Key Points
C-terminal cystine knot monomers in VWF are highly elongated and form antiparallel dimers.
Three disulfides across the dimer interface flanked by the cystine knots in each monomer form a highly force-resistant structure.
Abstract
The C-terminal cystine knot (CK) (CTCK) domain in von Willebrand factor (VWF) mediates dimerization of proVWF in the endoplasmic reticulum and is essential for long multimers required for hemostatic function. The CTCK dimer crystal structure reveals highly elongated monomers with 2 β-ribbons and 4 intra-chain disulfides, including 3 in the CK. Dimerization buries an extensive interface of 1500 Å2 corresponding to 32% of the surface of each monomer and forms a super β-sheet and 3 inter-chain disulfides. The shape, dimensions, and N-terminal connections of the crystal structure agree perfectly with previous electron microscopic images of VWF dimeric bouquets with the CTCK dimer forming a down-curved base. The dimer interface is suited to resist hydrodynamic force and disulfide reduction. CKs in each monomer flank the 3 inter-chain disulfides, and their presence in β-structures with dense backbone hydrogen bonds creates a rigid, highly crosslinked interface. The structure reveals the basis for von Willebrand disease phenotypes and the fold and disulfide linkages for CTCK domains in diverse protein families involved in barrier function, eye and inner ear development, insect coagulation and innate immunity, axon guidance, and signaling in extracellular matrices.
Introduction
Von Willebrand factor (VWF) is a mosaic of domains with many types of binding sites that crosslinks platelets to one another and to the vessel wall in hemostasis and thrombosis.1,2 Long length is key to VWF’s function as a sensor for flow changes at sites of hemostasis. Polymerization is mediated by specialized N- and C-terminal domains. Dimerization through a C-terminal cystine knot (CK) (CTCK) domain3-6 occurs first in the endoplasmic reticulum and is followed later in biosynthesis by N-terminal linkage through the D3 domain as VWF dimers assemble into helical tubules in the Golgi and Weibel-Palade bodies.7
Domains with CKs contain a unique motif of 3 disulfide bonds.8 Two closely spaced cysteines on each of 2 polypeptide segments disulfide link to form a ring through which a third disulfide linking other polypeptide segments crosses. The CK is actually a motif and not a domain; CKs are found in domains including knottins and cytokines that are otherwise completely unrelated.9
CTCK domains were called C-terminal or CT domains by Bork,10 who found them at the C termini of the CCN (cysteine-rich 61, connective tissue growth factor, nephroblastoma overexpressed) family of regulatory matricellular proteins and the Slit family of proteins that regulate axon guidance.4 Bork11 recognized lesser sequence homology to domains at the C termini of VWF, some gel-forming mucins, hemolectin, and norrie disease protein (norrin).4 Eight of 10 or 11 cysteines were invariant in CT domains. Finally, Bork4,5 found still weaker homology, but conservation of the spacing of 6 of the cysteines, in CK cytokines such as transforming growth factor β (TGFβ).
Twenty years has passed since the CTCK domain family was identified4 and suggested to be structurally related to TGFβ.5 However, despite presence in a diverse group of functionally important proteins mutated in disease, the CTCK domain has been recalcitrant to structural elucidation. Early work on VWF showed that at least one inter-chain disulfide was present in the C-terminal region.3 Later chemical and mutational studies characterized the 11 cysteines present in the VWF CTCK domain.12 Intra-chain connections were defined for 8 cysteines, and among the 3 remaining cysteines, either 1 or 3 were proposed to mediate inter-chain dimerization. Here, we report the crystal structure of the VWF CTCK dimer. Our structure defines the principles for assembly of proteins with functions as diverse as hemostasis (VWF), barrier function (mucins), axonal guidance (Slit), and regulation of growth, migration, and differentiation (the CCN family and Norrie disease protein).
Methods
Constructs and cell culture
Human VWF CTCK (residues 2720-2813 of pre-proVWF) was expressed in vector ET8.13 VWF sequence was fused C-terminal to the N-terminal signal peptide, His6 tag, and 3C protease site (LEVLFQGP). HEK293S N-acetylglucosaminyl transferase I-deficient cells14 were transfected using polyethyleneimine.15 Single colonies were selected in Dulbecco’s modified Eagle's medium and 5% fetal bovine serum with 1 mg/mL G418.16 This study was conducted in accordance with the Declaration of Helsinki.
Characterization of the CTCK domain
CTCK was secreted as a disulfide-linked dimer (supplemental Figure 1, available on the Blood Web site). Secretion was greatly decreased if the His6 tag and 3C protease site were not included in the expression construct. Expression in supernatant was semiquantitatively measured by western blotting with anti-VWF polyclonal antibody (Delta Biolabs, Gilroy, CA). One N-glycosylation site in CTCK sequence was confirmed by endoglycosidase H (Roche) digestion (supplemental Figure 1).
Purification and crystallization
CTCK in square bottle suspension culture Ex-CELL 293 serum-free medium supernatant was purified using Ni-NTA (Qiagen) and Hitrap Q (GE Healthcare) chromatography as described for other C-terminal fragments of VWF, including buffers, pH, and salt gradients,13 except that Hitrap Q was substituted for mono Q. The ion exchange eluate was concentrated and exchanged into 20 mM BisTris, pH 6.2, 0.15 M NaCl, and 3 mM EDTA and digested with a mass ratio of 1:400 of endoglycosidase H: CTCK for 48 hours at 22°C. The digest was concentrated and loaded onto a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 20 mM sodium acetate, pH 5.5, 0.15 M NaCl. Fractions containing CTCK dimer (supplemental Figure 1B) were concentrated to 20 mg/mL. Material was crystallized with the N-terminal His6 tag intact; material with the tag removed did not yield crystals.
Single crystals were obtained with a 1:1 volume to volume ratio well solution of 0.1 M bicine, pH 8.2 to 8.3, and 1.3 to 1.4 M ZnSO4 at 20°C in hanging drops. Crystallization trays were moved to 4°C for cryoprotection. Saturated LiSO4 was mixed with 80, 60, 40, 20, and 0% volume to volume ratio well solution, and individual crystals in cryo loops were passed through drops of these solutions in the same order and then plunge frozen in liquid N2. Anomalous diffraction data at zinc peak (1.28237 Å) were collected at General Medical Sciences/Cancer ID23-B beamline (Advanced Photon Source, Argonne National Laboratory, Argonne, IL). To minimize radiation damage, the beam position was vectorially scanned along the crystal during data collection.
Heavy atom location, phasing, model building, and refinement
Data from 2 isomorphous crystals were indexed and integrated separately with XDS and scaled together with XSCALE.17 Friedel pairs were kept unmerged. PHENIX.AUTOSOL18 used HySS19 for heavy atom location, PHASER20 to calculate phases using all reflections to 3.28 Å, and RESOLVE for density modification.21 One Zn was located on the dyad axis between each asymmetric unit (supplemental Figure 2). The figure of merit for phasing was 0.245. The solvent content of 88% (supplemental Figure 3) is unusually high and enabled us to calculate with solvent flattening an experimentally phased map of unusually high quality, which clearly showed the polypeptide chain and disulfide bridges. Initial chain building was with Arp/warp.22 A portion of CTCK β-strands 1, 4, 5, and 6 was recognized and used to superimpose models of TGFβ and human chorionic gonadotropin to help guide chain building. The sequence-to-structure register was guided by: 1) density for the sidechains of Tyr-2733, 2749, 2760, and 2795 and the N-acetyl glucosamine residue attached to Asn-2790; 2) the 4 intra-monomer disulfides previously defined12 ; and 3) the number of residues separating these cysteines in the polypeptide chain backbone.
Model building was with COOT.23 Refinement with PHENIX24 against anomalous data (unmerged Friedel pairs) and Hendrickson-Lattman coefficients from PHASER included atomic coordinates, individual atomic displacement parameters, and 5 translation libration screw groups. Newer versions of PHENIX (we used dev-1426) include the overall B factor in the individual atomic B factors, which increases the B factors reported in the coordinate file by about 50. MolProbity was used for model validation.25
Electron microscopy (EM) image processing
Results
Crystal structure of CTCK monomer
The VWF CTCK domain structure was solved using zinc single-wavelength anomalous diffraction at 3.28 Å. The experimental electron density, which was unusually good for this resolution, revealed the complete polypeptide chain path and all disulfide bridges and allowed us to refine the structure to a low R free of 24.8% (Table 1). The entire CTCK domain and an N-acetyl glucosamine residue attached to residue Asn-2790 are resolved (Figure 1A). The asymmetric unit contains only one monomer. When viewing the structure with molecular graphics, the crystallographic dimer must be generated by adding a symmetry-related monomer.
Crystal diffraction data and refinement statistics
| Data collection . | . |
|---|---|
| Wavelength (Å) | 1.28237 |
| Resolution range (Å) | 47-3.28 (3.37-3.28)* |
| Space group | P4332 |
| Unit cell | a=b=c=135.34 Å, α=β=γ=90° |
| Solvent content (%) | 88 |
| Total unique reflections† | 12294 |
| Redundancy‡ | 13.7 (10.7) |
| Completeness (%) | 99.99 (99.85) |
| I/ σ(I) | 10.87 (0.43) |
| Rsym‡ | 0.228 (6.097) |
| CC1/2 (%)§ | 99.8 (12.0) |
| Phasing and refinement | |
| Phasing figure of merit | 0.245 |
| Rwork / Rfree|| | 0.224 / 0.248 |
| Monomer / asymmetric unit | 1 |
| Nonhydrogen atoms | |
| Protein/ N-acetyl glucosamine /Zn/SO4/water | 721/14/0.5/25/6 |
| RMSD bonds (Å) | 0.003 |
| RMSD angles (°) | 0.46 |
| Ramachandran plot¶ | |
| (% favored/allowed/outliers) | 94.6/5.4/0.0 |
| Geometry and clash percentiles¶ | 100% / 100% |
| Protein data bank ID | 4NT5 |
| Data collection . | . |
|---|---|
| Wavelength (Å) | 1.28237 |
| Resolution range (Å) | 47-3.28 (3.37-3.28)* |
| Space group | P4332 |
| Unit cell | a=b=c=135.34 Å, α=β=γ=90° |
| Solvent content (%) | 88 |
| Total unique reflections† | 12294 |
| Redundancy‡ | 13.7 (10.7) |
| Completeness (%) | 99.99 (99.85) |
| I/ σ(I) | 10.87 (0.43) |
| Rsym‡ | 0.228 (6.097) |
| CC1/2 (%)§ | 99.8 (12.0) |
| Phasing and refinement | |
| Phasing figure of merit | 0.245 |
| Rwork / Rfree|| | 0.224 / 0.248 |
| Monomer / asymmetric unit | 1 |
| Nonhydrogen atoms | |
| Protein/ N-acetyl glucosamine /Zn/SO4/water | 721/14/0.5/25/6 |
| RMSD bonds (Å) | 0.003 |
| RMSD angles (°) | 0.46 |
| Ramachandran plot¶ | |
| (% favored/allowed/outliers) | 94.6/5.4/0.0 |
| Geometry and clash percentiles¶ | 100% / 100% |
| Protein data bank ID | 4NT5 |
Statistics for the highest-resolution shell are shown in parentheses.
Friedel pairs are treated as separate reflections.
Rsym = ΣhklΣi|Ii−<I>|/ΣhklΣiIi, where Ii and <I> are the ith and mean measurement of the intensity of reflection hkl.
Pearson’s correlation coefficient between average intensities of random half-data sets of the measurements for each unique reflection.38
Rwork = Σhkl||Fobs|−|Fcalc||/|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is the crossvalidation R factor computed for the 8% (929) test set of unique reflections.
Ramachandran, geometry, and clash values were reported by MOLPROBITY.25
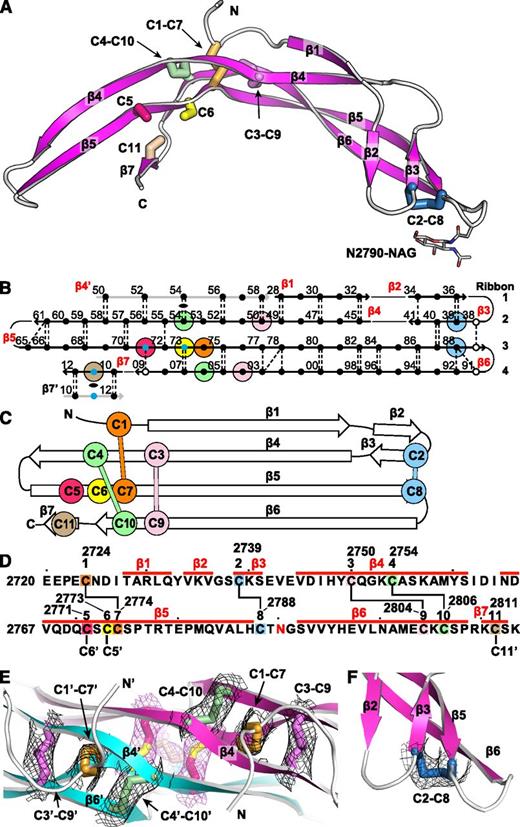
The VWF CTCK monomer crystal structure. Cysteines are colored consistently throughout the panels here and in other figures. (A) Ribbon diagram with cysteines shown in stick and the N-linked carbohydrate in thin stick. (B) β-Sheet diagram. Each backbone hydrogen bond is shown as a dashed line. Residues 2728 to 2812 are numbered as in pre-proVWF, except the first 2 digits are omitted for clarity. β4′ and β7′ from the other monomer are in gray. Residues in β-sheet framework and loops are shown in solid and open circles, respectively. Cysteines are surrounded with larger colored circles and those in inter-chain disulfides are also marked with small blue circles. The dyad axis at the β4 and β7 strands is marked with a lens. (C) Topology diagram. (D) Sequence, secondary structure, and disulfide connectivity. Cysteines are shown both in order C1 to C11 and with their residue numbers. (E-F) Experimental electron density after solvent flattening at disulfides. Density at 1σ around cysteine sidechains is shown as mesh in black around intra-chain disulfides and in purple around inter-chain disulfides. (E) The CKs in each monomer. (F) The C2-C8 disulfide.
The VWF CTCK monomer crystal structure. Cysteines are colored consistently throughout the panels here and in other figures. (A) Ribbon diagram with cysteines shown in stick and the N-linked carbohydrate in thin stick. (B) β-Sheet diagram. Each backbone hydrogen bond is shown as a dashed line. Residues 2728 to 2812 are numbered as in pre-proVWF, except the first 2 digits are omitted for clarity. β4′ and β7′ from the other monomer are in gray. Residues in β-sheet framework and loops are shown in solid and open circles, respectively. Cysteines are surrounded with larger colored circles and those in inter-chain disulfides are also marked with small blue circles. The dyad axis at the β4 and β7 strands is marked with a lens. (C) Topology diagram. (D) Sequence, secondary structure, and disulfide connectivity. Cysteines are shown both in order C1 to C11 and with their residue numbers. (E-F) Experimental electron density after solvent flattening at disulfides. Density at 1σ around cysteine sidechains is shown as mesh in black around intra-chain disulfides and in purple around inter-chain disulfides. (E) The CKs in each monomer. (F) The C2-C8 disulfide.
The CTCK monomer adopts a highly elongated β-strand structure with dimensions of 6 × 3 × 2 nm (Figure 1A). The monomer is formed by 4 ribbons that alternate in direction as they go from one end of the monomer to the other. Ribbons (Figure 1B, right) are defined as going in a single direction from one end of a monomer to another and may contain one or more β-strands. Ribbon 1 contains an N-terminal segment and the β1 and β2 strands, ribbon 2 contains the β3 and β4 strands, ribbon 3 is comprised only of the extremely long β5 strand, and ribbon 4 contains the β6 and β7 strands (Figure 1B). The 4 ribbons hydrogen bond into 2 β-ribbons. The 2 β-ribbons are also knit together by backbone hydrogen bonds of β3 and β4 with opposite ends of the β5 strand (Figure 1B). β-Ribbons contain only 2 antiparallel β-strands, and this building unit enables considerable twisting over the long lengths of the β4, β5, and β6 strands (Figure 1A).
Intra-chain disulfides
The experimental electron density defines the connectivity of 4 intra-chain disulfides (Figure 1E-F) and completely agrees with chemical determination.12 In the descriptions below and in the figures, we refer to the cysteines by their order in the CTCK domain, eg, C1 to C11, or by their pre-pro VWF sequence number, for example, Cys-2724 to Cys-2811 (Figure 1D).
The CK ties together the 4 ribbons at the middle of the long axis of the monomer (Figure 1A). Cys-2750 (C3) and Cys-2754 (C4) in ribbon 2 disulfide bond to Cys-2804 (C9) and Cys-2806 (C10), respectively, in ribbon 4 (Figure 1A-D). The C3-C9 and C4-C10 disulfides thus link β4 and β6, which run parallel to one another and do not hydrogen bond to one another. Linkage of C3 and C4, 5 residues apart in β4, to C9 and C10, 3 residues apart in β6, forms an 8-residue ring (Figure 1D). A third disulfide penetrates this ring, links Cys-2724 (C1) in ribbon 1 to Cys-2774 (C7) in ribbon 3 (Figure 1E), and thus secures the CK. The fourth intra-chain Cys-2739 to Cys-2788 (C2-C8) disulfide further links the 2 β-ribbons at their tips, distal from the dimer interface (see below).
Relation to CK cytokine monomers
Searches for structural homologs of the CTCK monomer using DALI26 confirmed the uniqueness of the CTCK domain and its relationship to CK cytokines (supplemental Figure 5). CK cytokines usually contain a CK and 2 β-ribbons. However, the paths taken by each β-ribbon, the length of the β-ribbons, hydrogen bonding between the 2 β-ribbons, and presence of α-helices are highly variable. The cytokine members of the family all dimerize, whereas others such as sclerostin are monomers. Some contain N-terminal prodomains such as TGFβ; however, the functional forms of all previously described structural relatives function with only a single domain in their monomers; ie, none are mosaic proteins. The CTCK domain has not been previously structurally characterized, in agreement with the absence of any of the mosaic proteins described by Bork4 in structural homology searches.
Dimerization of CTCK
Each CTCK monomer interacts across three-quarters of its long axis to form the CTCK dimer (Figure 2). The twisted β4/β5 ribbon extends deeply into the other monomer in the dimer and forms extensive main chain hydrogen bonds (Figure 1B) and hydrophobic interactions (Figure 3A). The buried interface is quite large, covers 32% of the surface (1520 Å2) of each monomer, and includes inter-chain disulfides, backbone β-sheet hydrogen bonds, and sidechain complementarity.
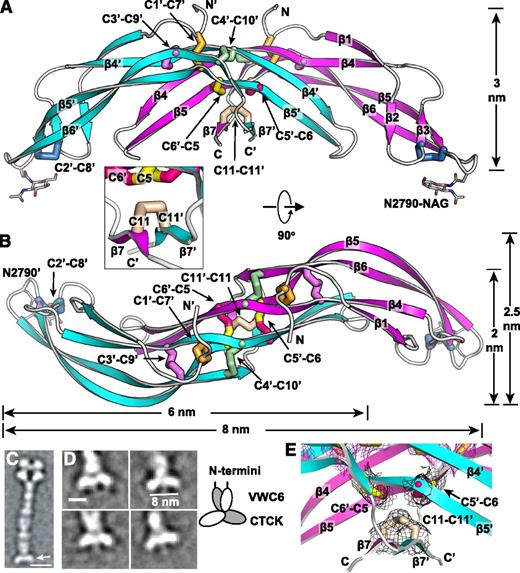
The VWF CTCK dimer crystal structure. (A-B) Two ribbon diagram views with each monomer in a different color and an inset showing another view of the β7 strands. Disulfides are shown in stick and colored as in Figure 1. (C-D) EM class averages with data from.13 (C) The VWF dimeric bouquet (arrow) in a C-terminal fragment extending from the A3 to the CTCK domain. (D) Class averages of the same fragment, recalculated with centering on the CTCK domain. Scale bars are 10 nm in A and 5 nm in D. A schematic interpretation is shown to right. (E) Experimental electron density around disulfides as in Figure 1E-F. Mesh is contoured at 1σ around cysteine sidechains and is shown in black for inter-chain disulfides and in purple for intra-chain disulfides.
The VWF CTCK dimer crystal structure. (A-B) Two ribbon diagram views with each monomer in a different color and an inset showing another view of the β7 strands. Disulfides are shown in stick and colored as in Figure 1. (C-D) EM class averages with data from.13 (C) The VWF dimeric bouquet (arrow) in a C-terminal fragment extending from the A3 to the CTCK domain. (D) Class averages of the same fragment, recalculated with centering on the CTCK domain. Scale bars are 10 nm in A and 5 nm in D. A schematic interpretation is shown to right. (E) Experimental electron density around disulfides as in Figure 1E-F. Mesh is contoured at 1σ around cysteine sidechains and is shown in black for inter-chain disulfides and in purple for intra-chain disulfides.
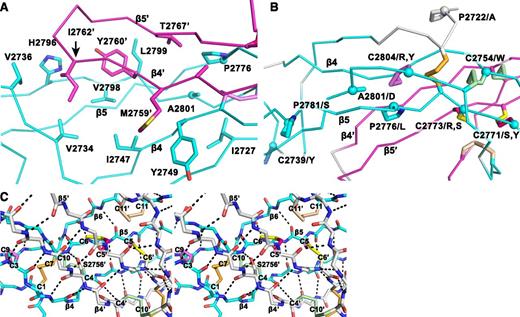
Structural details of the VWF CTCK dimer. (A) The hydrophobic pocket underlying the β4 and β5 strands and the β4-β5 loop in the dimer interface. Sidechains that interact across the interface are shown in stick. A2801 and P2776, involved in VWD mutations, are shown with sphere Cα atoms. (B) VWD mutations. Mutated residues are marked with Cα spheres and their native sidechains are shown in stick (some also appear in A). Disulfide bonds are shown, whether or not their cysteines are mutated in VWD. (C) Stereoview showing the high density of backbone hydrogen bonds (dashed) and disulfides in the dimerization interface. The view includes all inter-chain disulfides and the CK in each monomer.
Structural details of the VWF CTCK dimer. (A) The hydrophobic pocket underlying the β4 and β5 strands and the β4-β5 loop in the dimer interface. Sidechains that interact across the interface are shown in stick. A2801 and P2776, involved in VWD mutations, are shown with sphere Cα atoms. (B) VWD mutations. Mutated residues are marked with Cα spheres and their native sidechains are shown in stick (some also appear in A). Disulfide bonds are shown, whether or not their cysteines are mutated in VWD. (C) Stereoview showing the high density of backbone hydrogen bonds (dashed) and disulfides in the dimerization interface. The view includes all inter-chain disulfides and the CK in each monomer.
Backbone hydrogen bonds across the symmetry axis extend the β-ribbons. Long β4, β5, and β6 strands that run through the dimer interface link into a super β sheet, where β4 mates with β4′ in the other monomer across the dyad axis (Figure 1B). Residues contributed by the other monomer are shown in gray in Figure 1B. β4 strand residues 2750-2758 form antiparallel hydrogen bonds to residues 2750-2758 in β4′ (Figures 1B and 3A). Furthermore, a short β-ribbon is formed by β7, which interacts only with the symmetry-related β7′ strand (Figures 1B and 2A).
Despite the long and narrow shape of the dimer interface, it contains significant burial of hydrophobic residues. Where the edges of the β4 strands meet to hydrogen bond across the dimer interface, Met-2759 forms hydrophobic interactions to Tyr-2749 across the interface (Figure 3A). Continuing along, Tyr-2760 and Ile-2762 tuck the end of the β4- β5 loop securely into a hydrophobic cavity on the other monomer, and Val-2767 in the β5 strand tucks into the same cavity (Figure 3A).
The structure shows that the 3 previously unassigned cysteines, Cys-2771, Cys-2773, and Cys-2811, all form inter-chain disulfides (Figure 2A-B, E). Cys-2771 and 2773 each reside in β-strand 5 and form reciprocal C2771-C2773′ and C2773-C2771′ disulfides. These β5-β5 inter-chain disulfides complement the extensive intra-chain hydrogen bonds formed by β5 at the interface between the 2 β-ribbons in each monomer (Figure 1B). Cys-2811 is at the center of the short, 3-residue β7 strand, and disulfide bonds to its Cys-2811′ counterpart in the β7′ strand to further secure the C-terminus of the CTCK domain.
In a highly force-resistant structure, the 3 inter-chain disulfides are sandwiched between the 3 CK disulfides in each monomer (Figure 2B). Thus, 9 pairs of disulfides all lie within an ellipsoid only 2.5 nm long and 1 to 1.5 nm in diameter. Of these 18 cysteines, all but 2 lie within β-strands. Furthermore, Cys-2771 and Cys-2773 that form the reciprocal inter-chain β5- β5′ disulfides lie adjacent to Cys-2774 in β5, which forms the loop-penetrating CK disulfide. The density of disulfide and β-sheet backbone crosslinks in this region is most remarkable (shown in stereo in Figure 3C). Adding to this, Ser-2756 in the β4-strand forms an unusually strong 2.4 Å sidechain hydrogen bond to the backbone between Cys-2773 and Cys-2774 (Figure 3C). Notably, this highly crosslinked region lies in the center of the CTCK dimer immediately below the monomers’ N termini (Figure 2A), which bear all of the elongational force transmitted through VWF concatemers as a consequence of hydrodynamic flow.
Domains in the C-terminal portion of VWF dimers zip up into dimeric bouquet-like structure at acidic pH values found in the Golgi and Weibel-Palade bodies (Figure 2C).13 The CTCK dimer forms the slightly curved base at the C-terminal end of these dimers (Figure 2C arrow). The curved shape of the CTCK dimer crystal structure in the orientation shown in Figure 2A, and its long dimension of 8 nm, agree perfectly with its shape and dimension in negatively stained EM class averages (Figure 2D). Note that in the orientation of Figure 2A the N terminus of each CTCK monomer points vertically in an optimal orientation to connect to the C terminus of the VWC6 module in dimeric bouquets.
VWD mutations
VWD mutations in the CTCK domain can cause quantitative decrease in multimers (type 1), complete deficiency (type 3), or selective loss of longer multimers (type 2).27 In agreement with the importance of disulfide bonds to CTCK domain structure, 8 of 12 documented mutations27 occur in cysteines, and these cysteines contribute to all classes of disulfides, ie, the C2-C8 and CK intra-chain disulfides and the inter-chain disulfides (Figure 3B).
Interestingly, C2771S, C2771Y, C2773R, and C2773S are all type 2 mutations. These cysteines form the reciprocal Cys-2771-Cys-2773′ and Cys2771′-Cys-2773 inter-chain disulfides. Mutation of either of these cysteines has the interesting property of disrupting not 1 but 2 of the 3 inter-chain disulfides, explaining the type 2 phenotype of selective loss of longer VWF concatemers. In contrast, mutations of cysteines forming intra-chain disulfides, C2739Y, C2754W, C2804R, and C2804Y, cause complete deficiency of VWF (type 3).
Mutations of 4 non-cysteine residues, including 3 prolines, cause type 1 and 2 VWD. The A2801D mutation introduces an aspartic acid sidechain into the center of the hydrophobic core of the dimer interface that cradles the β4-β5 ribbon and its loop (Figure 3A-B). This mutation should not disrupt monomer structure and in agreement causes type 2 VWD. The P2776L mutation affects a Pro that is on the edge of the same hydrophobic cavity (Figure 3B) and helps mediate a twist in the β5-β6 ribbon, where its hydrogen bond pattern is disrupted (Figure 1B). P2776L causes type 1 VWD. P2772A is in the linker between the VWC6 and CTCK domains (Figure 3B). Only 4 residues, in a Glu-Glu-Pro-Glu sequence, intervene between the last Cys of VWC6 and first Cys of CTCK. P2772A is likely to affect the interaction between these domains and also causes type 1 VWD. Pro 2781, like Pro 2776, is in a region of the β5-β6 ribbon where hydrogen bonding is temporarily interrupted (Figure 1B). The P2781S mutation may disrupt this ribbon and causes type 2 VWD.
Discussion
Our crystal structure reveals the fold and mechanism of dimerization for the CTCK domain family.4 The structure provides insights into how the CTCK domain in VWF mediates formation of dimers in the endoplasmic reticulum. The dimerization interface is heavily reinforced with hydrogen bonds and 3 disulfide bonds and is sandwiched between the similarly heavily reinforced CK regions in each monomer. The inter-chain disulfide bond linkage is in agreement with 1 of 2 models previously proposed based on exclusion of intra-chain linked cysteines, structural models, mutation, and symmetry arguments.12 The structure explains VWD mutation phenotypes. Mutations of intra-chain linked cysteines cause complete VWF deficiency and mutations of inter-chain linked cysteines cause selective deficiency of large multimers. These findings suggest that intra-chain disulfide linkage in monomers precedes and is required for subsequent inter-chain linkage in dimers.
We propose that the highly crosslinked dimer interface in CTCK is a specialization for force resistance. Most investigators are familiar with the concept that DNA is sufficiently long for its covalent bonds to be broken in the shear flow of a pipette. Similarly, VWF is sufficiently long to have significant hydrodynamic force exerted on it. Even free in flow, the hydrodynamic force on VWF is sufficient to unfold the A2 domain.28 When bound to the vessel wall and platelets, the force on VWF would be much greater. An elementary principle of proteins, first demonstrated by Anfinsen,29 is that protein folding favors specific disulfide formation. Thus, disulfides that are buried in a protein core remain intact as disulfides even in the presence of reducing agents. However, reduction occurs when denaturants are added or tensile force is applied.30 In plasma, the concentrations of glutathione/oxidized glutathione and cysteine/cystine are 0.14/2.8 and 10/40 μM, respectively.31 When force is applied across a disulfide bond, the kinetics of its reduction greatly increase.30 Reduction of the inter-chain disulfides in the CTCK domain (or D3 domain) in a force-elongated VWF concatemer would have disastrous consequences, because the 2 half-molecules would separate in flow and have little chance of finding one another and reannealing after force subsided. In contrast, reduction of a long-range disulfide in an internal domain of VWF such as A1 could easily be subsequently reversed by refolding and oxidation.
We thus reemphasize the highly reinforced structure of the CTCK dimer. Ten inter-chain backbone hydrogen bonds link the β4 strands in each monomer to form a β6-β5-β4-β4′-β5′-β6′ supersheet, and 4 hydrogen bonds further link the β7 and β7′ strands. These interactions are enhanced by burial of each β4-β5 ribbon and its loop in a hydrophobic pocket in the other monomer. All but one of 6 CK cysteines, and all 3 cysteines that form inter-chain disulfides, are present in β-strands. This is important, because β-structures are rigid compared with α-helices and loops and thus better suited for force resistance. If a protein deforms in response to force, it is more readily elongated and unfolded. A term corresponding to compliance appears in an exponent along with force and determines how much force exponentiates the rate of unfolding.32 Finally, the β-structure in the inter-chain disulfide region is continuous with that in the CK region of each monomer, which immediately flanks each side of, and further reinforces, the inter-chain disulfide region. Having each looked at a large number of extracellular protein modules, we have never seen a domain of a soluble extracellular or membrane protein, let alone a dimer interface, so bristling with disulfide bonds and backbone hydrogen bonds (Figures 1B and 3C). The only other such heavily reinforced proteins that come to mind are incorporated into the extracellular matrix, such as the noncollagenous NC1 domain of collagen IV, which mediates collagen crosslinking into mechanically tough sheets.33
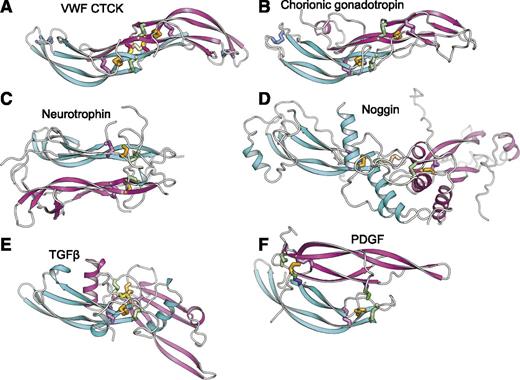
The uniqueness of the CTCK dimerization interface is further emphasized by comparison with CK cytokines (Figure 4). CK dimerization mechanisms are diverse with 5 different types of orientations (Figure 4B-F). Chorionic gonadotropin and follicle stimulating hormone dimerize with their monomers antiparallel, the same spacing between the CKs as in CTCK, and are the only other group where monomer β-ribbons form a dimeric super β-sheet as in CTCK (Figure 4B). However, this CK cytokine subfamily completely lacks inter-chain disulfides.
Diverse dimerization mechanisms among VWF CTCK and its relatives. Dimer structures are superimposed on the CTCK monomer in cyan and then aligned with it horizontally and vertically on the page. Disulfide bonds are shown in stick and colored as in Figure 1. (A) VWF CTCK. (B) Human chorionic gonadotropin (PDB code 1QFW). (C) Neurotrophin-4 (PDB 1HCF). (D) Noggin (PDB 1M4U). (E) TGFβ2 (PDB 1TFG). (F) PDGF (PDB 1PDG).
Diverse dimerization mechanisms among VWF CTCK and its relatives. Dimer structures are superimposed on the CTCK monomer in cyan and then aligned with it horizontally and vertically on the page. Disulfide bonds are shown in stick and colored as in Figure 1. (A) VWF CTCK. (B) Human chorionic gonadotropin (PDB code 1QFW). (C) Neurotrophin-4 (PDB 1HCF). (D) Noggin (PDB 1M4U). (E) TGFβ2 (PDB 1TFG). (F) PDGF (PDB 1PDG).
Members of the TGFβ superfamily dimerize in a similar orientation but have α-helices at the dimer interface (Figure 4E). There is one inter-chain disulfide, mediated by the equivalent of VWF Cys-2773. However, in the TGFβ family this cysteine dimerizes with the identical cysteine in the counterpart monomer (Figure 5) rather than to a cysteine 2 residues away in the same β-strand as in VWF. Therefore, the dimer interface slides in the TGFβ family, so the CKs in the 2 monomers are 2 β-ladder positions closer than in CTCK (Figure 4E).
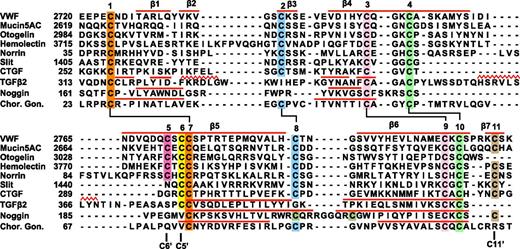
Sequence alignment of VWF CTCK to CTCK domains from other proteins and structural alignment to related proteins. Sequence alignment with MAFFT was with the G-INS-i strategy.39 Structural alignment was with SSM40 ; the alignment was then condensed, closing gaps in loop regions and preserving alignment in β-strands (overlined) and in cysteines. Sequence accessions are murine mucin5AC, GI:114431224; zebrafish otogelin, GI:326669509; Drosophila hemolectin, GI:24663920; human norrin, GI:4557789; Drosophila Slit, GI:17136482; human connective tissue growth factor, GI:49456477; human TGFβ2 GI:48429157; human noggin, GI:1117817; and human chorionic gonadotropin β-subunit, GI:132566538.
Sequence alignment of VWF CTCK to CTCK domains from other proteins and structural alignment to related proteins. Sequence alignment with MAFFT was with the G-INS-i strategy.39 Structural alignment was with SSM40 ; the alignment was then condensed, closing gaps in loop regions and preserving alignment in β-strands (overlined) and in cysteines. Sequence accessions are murine mucin5AC, GI:114431224; zebrafish otogelin, GI:326669509; Drosophila hemolectin, GI:24663920; human norrin, GI:4557789; Drosophila Slit, GI:17136482; human connective tissue growth factor, GI:49456477; human TGFβ2 GI:48429157; human noggin, GI:1117817; and human chorionic gonadotropin β-subunit, GI:132566538.
In noggin, a long helix-loop-helix N-terminal addition to the CK domain contributes most of the dimerization interface (Figure 4D). A 2-residue insertion between knot cysteines C3 and C4 (Figure 5) disrupts the hydrophobicity of the inter-monomer interface. Nonetheless, noggin achieves an inter-monomer orientation not dissimilar from CTCK and a cysteine in a similar sequence position to C11 in CTCK (Figure 5) mediates the inter-chain disulfide (Figure 4D).
Dimer interfaces in other CK cytokine subfamilies differ radically. Neurotrophins and ovulation-inducing factor form parallel dimers and have no inter-chain disulfides (Figure 4C). Platelet-derived growth factor and VEGF dimerize over a completely different antiparallel interface with 2 complementary interchain disulfides, distal from the center of the dimer, with no correspondence to the interchain disulfides in CTCK (Figure 4F).
How do other CT family members compare with CTCK in VWF?
The VWF CTCK subfamily includes hemolectin, norrin, mucins, and otogelin (Figure 5). These are predicted to dimerize similarly to VWF CTCK. All contain sequence signatures for the long β-strands present in CTCK, all contain C5 and C6 in β5 that form the reciprocal inter-chain disulfides, all have C2 and C8 that form the intrachain disulfide unique to CTCK domains, and most contain C11 that forms the C11-C11′ inter-chain disulfide (Figure 5). Differences among the CTCK family are easily interpretable from our structure-sequence alignment.
The hemolectin/hemocytin proteins in insects are secreted by hemocytes into hemolymph.34 They contain all of the modules found in VWF, together with discoidin domains that recognize carbohydrates, and function in both clotting and innate immune defense. The insertion in hemolectin (Figure 5) will extend the β2 and β3 strands and/or their loop, which lie distal from the dimer interface.
Mucins are also evolutionarily ancient and function in development in the ear (otogelin) as well as in mucous barriers. Many gel-forming mucins contain the CTCK domain. In otogelin, lack of the β7 strand and its inter-chain C11 cysteine (Figure 5) are easily accommodated structurally. Like VWF, mucins with CTCK domains contain N-terminal D domains and are assembled into multimers, although these may be net-like hexagonal arrays rather than linear.11,35 Mucins may also bear high forces, both as a consequence of strong repulsion between their densely packed O-linked glycans and of their high viscosity.
Norrin uniquely contains only the CTCK module. The insertion in norrin lengthens the β4 and β5 strands and/or their loop and will extend the region of overlap of the monomers at the dimer interface. Remarkably, norrin forms multimers rather than dimers, and mutation of the cysteine equivalent to C6, which disulfide bonds across the dimer interface to C5 in VWF, results in dimers.36 If the C6-C5′ and C11-C11′ interchain disulfides were among different rather than identical monomers in norrin, ie, C6-C5′ and C11-C11′′, then multimers could form. Norrin signals in multiple developmental pathways by binding frizzled4. The binding site has been mapped by alanine scanning mutations and modeled using a BMP2 dimer.37 Our VWF CTCK dimer structure provides an improved modeling template. Mutations in norrin define a contiguous frizzled4 binding site in ribbon 1, the β3-β4 loop, the C-terminus of β5, and the middle of β6 (supplemental Figure 6).
The Slit and CCN families regulate axon guidance and diverse adhesive and signaling activities in the extracellular matrix, respectively. Exemplified by Slit1 and connective tissue growth factor in Figure 5, they are predicted to have structures intermediate between the VWF CTCK domain and TGFβ. Slit and CCN family members lack cysteine C5, and thus their dimerization interface is predicted, like that in TGFβ, to slide 2 residues relative to CTCK. This is consistent with shortening of β4 and β5 in Slit and CCN, because these tuck into the dimer interface, and sliding leaves less room for this interface. Slit, connective tissue growth factor, and their relatives share the C2, C8, and C11 cysteines with VWF CTCK and not with CK cytokine family members and therefore are predicted to share the C2-C8 intrachain and C11-C11′ interchain disulfides with VWF. Thus, Slit and CCN members appear to have a dimerization interface that is a hybrid of the interface found in the VWF CTCK domain and TGFβ superfamily. This may require future adjustment of CTCK domain nomenclature to split CTCK domains into 2 subfamilies.
In conclusion, the CTCK domain structure provides specific insights into an important dimerization and force-bearing module in VWF and a more general overview of a dimerization module in proteins with surprisingly diverse functions.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.-F.Z. performed research, analyzed data, designed research, and wrote the manuscript; and T.A.S. analyzed data, designed research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy A. Springer, Program in Cellular and Molecular Medicine, Children’s Hospital Boston and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115; e-mail: timothy.springer@childrens.harvard.edu.