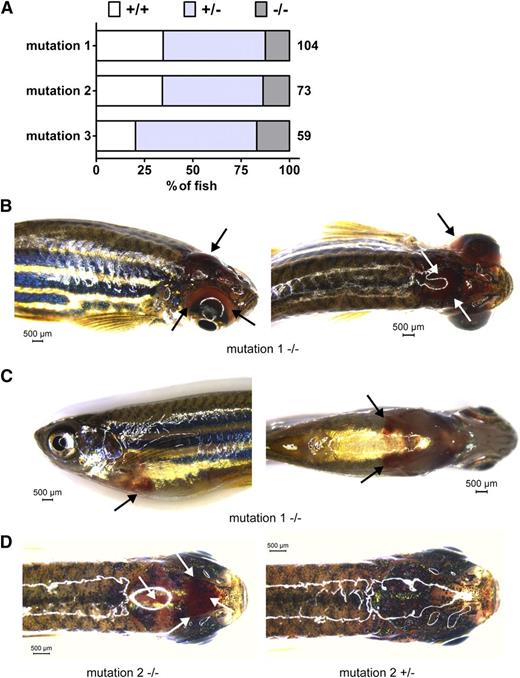
Survival and bleeding of afibrinogenemic zebrafish. A generation of adult zebrafish from mating of heterozygous animals from each mutant line was genotyped for exon 2 of the fga gene aged 4 to 5 months. The percentage of fish for each fga genotype, wild-type (+/+), heterozygous (+/−), and homozygous mutant fish (−/−) is represented in A. The total number of live animals genotyped for each line in this generation is shown to the right of each bar. Compared with an expected Mendelian ratio, homozygous mutant fish (−/−) are underrepresented and wild-type (+/+) fish overrepresented in animals from heterozygous mutation 1 and 2 crosses. In B-D, unprompted bleeding events from mutation 1 and 2 fish are represented. Black and white arrows highlight sites of bleeding. B shows a cephalic and ocular bleed with right side (left) and dorsal views (right). C shows a ventral hemorrhage with left side (left) and ventral views (right), and D shows a dorsal view of a cephalic bleed (left) with a nonbleeding fish for comparison (right). Scale bars are given in each image. Images were acquired using a Leica MZ16FA stereomicroscope equipped with a DFC420 digital camera and the Leica Application Suite software V4.2. A ×1.0 Planapochromatic objective was used. The zoom magnification was between 0.71 and 1.48 with a visual magnification between 7.1 and 14.8. The numerical aperture was 0.020 (B-C) or 0.040 (D). Color was set to a saturation setting of 1.7 to 2.0 and γ at 0.99. Images were taken at room temperature.
Survival and bleeding of afibrinogenemic zebrafish. A generation of adult zebrafish from mating of heterozygous animals from each mutant line was genotyped for exon 2 of the fga gene aged 4 to 5 months. The percentage of fish for each fga genotype, wild-type (+/+), heterozygous (+/−), and homozygous mutant fish (−/−) is represented in A. The total number of live animals genotyped for each line in this generation is shown to the right of each bar. Compared with an expected Mendelian ratio, homozygous mutant fish (−/−) are underrepresented and wild-type (+/+) fish overrepresented in animals from heterozygous mutation 1 and 2 crosses. In B-D, unprompted bleeding events from mutation 1 and 2 fish are represented. Black and white arrows highlight sites of bleeding. B shows a cephalic and ocular bleed with right side (left) and dorsal views (right). C shows a ventral hemorrhage with left side (left) and ventral views (right), and D shows a dorsal view of a cephalic bleed (left) with a nonbleeding fish for comparison (right). Scale bars are given in each image. Images were acquired using a Leica MZ16FA stereomicroscope equipped with a DFC420 digital camera and the Leica Application Suite software V4.2. A ×1.0 Planapochromatic objective was used. The zoom magnification was between 0.71 and 1.48 with a visual magnification between 7.1 and 14.8. The numerical aperture was 0.020 (B-C) or 0.040 (D). Color was set to a saturation setting of 1.7 to 2.0 and γ at 0.99. Images were taken at room temperature.
fga mutants are afibrinogenemic
We collected blood samples from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) animals from each mutant line and subjected diluted plasma to western blotting using antibodies that recognize zebrafish fibrinogen Aα, Bβ, and γ chains and ceruloplasmin as a control plasma protein. No fibrinogen was detected in the blood of homozygous mutants (Figure 1B), demonstrating that the mutations described render zebrafish afibrinogenemic. We dissected liver tissue to analyze fibrinogen mRNA expression by RT-PCR. Expression of fga is reduced in −/− animals, presumably by nonsense-mediated decay (Figure 1C). DNA sequencing of the fga cDNA confirmed the presence of wild-type and mutated transcripts in +/+ and −/− animals for each mutant line (not shown).
Effects on survival
Compared with a Mendelian ratio of 0.25 +/+: 0.5 +/−: 0.25 −/− genotypes, −/− fish were underrepresented for mutations 1 and 2 in the live genotyped fish (Figure 2A). Thirteen of 104 fish were mutation 1 homozygous mutants compared with 36/104 wild types. Ten of 73 fish were mutation 2 homozygous mutants, compared with 25/73 wild types. This implies survival of only ∼40% of the homozygotes from heterozygous in-crossings. Concordant with this data, homozygous mutants were overrepresented in dead fish recovered from the mutation 1 and 2 groups (supplemental Data). This demonstrates reduced survival with 2 of the homozygous fga mutations, although the effect was not seen with mutation 3 in the generation described. Analysis of further generations will determine whether mutation 3 confers modified survival compared with mutations 1 and 2.
A bleeding phenotype
Criteria for a model of human afibrinogenemia include the absence of circulating fibrinogen (Figure 1B) and a variable bleeding phenotype. We sporadically observed bleeding in fish from the lines described and suspected catastrophic bleeding events to explain fatalities of mutant fish. Figure 2B-D shows examples of unprompted bleeding found in adult mutant fish. Images were taken prior to genotyping. These bleeding events were variable; cephalic (Figure 2B,D), ocular (Figure 2B), and ventral hemorrhaging (Figure 2C) are represented. Such incidents were observed in <10% of the expected number of mutation 1 and 2 homozygotes. However, if lethal bleeding events explain the underrepresentation of the −/− genotype in live adult fish, compared with +/+ siblings, such events occurred in ∼60% of mutation 1 and 2 homozygotes. We did not detect severe bleeding in live mutation 3 homozygous mutants in the generation described, in accordance with our survival data.
We looked for bleeding in 3- and 5-day-old mutant larvae by staining with o-dianisidine to detect heme. We observed pools of apparent blood extravasation in 3-day-old mutant larvae, but these events were rare and also seen in wild types (see supplemental Data for images and bleeding frequencies). Either larval mutants do not suffer from additional bleeding events compared with wild types, or with the numbers of larvae analyzed we have not detected it in the current study. Our data imply that severe bleeding as a phenotypic end point for further study will require large numbers of adult or larval afibrinogenemic zebrafish.
In summary, we describe zebrafish lacking circulating fibrinogen, presenting a bleeding phenotype and reduced survival. Unprompted bleeding was rare, but the phenotypic similarity with human congenital afibrinogenemia validates the use of the zebrafish to search for modifier genes or agents that by functional conservation could affect the human disease. Our findings also infer that the zebrafish can be used as a model for other congenital bleeding disorders.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by a Swiss National Science Foundation grant to M.N.-A. (reference: 31003A-134967).
Authorship
Contribution: R.J.F. designed the study, performed zebrafish experiments and wrote the paper; C.D.S. performed experiments; and M.N.-A. directed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marguerite Neerman-Arbez, Department of Genetic Medicine and Development, University of Geneva Medical Centre, 1, rue Michel-Servet, 1211 Geneva 4, Switzerland; e-mail: marguerite.neerman-Arbez@unige.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal