Key Points
Chromosome copy-number alterations that may affect immune surveillance and the NF-κB and p53 pathways are more frequent in tFL than FL.
Abnormalities involving chromosomes 6 and X are predictive of overall survival in FL.
Follicular lymphoma (FL), the second most common type of non-Hodgkin lymphoma in the western world, is characterized by the t(14;18) translocation, which is present in up to 90% of cases. We studied 277 lymphoma samples (198 FL and 79 transformed FL [tFL]) using a single-nucleotide polymorphism array to identify the secondary chromosomal abnormalities that drive the development of FL and its transformation to diffuse large B-cell lymphoma. Common recurrent chromosomal abnormalities in FL included gains of 2, 5, 7, 6p, 8, 12, 17q, 18, 21, and X and losses on 6q and 17p. We also observed many frequent small abnormalities, including losses of 1p36.33-p36.31, 6q23.3-q24.1, and 10q23.1-q25.1 and gains of 2p16.1-p15, 8q24.13-q24.3, and 12q12-q13.13, and identified candidate genes that may be driving this selection. Recurrent abnormalities more frequent in tFL samples included gains of 3q27.3-q28 and chromosome 11 and losses of 9p21.3 and 15q. Four abnormalities, gain of X or Xp and losses of 6q23.2-24.1 or 6q13-15, predicted overall survival. Abnormalities associated with transformation of the disease likely impair immune surveillance, activate the nuclear factor–κB pathway, and deregulate p53 and B-cell transcription factors.
Introduction
Follicular lymphoma (FL) arises from germinal center (GC) B cells and 80% to 90% of FLs have the t(14;18) translocation, which places BCL2 under control of the immunoglobulin heavy-chain enhancers, leading to its constitutive expression and protecting the cell from apoptosis. This abnormality alone is not enough to lead to FL1 but likely provides a survival advantage in the GC microenvironment, where the cell can accumulate additional abnormalities. Identifying these secondary abnormalities will help elucidate how a preneoplastic clone evolves into malignant lymphoma.
Although FL is usually indolent, 40% of patients will develop diffuse large B-cell lymphoma (DLBCL) with markedly worsened prognosis.1 What drives this transition between indolent and aggressive disease is poorly defined but may be clarified by analyzing alterations that differ between FL and transformed FL (tFL) specimens.
Cytogenetic studies and smaller array-based comparative genomic hybridization studies have identified large, recurrent abnormalities, including gains of chromosomes 7, 12, 18, and X, and losses of 6q and 1p.2,,,,-7 However, few large, high-resolution studies have been reported.2,,,,-7 Thus, we performed array-based copy-number analysis using the high-resolution GeneChip Human Mapping 250K Nsp single-nucleotide polymorphism array (Affymetrix) on 225 FL tumor samples and 84 tFL tumors to identify abnormalities that contribute to FL and influence survival. Additionally, we identified abnormalities that were significantly more common in tFL than FL samples and which may contribute to transformation and disease progression.
Materials and methods
Clinical samples
Samples (225 FL and 84 tFL) were obtained from the Lymphoma/Leukemia Molecular Profiling Project (LLMPP), or the University of Nebraska Medical Center (UNMC) Pathology/Oncology Database. This study was approved by the UNMC institutional review board and conducted in accordance with the Declaration of Helsinki. Diagnoses were confirmed by a panel of LLMPP hematopathologists. tFL was defined as DLBCL that occurred in a patient diagnosed with FL. Most tFLs were diagnosed concurrently or subsequently to the FL diagnosis; however, for 8 cases the DLBCL preceded discovery of the FL, which was likely present previously but undetected. For 44 paired cases, clonal relationship was confirmed by comparing the genetic profile to confirm there was close similarity between pairs. When only the DLBCL sample was available and the BCL2 status was unknown, BCL2 rearrangement was assessed by polymerase chain reaction (PCR) to determine whether the frequency of t(14;18) positivity expected for an FL data set was observed in the tFL data set (see supplemental Methods and supplemental Table 6 for details, available on the Blood Web site). Fifty-five percent of samples were confirmed to be t(14;18)-positive by PCR of the major breakpoint region, minor cluster region, and intermediate cluster region, which is similar to previously published results on t(14;18) PCR positivity of FLs (∼60%)8 and much higher than what is expected in de novo DLBCL.9
DNA CNAs and aUPD analysis
Two hundred fifty nanograms of genomic DNA (gDNA) isolated using the AllPrep DNA/RNA kit (Qiagen) was processed as recommended (Affymetrix). Copy-number abnormalities (CNAs) were identified as described10 with modifications (supplemental Figure 1). Single-nucleotide polymorphism genotypes and probe-intensity log2 ratios, generated using Genotyping Console 4.1 software (Affymetrix), were analyzed for CNAs using the DNAcopy package11 (Bioconductor); ≥4 copies were considered an amplification. Acquired uniparental disomy (aUPD) was analyzed using AsCNAR (CNAG program, Version 3.3.0.0, http://www.genome.umin.jp/CNAG_DLpage/files/CNAGdownload_list.html.),12 which identifies aUPD in samples with contaminating normal tissue. Recurring CNAs (rCNAs) were aligned to determine the minimal common region (MCR), and genes within MCRs were identified. rCNA frequencies for the FL and tFL groups were compared using the Fisher exact test without correcting for multiple tests.
Analysis of co-occurrence of rCNAs
A Monte Carlo simulation was performed to identify significantly positively/negatively associated rCNAs (supplemental Methods).
Gene expression analysis
Gene expression profiling (GEP) data were generated at UNMC or as part of the Strategic Partnering to Evaluate Cancer Signatures (SPECS)/LLMPP initiatives using HG-U133 A+B (112 FL) or HG-U133Plus2.0 arrays (60 FL, 46 tFL) (Affymetrix). Gene expression was analyzed for the most frequent, small rCNAs (rCNA122 and rCNA442). GEP data from tFL cases were used to classify samples as GCB or ABC-like as previously described.8
Clinical correlations
Clinical data were obtained for 149 FL cases and 21 tFL cases. The median follow-up of the 77 FL patients alive at last contact was 7.0 years (range, 0-28 years). Univariate Cox regression models were used to assess whether rCNAs that occurred in ≥10 FL cases predicted overall survival (OS). The Kaplan-Meier method was used to estimate OS distributions, and OS distributions were compared between groups with the log-rank test.
Results
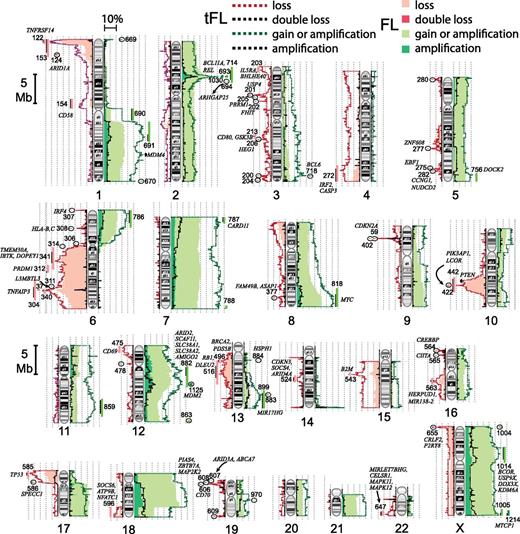
We included 198 FL and 79 tFL arrays of sufficient quality in the study after preliminary analysis. Supplemental Figure 1 shows the CNA data-analysis pipeline. After removing known germline copy-number variations (CNVs) using the Database of Genomic Variants from the Centre for Applied Genomics, 959 CNAs occurred in ≥2 tumors (supplemental Table 1A-C). Four hundred sixty-two CNAs were present in ≥2.5% of FL or tFL samples, and these were considered recurring acquired CNAs (rCNAs). FL cases averaged 7.2 rCNAs, and tFLs, 10.6. Figure 1 illustrates the frequency of all gains and losses along the chromosomes and shows genes of interest based on known function or known mutations in lymphoid malignancies.
Ideograms with the frequency of gains, amplifications, losses, or double losses found in FL or tFL tumors. Common rCNAs and candidate genes are noted.
Ideograms with the frequency of gains, amplifications, losses, or double losses found in FL or tFL tumors. Common rCNAs and candidate genes are noted.
Chromosomal abnormalities
Supplemental Table 1A summarizes abnormalities found in >2.5% of FLs or tFLs that involve whole chromosomes or an arm. Our study confirmed many abnormalities identified previously.2,,,-6 Whole-chromosome/whole-arm gains of 18 (31%), X (23%), 7 (21%), 21 (11.5%), 12 (11%), 5 (8.5%), 8 (8%), and 2 (8%) were among the most frequent in FL. Gains of chromosomes 7, 8, and 21 were 1.5 to 1.8 times more frequent in tFL (supplemental Table 1A). Frequent chromosome arm rCNAs included gain of 1q, 6p, 9q, and 17q, and loss of 6q and 17p. 17p loss, which contains TP53, was twice as frequent in tFL as in FL, similar to previous observations.1
Small abnormalities
Small, recurrent abnormalities can indicate genes critical in the encompassing larger abnormalities. Eighty-seven rCNAs were found at >5% frequency in tFL or FL (supplemental Table 1B). Eighteen rCNAs occurred in ≥10% of FL or tFL cases, and rarer rCNAs were often identified within more common, larger abnormalities. Our findings confirm the frequent loss2,3 on the tip of 1p, (1p36.31-36.33) (rCNA122) which includes several possible tumor suppressor genes (TSGs) and cancer-related genes such as TNFRSF14, the H3K9me1 methyltransferase PRDM16,13 and the p53-family member TP73. Copy loss and mutation of TNFRSF14 have been reported in FL.14,15 In 2.5% of FL and 3.75% of tFLs there was a double deletion at 1p36.32 (rCNA1) that encompassed 16 genes, including TP73 and PRDM16; this core deletion extended within 9 kb of TNFRSF14 and may affect its regulation. GEP analysis of genes within rCNA122 identified 6 genes with decreased expression in FL cases with the deletion (supplemental Table 2). Two of these genes were differentially expressed in positive and negative tFL cases (TNFRSF14 and C1orf174); however, only TNFRSF14 appears to be expressed in normal B cells.
6q loss was very common, and smaller deletions within the arm may suggest which genes are important. The most frequent small losses identified were 6q13-q15, 6q23.3-q24.1, and 6q23.2-q25.1 (rCNA341/rCNA304/rCNA340). Thus, multiple subregions likely drive the 6q loss. As previously reported,6 a homozygous deletion on 6q23.3 (rCNA37) in 6% of FL and 10% of tFL encompassed the nuclear factor–κB (NF-κB) regulator TNFAIP3; however, in our study, the peak of double loss lies directly over TNFAIP3 (supplemental Figure 2), which thus likely primarily drives the 6q23.3 homozygous loss and other small,TNFAIP3-containing losses. Critical genes within 6q13-q15 are less clear, but candidates include IBTK, TMEM30A, and DOPEY1.
A loss of 10q23.1-q25.1 (rCNA442) observed in 15% of FLs and tFLs includes the TSGs FAS and PTEN and occurs early in FL (see Figure 2 ). This region includes a prominent smaller peak of loss in the tFL cases centered on 2 genes: PIK3AP1, which plays a role in the activation of phosphoinositide 3-kinase in B cells, and the transcriptional corepressor LCOR (Figure 1, rCNA422). One tFL and 2 FL cases had a double loss that included only LCOR. Thirty-one genes within rCNA442, including PIK3AP1, LCOR, and PTEN, had significantly reduced expression in rCNA442-deleted FL cases (supplemental Table 2). We also found gains/amplifications on 2p16.1-p15 (Figure 1, rCNA963/rCNA1030) that include REL and BCL11A, 8q24.13-q24.3 (rCNA818), that include MYC, and 12q12-q13.13, which contains several potential target genes including ARID2, SCAF11, SLC38A1, SLC38A2, and AMIGO2 (Table 1; Figure 1).
Small rCNAs with candidate genes
| rCNA ID . | Locus . | FL frequency . | tFL frequency . | tFL:FL ratio . | No. of genes . | Interesting genes . |
|---|---|---|---|---|---|---|
| 669 | 1p36.33+ | 0.025 | 0.076 | 3.008 | 56 | TNFRSF14 |
| 122 | 1p36.33-p36.31− | 0.247 | 0.241 | 0.972 | 51 | TNFRSF14*, PRDM16 |
| 1 | 1p36.32− (×2) | 0.025 | 0.038 | 1.504 | 16 | PRDM16, TP73 |
| 124 | 1p36.11-p35.3− | 0.045 | 0.051 | 1.114 | 40 | ARID1A |
| 674 | 1p22.3+ | 0.010 | 0.025 | 2.506 | 7 | BCL10 |
| 1019 | 1p22.3+ (×2) | 0.005 | 0.025 | 5.013 | 5 | BCL10 |
| 125 | 1p13.1− | 0.030 | 0.038 | 1.253 | 7 | CD58* |
| 714 | 2p16.3-p14+ | 0.030 | 0.051 | 1.671 | 72 | REL, BCL11A |
| 693 | 2p16.1-p15+ | 0.106 | 0.190 | 1.790 | 13 | REL, BCL11A |
| 1030 | 2p16.1-p15+ (×2) | 0.086 | 0.114 | 1.327 | 8 | REL, BCL11A |
| 163 | 2q11.2− | 0.015 | 0.038 | 2.506 | 1 | AFF3 |
| 718 | 3q27.3-q28+ | 0.020 | 0.114 | 5.639 | 5 | BCL6 |
| 275 | 5q33.3− | 0.045 | 0.051 | 1.114 | 1 | EBF1* |
| 307 | 6p25.3− | 0.020 | 0.114 | 5.639 | 9 | IRF4 |
| 308 | 6p21.33-p21.32− | 0.030 | 0.089 | 2.924 | 52 | HLA-C, HLA-B |
| 769 | 6p21.2-p21.1+ | 0.030 | 0.013 | 0.418 | 78 | CCND3 |
| 342 | 6q16.3-q22.33− | 0.035 | 0.038 | 1.074 | 127 | PRDM1 |
| 312 | 6q21− | 0.020 | 0.051 | 2.506 | 10 | PRDM1 |
| 340 | 6q23.2-q25.1− | 0.056 | 0.101 | 1.823 | 102 | TNFAIP3, PERP |
| 304 | 6q23.3-q24.1− | 0.081 | 0.127 | 1.566 | 19 | TNFAIP3, PERP |
| 37 | 6q23.3− (×2) | 0.056 | 0.101 | 1.823 | 1 | TNFAIP3 |
| 787 | 7p22.3-p21.3+ | 0.025 | 0.076 | 3.008 | 89 | CARD11, RNF216 |
| 1101 | 8q24.12-q24.3+ (×2) | 0.030 | 0.038 | 1.253 | 109 | MYC |
| 818 | 8q24.13-q24.3+ | 0.106 | 0.114 | 1.074 | 174 | MYC |
| 402 | 9p21.3− | 0.051 | 0.177 | 3.509 | 5 | CDKN2A, CDKN2B |
| 59 | 9p21.3− (×2) | 0.030 | 0.114 | 3.759 | 4 | CDKN2A, CDKN2B |
| 407 | 9q34.3− | 0.025 | 0.000 | 0.000 | 3 | EHMT1 |
| 422 | 10q24.1− | 0.040 | 0.114 | 2.820 | 7 | LCOR* |
| 447 | 11q14.1-q14.2− | 0.015 | 0.025 | 1.671 | 5 | EED |
| 1146 | 12p11.22-q13.11+ (×2) | 0.025 | 0.013 | 0.501 | 49 | ARID2, DDX11, ALG10, KIF21A |
| 866 | 12q12-q13.12+ | 0.020 | 0.038 | 1.880 | 47 | ARID2, HDAC7, RPAP3 |
| 1145 | 12q13.2-q21.1+ (×2) | 0.040 | 0.025 | 0.627 | 127 | MDM2,TBK1, CDK4, STAT6 |
| 867 | 12q13.3+ | 0.025 | 0.025 | 1.003 | 18 | STAT6 |
| 1125 | 12q15+ (×2) | 0.020 | 0.063 | 3.133 | 22 | MDM2 |
| 864 | 12q15+ | 0.030 | 0.025 | 0.835 | 17 | MDM2 |
| 496 | 13q12.3-q13.2− | 0.015 | 0.051 | 3.342 | 17 | |
| 543 | 15q21.1− | 0.040 | 0.114 | 2.820 | 4 | B2M* |
| 564 | 16p13.3− | 0.025 | 0.051 | 2.005 | 8 | CREBBP* |
| 565 | 16p13.13− | 0.015 | 0.051 | 3.342 | 4 | CIITA* |
| 585 | 17p13.3-p13.1− | 0.071 | 0.152 | 2.148 | 160 | TP53 |
| 966 | 18q21.2-q21.33+ | 0.025 | 0.038 | 1.504 | 43 | MALT1, BCL2 |
| 606 | 19p13.3-p13.2− | 0.045 | 0.114 | 2.506 | 23 | CD70* |
| rCNA ID . | Locus . | FL frequency . | tFL frequency . | tFL:FL ratio . | No. of genes . | Interesting genes . |
|---|---|---|---|---|---|---|
| 669 | 1p36.33+ | 0.025 | 0.076 | 3.008 | 56 | TNFRSF14 |
| 122 | 1p36.33-p36.31− | 0.247 | 0.241 | 0.972 | 51 | TNFRSF14*, PRDM16 |
| 1 | 1p36.32− (×2) | 0.025 | 0.038 | 1.504 | 16 | PRDM16, TP73 |
| 124 | 1p36.11-p35.3− | 0.045 | 0.051 | 1.114 | 40 | ARID1A |
| 674 | 1p22.3+ | 0.010 | 0.025 | 2.506 | 7 | BCL10 |
| 1019 | 1p22.3+ (×2) | 0.005 | 0.025 | 5.013 | 5 | BCL10 |
| 125 | 1p13.1− | 0.030 | 0.038 | 1.253 | 7 | CD58* |
| 714 | 2p16.3-p14+ | 0.030 | 0.051 | 1.671 | 72 | REL, BCL11A |
| 693 | 2p16.1-p15+ | 0.106 | 0.190 | 1.790 | 13 | REL, BCL11A |
| 1030 | 2p16.1-p15+ (×2) | 0.086 | 0.114 | 1.327 | 8 | REL, BCL11A |
| 163 | 2q11.2− | 0.015 | 0.038 | 2.506 | 1 | AFF3 |
| 718 | 3q27.3-q28+ | 0.020 | 0.114 | 5.639 | 5 | BCL6 |
| 275 | 5q33.3− | 0.045 | 0.051 | 1.114 | 1 | EBF1* |
| 307 | 6p25.3− | 0.020 | 0.114 | 5.639 | 9 | IRF4 |
| 308 | 6p21.33-p21.32− | 0.030 | 0.089 | 2.924 | 52 | HLA-C, HLA-B |
| 769 | 6p21.2-p21.1+ | 0.030 | 0.013 | 0.418 | 78 | CCND3 |
| 342 | 6q16.3-q22.33− | 0.035 | 0.038 | 1.074 | 127 | PRDM1 |
| 312 | 6q21− | 0.020 | 0.051 | 2.506 | 10 | PRDM1 |
| 340 | 6q23.2-q25.1− | 0.056 | 0.101 | 1.823 | 102 | TNFAIP3, PERP |
| 304 | 6q23.3-q24.1− | 0.081 | 0.127 | 1.566 | 19 | TNFAIP3, PERP |
| 37 | 6q23.3− (×2) | 0.056 | 0.101 | 1.823 | 1 | TNFAIP3 |
| 787 | 7p22.3-p21.3+ | 0.025 | 0.076 | 3.008 | 89 | CARD11, RNF216 |
| 1101 | 8q24.12-q24.3+ (×2) | 0.030 | 0.038 | 1.253 | 109 | MYC |
| 818 | 8q24.13-q24.3+ | 0.106 | 0.114 | 1.074 | 174 | MYC |
| 402 | 9p21.3− | 0.051 | 0.177 | 3.509 | 5 | CDKN2A, CDKN2B |
| 59 | 9p21.3− (×2) | 0.030 | 0.114 | 3.759 | 4 | CDKN2A, CDKN2B |
| 407 | 9q34.3− | 0.025 | 0.000 | 0.000 | 3 | EHMT1 |
| 422 | 10q24.1− | 0.040 | 0.114 | 2.820 | 7 | LCOR* |
| 447 | 11q14.1-q14.2− | 0.015 | 0.025 | 1.671 | 5 | EED |
| 1146 | 12p11.22-q13.11+ (×2) | 0.025 | 0.013 | 0.501 | 49 | ARID2, DDX11, ALG10, KIF21A |
| 866 | 12q12-q13.12+ | 0.020 | 0.038 | 1.880 | 47 | ARID2, HDAC7, RPAP3 |
| 1145 | 12q13.2-q21.1+ (×2) | 0.040 | 0.025 | 0.627 | 127 | MDM2,TBK1, CDK4, STAT6 |
| 867 | 12q13.3+ | 0.025 | 0.025 | 1.003 | 18 | STAT6 |
| 1125 | 12q15+ (×2) | 0.020 | 0.063 | 3.133 | 22 | MDM2 |
| 864 | 12q15+ | 0.030 | 0.025 | 0.835 | 17 | MDM2 |
| 496 | 13q12.3-q13.2− | 0.015 | 0.051 | 3.342 | 17 | |
| 543 | 15q21.1− | 0.040 | 0.114 | 2.820 | 4 | B2M* |
| 564 | 16p13.3− | 0.025 | 0.051 | 2.005 | 8 | CREBBP* |
| 565 | 16p13.13− | 0.015 | 0.051 | 3.342 | 4 | CIITA* |
| 585 | 17p13.3-p13.1− | 0.071 | 0.152 | 2.148 | 160 | TP53 |
| 966 | 18q21.2-q21.33+ | 0.025 | 0.038 | 1.504 | 43 | MALT1, BCL2 |
| 606 | 19p13.3-p13.2− | 0.045 | 0.114 | 2.506 | 23 | CD70* |
Confirmed by PCR.
Acquired uniparental disomy
A low frequency of aUPD (copy-neutral loss of heterozygosity [LOH]) was detected along most of the chromosomes, probably due to whole-chromosome aUPD, but we identified several regions of frequent aUPD (supplemental Figure 3A; supplemental Table 3), confirming observations from smaller studies.6,14,16,17 A region of copy-number loss on 1p frequently had aUPD in FL and tFL (supplemental Figure 3B). A previous study16 found 1p36 aUPD to negatively correlate with OS; however, we did not observe this, nor was aUPD at TNFRSF14 predictive of OS (data not shown). Frequent aUPD also occurred on 6p, 12q, and 16p in FL and tFL cases, and tFLs had frequent aUPD on chromosomes 9, 12p, 16q, and 17.
Temporal order of abnormalities in FL
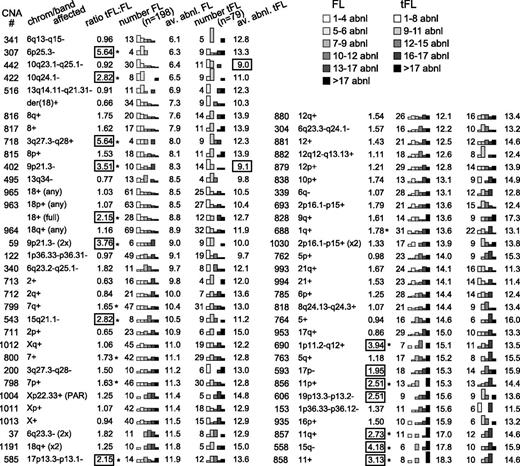
To model the progression of rCNAs with FL disease progression, the average number of rCNAs in a tumor with a specific rCNA was calculated (Figure 2, av.abnl.FL) and ranged from 6.1 to 18.3 for FL. Abnormalities that occur early in the disease would be expected to occur in tumors with both high and low numbers of other abnormalities, whereas abnormalities that occur later in disease would be expected to occur in cases with a high number of abnormalities.18 Therefore, early CNAs would be expected to have a lower average number of other abnormalities than CNAs that arise later. The suggested temporal orders of these rCNAs are presented in Figure 2. Their significance will be addressed in the “Discussion” section.
Number of additional abnormalities associated with each CNA. The average numbers of abnormalities per tumor for tumors with a specific CNA were analyzed in FL and tFL (av. abnl. FL, av. abnl. tFL) and are presented in ascending order according of the av. abnl. FL. The distribution of the number of cases falling within the noted ranges of abnormalities is displayed with bar graphs. The number of cases with an rCNA in FL and tFL (number FL, number tFL) and the ratio of the frequency in tFL to FL are shown. rCNAs that are ∼2 or more times as frequent in tFL are boxed, and frequencies that differ significantly between FL and tFL by the Fisher exact test are marked with an asterisk (*).
Number of additional abnormalities associated with each CNA. The average numbers of abnormalities per tumor for tumors with a specific CNA were analyzed in FL and tFL (av. abnl. FL, av. abnl. tFL) and are presented in ascending order according of the av. abnl. FL. The distribution of the number of cases falling within the noted ranges of abnormalities is displayed with bar graphs. The number of cases with an rCNA in FL and tFL (number FL, number tFL) and the ratio of the frequency in tFL to FL are shown. rCNAs that are ∼2 or more times as frequent in tFL are boxed, and frequencies that differ significantly between FL and tFL by the Fisher exact test are marked with an asterisk (*).
Associated rCNAs
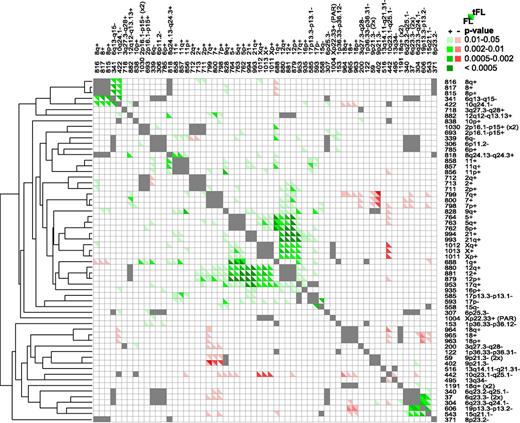
A Monte Carlo simulation was performed to determine whether rCNAs occur together more/less than expected by chance. Several sets of rCNAs occurred together (Figure 3), such as gains of chromosomes 5, 7, 12, and X in FL. Notably, in FL and tFL, 6q23.3 (rCNA37/rCNA304/rCNA340) frequently was lost with 19p13.3-p13.2 (rCNA606) and 15q21.1 (rCNA543). The 6q loss encompasses TNFAIP3, while the likely targets on 19p and 15q are CD70 and B2M, respectively. In tFL, the 6q13-15 loss, which includes IBTK, DOPEY1, and TMEM30A (rCNA341), was strongly associated with a small loss on 10q24.1 (rCNA422) encompassing PIK3AP1 and LCOR and chromosome 8 gain (rCNA815/rCNA816/rCNA817).
Associated CNAs. A Monte Carlo simulation was performed to determine whether pairs of CNAs were positively or negatively associated. rCNA pairs on the same chromosome were not analyzed and are marked in gray. Significantly associated pairs of CNAs (P ≤ .05) are denoted in shades of green (positive association) or red (negative association). The lower left of a box represents FL cases and the upper right represents tFL cases. The results are ordered according to a hierarchical cluster tree generated using Cluster 3.0.
Associated CNAs. A Monte Carlo simulation was performed to determine whether pairs of CNAs were positively or negatively associated. rCNA pairs on the same chromosome were not analyzed and are marked in gray. Significantly associated pairs of CNAs (P ≤ .05) are denoted in shades of green (positive association) or red (negative association). The lower left of a box represents FL cases and the upper right represents tFL cases. The results are ordered according to a hierarchical cluster tree generated using Cluster 3.0.
MicroRNAs
Several rCNAs included microRNA (miRNA) candidates (supplemental Table 1B). We found gains/amplifications on 13q (Figure 1, rCNA898/rCNA899/rCNA1150), including the miR-17∼92 cluster (MIR17HG) in 7% of FL cases and 15% of tFL cases. The miR-17∼92 cluster has been shown to have oncogenic activities.19,20 A more centromeric loss on 13q (rCNA513), occurring early in FL, affects RB1 and the miR-16-1 and miR-15a–encoding DLEU2. Additionally, 2 highly expressed miRNAs in normal GCB cells, miR-28 and miR-138, had decreased expression in lymphomas (Leich et al21 and data not shown). A 3q27.3-q28 loss (rCNA200) in 5% of FL and 7.5% of tFL includes 3 genes, 1 of which is LPP, the miR-28 host gene. MiR-138 is encoded by 2 loci, and it is unclear which is expressed in B cells. A prominent peak of loss on chromosome 16 that includes miR-138-2 (Figure 1) occurs in 5% of FLs and 10% of tFLs.
Transformation-related rCNAs
rCNAs that are associated with transformation are expected to occur more frequently in the tFL data set, and abnormalities that promote rapid transformation would be associated with few abnormalities per tumor, as the tumor did not have time to accumulate rCNAs before transformation. Fifteen rCNAs illustrated in Figure 2 were ≥2 times as frequent in tFL as FL, with 13 reaching significance (P < .05, Fisher exact test). Seven of the rCNAs were early-to-intermediate abnormalities, as the average number of abnormalities in the tumors was relatively low (av.abn.FL ≤ 10.5). Two rCNAs were over 5 times as frequent in tFL as in FL. The genes likely to be targeted by these rCNAs are noted in Table 1.
De novo DLBCL and tFL
tFLs usually present as GCB-DLBCLs. Thus, we compared the frequency of abnormalities in tFLs to those found in de novo GCB-type DLBCL cases10,22 (supplemental Figure 4). GCB-DLBCL frequently has a small loss on 1p and gain of REL on 2p, and our tFL cases showed similar frequencies. A gain on 13q including miR-17∼92, while frequent in tFL, is more common in GCB-DLBCL, and a loss at the tip of 13q thought to be driven by ING110 is over twice as common in GCB-DLBCL as in tFL (supplemental Figure 4). Another striking difference is a 10q loss, apparently driven by PTEN in GCB-DLBCL, but in tFL the loss centers on LCOR and PIK3AP1. A 6q arm loss occurs in almost 40% of ABC-DLBCL cases and only around 15% GCB-DLBCL.10,22 Only 15% of tFLs have the 6q loss; however, a small region of 6q including TNFAIP3 is lost about twice as frequently in tFL (40%) as in de novo DLBCL (supplemental Figure 4). Additionally, a 9p loss including CDKN2A and CDKN2B occurs in 30% of ABC-DLBCL and only 5% of GCB-DLBCL,10,22 whereas tFL has an intermediate frequency (18%).
tFL cases with available GEP data were classified according to GCB- or ABC-like expression patterns as described.10 Although most tFL classify as GCB type, as expected, 29% classify as ABC-like or unclassifiable. The frequencies of 14 rCNAs differ significantly between the GCB and non-GCB groups (supplemental Table 1A-B). Gain of chromosome 3, 3p, or 3q was more frequent in the non-GCB tFL group. Several small rCNAs were also more common in the non-GCB tFL group: for example, gain of 1q44 (rCNA670), and losses of 1p13.1 (rCNA125) and 1p36.11-p35.3 (rCNA124), which include CD58 and ARID1A, respectively, and loss/double-loss of 6q that includes the NF-κB negative regulator TNFAIP3 (rCNA304/rCNA37), consistent with the importance of constitutive NF-κB to ABC-DLBCL. Furthermore, loss of 19p13.3-p13.2, which includes CD70, tended to co-occur with rCNA304 and 37 and is more common in non-GCB–type tFL cases although this does not quite reach a P < .05 significance (supplemental Table 1B). Conversely, gains of whole chromosomes or arms of 7 and 12 were more frequent in GCB-type tFL cases.
Clinical correlations
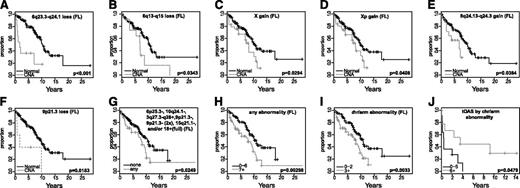
Several rCNAs predicted survival in univariate analysis at P < .05 significance (supplemental Table 4) and by the Cox regression model. We also examined the association of the presence of early/intermediate abnormalities that were ≥2 times as frequent in tFLs, the number of abnormalities present, and the types of rCNAs with survival. The results are shown in Figure 4 and in the “Discussion” section.
Correlation of CNAs with OS. (A-G) Kaplan-Meier curves comparing OS of FL patients with or without specific CNAs. (H-J) Kaplan-Meier curves comparing OS of FL (H-I) or tFL patients (J) with different numbers of chromosomal abnormalities.
Correlation of CNAs with OS. (A-G) Kaplan-Meier curves comparing OS of FL patients with or without specific CNAs. (H-J) Kaplan-Meier curves comparing OS of FL (H-I) or tFL patients (J) with different numbers of chromosomal abnormalities.
Discussion
FL is indolent, with relatively long survival; however, some FLs transform and have a worse prognosis.1 Identifying rCNAs associated with transformation would be beneficial for identifying patients who are more likely to transform and need to be managed differently or monitored more closely. We cataloged rCNAs that occur in FL and tFL and identified abnormalities that appear to occur early in the disease, are associated with poor outcome, or are associated with transformation. For 44 patients, paired FL and tFL biopsies were studied. Although CNA analysis showed the paired samples to be clonally related, the tFL generally did not appear to evolve directly from the FL we studied, as many of the pairs had abnormalities unique to the FL. This is in keeping of the clonal evolution models we and others have proposed.4,7
FL cells retain much of the normal GCB-cell phenotype, including apparent dependence upon T follicular helper (TFH) cells and follicular dendritic cells (FDCs). As long predicted,23,24 recent work25 has confirmed that normal GCB cells recycle between a light zone (LZ), in which TFH cells provide stimulation, and a dark zone (DZ) in which antigen-selected cells undergo a burst of proliferation and activation-induced cytidine deaminase-dependent somatic hypermutation. Cells stimulated in the LZ show high NF-κB activity, MYC26,27 and miR-155 expression,28 and decreased BCL6 functional activity.29 They initiate cell division and migrate to the DZ, where activation-induced cytidine deaminase is upregulated and high BCL6 activity represses not only miR-15530 and MYC, but also CDKN1A and -B, allowing the proliferative burst to continue. Whereas DLBCL, whether de novo or arising by transformation of FL, may center on either a BCL6-dependent DZ-like (GCB-subtype) or NF-κB/MYC-dependent LZ-like (ABC-subtype–like) proliferative strategy, the CNAs and mutations that occur in FL may enhance either phase of this complex recycling process, and rCNAs predominantly associated with either ABC- or GCB-DLBCL are both found in FL.
In addition to t(14;18), several early abnormalities are likely involved in establishing an FL by cooperating to disrupt critical regulatory pathways. These early rCNAs, shown in Figure 2, included likely target genes listed in Table 1. Gain of the full chromosome 18 was common but occurred later than der(18)t(14;18) (Figure 2, av.abnl.FL: 8.8 vs 7.3). Loss at the most distal segments of 1p (rCNA122) also occurred relatively early (av.abnl = 9.1). rCNAs associated with few other abnormalities can be seen in both FL and in tFL, and several, such as the loss on 9p21.1 (rCNA402), had a high tFL:FL ratio and thus may arise early but induce rapid transformation. Indeed, genes within these early abnormalities regulate pathways whose disruption could contribute to lymphomagenesis, including the NF-κB, p53, and RB pathways, and B-cell differentiation.
Many abnormalities identified are found at similar frequencies in FL and tFL. Only 4 rCNA found in >10% of FLs were smaller than a chromosome arm: loss of 1p36.33-p36.31 (CNA122) and 10q23.1-q25.1 (CNA442), and gain of 2p16.1-p15 (CNA693) and 8q24.13-q24.3 (CNA818). One striking and frequent (∼50% incidence) abnormality in FL is the occurrence of copy-number loss of the distal 1p (CNA122) or aUPD at 1p36.33-p36.3. TNFRSF14 is the likely candidate gene in this region, and it is frequently mutated with a resultant loss of surface expression.14,15 The aUPD extends more broadly along 1p, and a less frequent copy loss (∼10%) contains the tumor suppressor ARID1A (rCNA124), which may also be a target gene. ARID1A interacts with SWItch/Sucrose NonFermentable (SWI/SNF) chromatin-remodeling complexes and affects CDKN1A and SMAD3 transcription through p53.31 Loss of 6q13-15 (rCNA304), 6q23.3-24.1 (rCNA341) or 9p21.3 (rCNA402), or gain of X, Xp, or 8q24.13-q24.3 (rCNA818) predict poor OS in FL. Although the target genes in rCNA341 and rCNA402 are likely TNFAIP3 and CDKN2A, respectively, the driver genes in the other rCNAs are less clear. Xp, for example, is a large region, but candidates include BCOR, USP9X, DDX3X, and KDM6A. USP9X deubiquitinates and thus stabilizes the anti-apoptotic protein MCL1.32 BCOR, a transcriptional corepressor, binds BCL6 to increase repression of its target genes.33 DDX3X, an RNA helicase, is recurrently mutated in chronic lymphocytic leukemia34 and FL (unpublished exome sequencing data, A.B., W.Z., and W.-C.C.) and can act as an oncogene.35 The H3K27 histone demethylase KDM6A is mutated in cancers including chronic myelomonocytic leukemia,36 and in breast cancer,37 increased expression confers poor prognosis. Thus, in large rCNAs such as Xp, multiple genes likely drive their selection and contribute to lymphomagenesis.
Deregulation of miRNAs by CNAs is another important and largely unexplored contributor to FL. Several rCNAs included miRNAs known to be deregulated in cancer such as miR-17∼92, miR-15 and -16a, and miR-28.19,20,38 The loss of miR-138-2, for example, may be of interest as one of the targets of miR-138 is CCND3 which is the d-cyclin used by GCB cells, and oncogenic mutation of CCND3 has been recently reported in Burkitt lymphoma.39
Abnormalities important for transformation appear to target several key pathways; most notably, disruption of the p53 pathway, activation of NF-κB, dysregulation of transcription factors, and evasion of immune surveillance. The p53 pathway is the most commonly deregulated pathway in cancer, and CNAs affecting it have recently been discussed in DLBCL37 (also see supplemental Discussion).40 MDM2, on chromosome 12, encoding a negative regulator of p53, is gained/amplified in 23% of FLs and 32% of tFLs (supplemental Figure 5A). MDM4, on chromosome 1, also regulates p53 and is gained/amplified in 17% of FLs and 30% of tFLs. The TP53 locus is lost in 12% of FLs and 22% of tFLs. 9p21.3 (CDKN2A and -B) loss, occurring in 6% of FLs and 18% of tFLs, deregulates both the p53 and RB pathways. tFL cases with this rCNA had few other abnormalities (av.abn.tFL ≤ 10), suggesting that FL tumors, after this deletion, quickly progress to tFL. Overall, 43% of FLs and 70% of tFLs had a CNA affecting at least one of the above. Additionally, FL cases with gains of MDM2 or MDM4 or loss of TP53 or CDKN2A, had worse survival than cases wild type at all these loci, consistent with a previous observation41 (supplemental Figure 5B).
NF-κB is activated in many cancers.42 Many rCNAs include genes affecting NF-κB (supplemental Table 1B). Gain/amplification including REL and deletion at 6q including TNFAIP3 are 2 of the most frequent abnormalities. Twenty-six percent of FLs and 34% of tFLs have loss of TNFAIP3, and 5% of FLs and 10% of tFLs have homozygous loss. Twenty-four percent of FLs and 30% of tFLs have a gain including REL. Many other rCNAs include genes whose products affect the NF-κB pathway, such as IBTK (rCNA341),43 PI3KAP1 and BLNK (rCNA442),44 TRAPPC9 (rCNA818),45 ARRB2,46 and TXNDC17 (rCNA585).47 Non-GCB tFL more frequently lose TNFAIP3 whereas the opposite is true for gain/amplification of REL. Alterations that drive NF-κB activation may shift the GEP closer to unclassifiable/ABC-DLBCL. REL-containing dimers are regulated differently from other NF-κB dimers48 and target a somewhat distinct set of genes49 including CD40 and TNFRSF13C (BAFFR), which are important in receiving microenvironmental stimuli.
The follicular architecture of FL is maintained by FDCs and TFH cells in the microenvironment.1 Upon transformation, this microenvironment is altered, but usually the GC phenotype is maintained. BCL6 is a master regulator of the GC phenotype, and its expression suppresses differentiation into plasma cells, whereas the expression of transcription factors (TFs) such as PRDM1 and IRF4 promotes differentiation. Thus, disruption of these critical TFs blocks the normal differentiation process. Several rCNAs included these important B-cell TFs. PRDM1 (6q) is lost in 20% of FL and tFL. We also observed IRF4 loss, more commonly in tFLs. BCL6 was gained in ∼5% of FLs but 3 times as many tFLs. Overall, 43% of tFLs had loss of IRF4, PRDM1, and/or gain of BCL6 indicating blocking differentiation may be critical, especially for tFL, which lacks the FL microenvironment.
We also detected that small deletions affected several other transcriptional regulators known to be mutated in FL or DLBCL, such as CREBBP (rCNA564), ETS1 (rCNA444), and EBF1 (rCNA275), but the frequencies of these rCNAs do not differ significantly between FL and tFL. EBF1 is required for normal B-cell differentiation50 and is profoundly downregulated in chronic lymphocytic leukemia.51 In contrast, ETS1 is often gained in DLBCL,52 so the loss in tFL may reflect biologic differences between tFL and de novo DLBCL.
Certain alterations in FL may promote help from TFH cells. 6p aUPD would result in loss of HLA class I and II haplotypes. Although class I molecules are important in CD8+ T-cell response, class II are important in B-cell/TFH interaction. Depending on the peptide presentation, aUPD could reduce immune surveillance on the one hand and increase B-cell help on the other. BTLA, whose ligand is TNFRSF14, is highly expressed on TFH cells53 and suppresses their production of interleukin-21 (IL-21).54 Mutation/deletion of TNFRSF14 is thus expected to enhance availability of IL-21 to lymphoma cells. Single-copy loss of negative regulators may allow otherwise subthreshold stimuli to promote proliferation and survival. Thus, loss of IBTK on 6q14, reducing inhibition of Bruton agammaglobulinemia tyrosine kinase, may enhance signaling from the B-cell receptor. Loss of PTEN (10q23) or TNFAIP3 (6q23.3), or gain of REL (2p15) or miR-17∼92, may enhance phosphatidylinositol-3-kinase or NF-κB signaling from TFH cells or FDCs. Additional mutations may constitutively activate these pathways, eliminating dependence on CD4+ T cells and FDCs and promoting transformation.
In contrast to the initial requirement for TFH cell support, evasion of the cytotoxic immune response likely becomes increasingly important for tumor cell survival during progression of the disease as mutations that provide immunogenic epitopes accumulate. We found frequent losses that affected genes (B2M, HLA-A, HLA-B, and HLA-C) encoding HLA class I proteins that present antigens to cytotoxic T cells. 6p aUPD would lead to loss of 1 HLA class I haplotype and thus could decrease interaction with CD8+ T cells. Additionally, recurrent losses including genes encoding proteins important for T-cell and NK-cell interaction and activation (eg, CD58 and CD70) were more common in tFLs; both CD58 and CD70 are known to be lost by multiple mechanisms in de novo DLBCL.55,56 It is high levels of expression of CD70, and not loss, however, that is associated with decreased overall survival in DLBCL,55 demonstrating the challenge of interpreting the consequence of abnormalities. Whether an abnormality is associated with prognosis likely depends on the combination of other abnormalities and the treatment provided. Nevertheless, our data suggest that evading immune surveillance is particularly important in transformed FL.
1p36.33-p36.31 or 1p36.33-p36.12 loss or loss at TNFRSF14 did not predict OS, in contrast to a previous report.3,14 We confirmed and refined the association of 6q deletion with poor survival3,7 and narrowed the regions (6q13-15 and 6q23.3-24.1, Figure 4A-B). Gain of X or Xp, a large gain on chromosome 8 that includes MYC, and a small loss on chromosome 9 that encompasses CDKN2A were associated with poor survival (Figure 4C-F). Seven early/intermediate abnormalities occurred significantly more frequently in tFL cases. FL patients with 1 or more of those abnormalities had significantly worse survival than patients who lacked all these abnormalities (Figure 4G), suggesting that those specific abnormalities may promote rapid progression to tFL or resistance to treatment. Several of these rCNAs have likely target genes (Table 1). We confirmed that FL with more complex abnormalities was associated with worse survival (Figure 4H-I), particularly for whole-chromosome or chromosome-arm abnormalities. Surprisingly, unlike FL, tFL cases with >4 such abnormalities had better survival (Figure 4J). With numerous abnormalities and lacking a supportive microenvironment, tFL may be more sensitive to aggressive treatment.
In conclusion, a comprehensive analysis of rCNAs in FL and tFL identified lesions that may be involved in transformation, and several abnormalities predict a poor outcome. Integration of these rCNAs with mutation and epigenetic studies will clarify which abnormalities drive the disease process, providing possible therapeutic targets.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Zhongfeng Liu for technical assistance.
This work was supported by the Lymphoma Research Foundation Follicular Lymphoma Initiative (W.-C.C.), National Institutes of Health Lymphoma SPORE P50CA136411-01-(NC1) (W.-C.C.), Oncosuisse grant OCS-1939-8-2006 (F.B.), and the Nelia et Amadeo Barletta Foundation (Lausanne, Switzerland) (F.B.).
Authorship
Contribution: K.E.D., W.-C.C., and T.W.M. designed research; K.E.D., C.L., A.B., J.I., and C.K. performed research; T.W.M., A.B., B.J.D., and K.E.D. analyzed data; L.M. Smith., G.W.W., T.W.M., W.Z., and A.B. performed statistical analyses; F.B., A. Rinaldi, J.F., K.F., D.D.W., T.C.G., B.J.D., R.D.G., A. Rosenwald, G.O., E.C., L.M.R., J.D., E.S.J., R.M.B., J.M.C., L.M. Staudt., and W.-C.C. provided materials, reviewed pathology results, or contributed clinical/aCGH/GEP data; and A.B., T.W.M., and W.-C.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wing-Chung Chan, Center for Research in Lymphoma and Leukemia, Department of Pathology and Microbiology, 983135 Nebraska Medical Center, Omaha, NE 68198-3135; e-mail: jchan@unmc.edu.
References
Author notes
A.B. and T.W.M. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal