Key Points
Targeted capture/next-generation sequencing is a powerful tool for the diagnosis of known and discovery of new IGH fusions in DLBCL.
IGH-mediated deregulation of IRF8 and EBF1 in DLBCL is characterized by induction of AID and BCL6, suppression of PRDM1, and antiapoptosis.
Abstract
The characterization of immunoglobulin heavy chain (IGH) translocations provides information on the diagnosis and guides therapeutic decisions in mature B-cell malignancies while enhancing our understanding of normal and malignant B-cell biology. However, existing methodologies for the detection of IGH translocations are labor intensive, often require viable cells, and are biased toward known IGH fusions. To overcome these limitations, we developed a capture sequencing strategy for the identification of IGH rearrangements at nucleotide level resolution and tested its capabilities as a diagnostic and discovery tool in 78 primary diffuse large B-cell lymphomas (DLBCLs). We readily identified IGH-BCL2, IGH-BCL6, IGH-MYC, and IGH-CCND1 fusions and discovered IRF8, EBF1, and TNFSF13 (APRIL) as novel IGH partners in these tumors. IRF8 and TNFSF13 expression was significantly higher in lymphomas with IGH rearrangements targeting these loci. Modeling the deregulation of IRF8 and EBF1 in vitro defined a lymphomagenic profile characterized by up-regulation of AID and/or BCL6, down-regulation of PRMD1, and resistance to apoptosis. Using a capture sequencing strategy, we discovered the B-cell relevant genes IRF8, EBF1, and TNFSF13 as novel targets for IGH deregulation. This methodology is poised to change how IGH translocations are identified in clinical settings while remaining a powerful tool to uncover the pathogenesis of B-cell malignancies.
Introduction
Chromosomal translocations that juxtapose the regulatory regions of the immunoglobulin heavy chain (IGH) to various oncogenes are emblematic of mature B-cell malignancies.1,2 These aberrant fusions characteristically lead to deregulated expression of the IGH partner genes, a well-defined driver event in these tumors’ pathogenesis. In addition, although not pathognomonic, a marked degree of specificity exists between the clinical/histological diagnosis and the IGH partner gene identified, eg, follicular lymphoma and BCL2, diffuse large B-cell lymphoma (DLBCL) and BCL6, mantle cell lymphoma and CCND1, and Burkitt lymphoma and myelocytomatosis viral oncogene homolog (MYC). Thus, characterization of IGH translocations can be of immediate clinical relevance.
Currently, conventional cytogenetics and fluorescence in situ hybridization (FISH) are the principal methodologies used to define these events.3 Both approaches are labor intense and time consuming and have their own unique limitations: the need for viable cells in the former and a bias toward a small number of known translocations in latter. Of note, as IGH rearrangements only rarely result in a fusion mRNA transcript, even more contemporary approaches such as exome and RNA sequencing (RNAseq) are of limited value in this realm.4 To overcome these limitations, we need a tool that can be deployed with the simplest of the materials, DNA, that is completely unbiased as to the identity of IGH partner gene/chromosome and that has the potential to be developed into a rapid and cost-effective clinical assay. Using DLBCL as a tumor model, we show here that IGH capture followed by next-generation sequencing is such a methodology.
Methods
Primary DLBCL samples and cell lines
Frozen biopsies from 78 untreated DLBCL patients were obtained from our local tumor bank, Department of Pathology, University of Texas Health Science Center at San Antonio. The clinical, pathological, and molecular features of this tumor collection were described previously5 and are listed in supplemental Table 1 on the Blood website. The use of these samples was approved by our Institutional Review Board, and this study was conducted in accordance with the Declaration of Helsinki. The DLBCL cell lines SU-DHL-5, SU-DHL-6, SU-DHL-7, OCI-Ly7, and OCI-Ly8 were cultured at 37°C in 5% CO2 in RPMI-1640 medium (Invitrogen) containing 10% (v/v) fetal bovine serum, whereas HEK-293 cells were maintained in Dulbecco’s modified Eagle media (Mediatech) with 10% fetal bovine serum, as described.6
Capture library design and fabrication
A targeted capture library directed at the IGH locus (chromosome 14:106,032,614-107,288,051; hg19) on chromosome 14q32 was designed using the SeqCap EZ Developer product from Roche/NimbleGen. To overcome the limitations imposed by the repetitive nature of this region, we allowed up to 5 close matches between the probes and the genome and the inclusion of nonunique probes throughout the design. This strategy significantly improved the coverage in the 106,032,614 to 106,350,668 region, which stretches from constant genes and switch sequences to the joining and diversity regions. The final design covered 94.8% of the target region at a 100-bp offset from the probes and contained 2.16 million probes (supplemental Figure 1).
Library preparation, target capture, and sequencing
High-molecular-weight DNA was isolated from 78 primary DLBCLs, 3 DLBCL cell lines (SU-DHL-6, OCI-Ly7, and OCI-Ly8), and a nonlymphoid cell line (HEK-293) using Gentra Puregene DNA purification kits (Qiagen). DNA libraries were created using the NEB DNA Library Prep Master Mix for Illumina, as described.7 DNA fragmentation and purification conditions were selected to yield a final library insert size of ∼400 bp to enable efficient capture of off-target translocation features. Paired-end sequencing was performed on the Illumina HiSequation 2000. Detailed methodology, including bioinformatics analysis and the sequence of all oligonucleotides used in this work, is described in the supplemental Methods. A diagrammatic representation of the sample preparation and data analysis pipelines is shown in supplemental Figure 2.
Polymerase chain reaction amplification of novel IGH rearrangements in DLBCLs
DNA from DLBCLs harboring the IGH-IRF8, IGH-EBF, and IGH-TNFSF13/EIF4A1 fusions was used for polymerase chain reaction (PCR) amplifications and Sanger sequencing validation of each rearrangement. Primers were anchored on each of the derivative chromosomes, as defined by the assembly of the read pairs obtained from the next-generation sequencing.
PCR for the detection of the t(14;18) translocation
A PCR approach was used to identify putative fusions between the joining segments of the IGH gene and 3 distinct breakpoint regions at the BCL2 locus, an assay that captures ∼80% of all BCL2/IgH rearrangements.8 Individual PCR reactions were implemented to detect breakpoints in the major breakpoint region, minor cluster region, and intermediate cluster region, as described.9 The quality and quantity of input DNA were confirmed by amplification of a genomic fragment of ∼400 bp mapping to a gene segment on human chromosome 2q37. Primer sequences are listed in the supplemental Materials.
Expression of novel IGH partner genes in DLBCLs
RNA was isolated from 21 nodal DLBCLs analyzed for the presence of IGH rearrangements and cDNA synthesized using SuperScript VILO cDNA synthesis kit (Life Technologies). Expression of IRF8, EBF1, TNFSF13, and EIF4A1 was determined by real-time reverse transcriptase (RT)-PCR. The expression of the target genes was normalized by a housekeeping control (TATA-binding protein), relative quantification was defined using the ΔΔCT method, and expression is reported as 2−ΔΔCT. All assays were performed in triplicate.
Stable expression of IRF8 and EBF1 in DLBCL cell lines
Full-length wild-type IRF8 and EBF1 cDNAs were PCR amplified, sequence verified, and cloned into the murine stem cell virus-enhanced green fluorescent protein retrovirus system. Retrovirus production, transduction, and enrichment of green fluorescent protein–positive populations by fluorescence-activated cell sorter were performed as we described.10 Stable ectopic expression of IRF8 and EBF1 was confirmed by western blot.
Western blots
Protein was isolated in 2% sodium dodecyl sulfate, 4% glycerol, 0.04 million Tris-Hcl, pH = 6.8, and 2 mM 2-mercaptoethanol and was detected using antibodies directed at IRF8, BCL6, AID, PRMD1, EBF1, PAX5, and β-actin, as detailed in supplemental Methods.
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed paraffin-embedded tissue sections and subjected to antigen retrieval (CC1, pH 8.0; Ventana Medical Systems) for 30 minutes in EDTA. The primary antibody, mouse monoclonal anti-human IRF8 (sc-365042; Santa Cruz Biotechnology), was used at a 1:50 dilution and incubated for 32 minutes at 37°C using an automated immunostainer (Benchmark XT; Ventana Medical Systems). Detection was performed with iVIEW detection (Ventana Medical Systems) using 3′3-diaminobenzidine as a chromogen. The slides were counterstained with hematoxylin. Positive staining was localized in the nuclei with a homogeneous pattern. All cases were scored semiquantitatively with respect to the percentage of positive neoplastic cells (0%-5%; 6%-25%; 26%-50%; 51%-75%; 76%-100%) and intensity of staining (negative; weak; moderate; strong).
Apoptosis assay
Apoptosis was measured in response to oxidative or starvation stresses, as described.10 In brief, DLBCL cell lines genetically modified to express IRF8, EBF1, or their isogenic controls were exposed to hydrogen peroxide (10 µM for SU-DHL-5, SU-DHL-7, and OCI-OCI-Ly7, 50 µM for OCI-Ly8) or starved in serum-free conditions (SU-DHL-7, OCI-Ly7, and OCI-Ly8) or 2% serum (SU-DHL-5). Cells were harvested at 24 or 48 hours, resuspended in binding buffer, stained with AnnexinV-PE (#556421; BD BioSciences), and analyzed by flow cytometry on a LSRII (Becton-Dickinson) instrument using the BD fluorescence-activated cell sorter Diva software. Three independent biological replicates, each performed in triplicate, were completed for each measurement.
Statistics
Analyses were performed using a two-tailed Student t test. P < .05 was considered significant. Data analyses were performed in the Prism software (version 5.02; GraphPad Software).
Results
Pilot validation of the capture/sequencing strategy
To test the effectiveness of this capture and downstream sequencing/analysis strategy, we performed a pilot assay with 3 DLBCL cell lines (SU-DHL-6, OCI-Ly7, and OCI-Ly8) with known IGH translocations and a non–B-cell sample (negative control). This initial investigation readily identified IGH-BCL2, IGH-MYC, and IGH-BCL6 fusions in DLBCL and yielded no putative IGH rearrangement in the non–B-cell sample (supplemental Table 2). In 1 cell line (OCI-Ly8), 2 fusions, IGH-BCL6 and IGH-MYC, were recovered, indicating that this methodology efficiently detects multiple rearrangements in a single sample.
Landscape of IGH rearrangements in DLBCLs
We applied the capture sequencing approach toward the entire collection of 78 well-characterized primary DLBCLs available in our group (supplemental Table 1). We conservatively hard-coded in our bioinformatics pipeline a threshold of 20 read-pairs spanning the putative translocation junction for a sample to classify for further analysis (supplemental Methods; supplemental Figure 2). Next, examining the ratio of informative junction-spanning read-pairs to the total read counts, we empirically defined a minimum of ∼6 million read-pairs is required to reach the 20 read-pairs threshold (see supplemental Methods for details). These values (20 read-pairs threshold and 6 million read-pairs) should not be considered absolute but rather as an annotative guideline on how to best discriminate between high confidence negative cases, and cases wherein there may simply be insufficient data for a sample to be analyzed with confidence. Seventy-two DLBCLs from our series (92%) fulfilled our criteria for further analysis. In this cohort, we identified 31 IGH fusions in 28 biopsies (Table 1; supplemental Table 3). Of the previously reported IGH fusions, IGH-BCL2 was the most prevalent aberration, followed by IGH-BCL6 and IGH-MYC (Table 1). In 3 independent tumors, 2 IGH partners were identified (supplemental Table 3). One of these cases (#5782, supplemental Table 3), included both BCL2 and MYC translocations, thus highlighting the ability of this platform to identify the “double-hit” DLBCLs, which associate with an inferior outcome.11 The frequency and partner locus distribution identified are in general agreement with karyotyping and FISH data in nodal DLBCLs.1,2,12,13 In all instances, the breakpoints on both sides of the translocation were mapped at the single nucleotide level (supplemental Tables 2 and 3).
Frequency and characteristics of the IGH rearrangements found by the capture/sequencing methodology in DLBCLs
| Group . | Cases (%) . | IGH locus bkp mapping (cases) . |
|---|---|---|
| No IGH fusion | 44 (61%) | |
| IGH fusion | 28* (39%) | DHJH (16); VHJH (1); DH (1); Sγ (7); Sµ (3); Sα (3) |
| IGH-BCL2 | 16† (22%) | DHJH (14); VHJH(1); Sα (1) |
| IGH-BCL6 | 6 (8.3%) | Sγ (4); Sμ (2) |
| IGH-MYC | 3 (4.1%) | Sα (2); DH (1) |
| IGH-CCDN1 | 2 (2.7%) | DHJH (2) |
| IGH-IRF8 | 2 (2.7%) | Sγ (1); Sμ (1) |
| IGH-EBF1 | 1 (1.4%) | Sγ (1) |
| IGH-EIF4A1/TNFSF13 | 1 (1.4%) | Sγ (1) |
| Total | 72 |
| Group . | Cases (%) . | IGH locus bkp mapping (cases) . |
|---|---|---|
| No IGH fusion | 44 (61%) | |
| IGH fusion | 28* (39%) | DHJH (16); VHJH (1); DH (1); Sγ (7); Sµ (3); Sα (3) |
| IGH-BCL2 | 16† (22%) | DHJH (14); VHJH(1); Sα (1) |
| IGH-BCL6 | 6 (8.3%) | Sγ (4); Sμ (2) |
| IGH-MYC | 3 (4.1%) | Sα (2); DH (1) |
| IGH-CCDN1 | 2 (2.7%) | DHJH (2) |
| IGH-IRF8 | 2 (2.7%) | Sγ (1); Sμ (1) |
| IGH-EBF1 | 1 (1.4%) | Sγ (1) |
| IGH-EIF4A1/TNFSF13 | 1 (1.4%) | Sγ (1) |
| Total | 72 |
Of 72 samples with adequate coverage, 51 are nodal DLBCLs, and 21 are extranodal tumors. The incidence of IGH fusions was 43% in the nodal subset and 28% in the extranodal group.
Three IGH-BCL2–positive samples had a second fusion (supplemental Table 3).
Finally, 2 independent biopsies displayed an IGH-CCND1 fusion, prompting us to review the morphology and immunophenotype of these cases, which had been diagnosed >20 years ago. For both cases, histology demonstrated sheets of medium to large cells with oval to slightly irregular nuclei, dispersed nuclear chromatin, ≥1 nucleoli, numerous mitoses, and a variable number of tingible body macrophages. Review of flow cytometric and immunohistochemical data showed that both cases expressed CD20, CD5, and BCL-2 and were CD10 negative. One case (#517), expressed surface IgM/IgD-κ, and the other (#1309) expressed BCL6 and surface IgM-λ and was CD23 negative. These morphologic and immunophenotypic features could be consistent with a blastoid/pleomorphic variant of mantle cell lymphoma or a DLBCL. However, in conjunction with the discovery of an IGH-CCND1 fusion, these 2 cases of CD5+ B-cell lymphoma would be best classified as blastoid/pleomorphic variants of mantle cell lymphoma.
Informative karyotyping or FISH was available for 23 of 72 DLBCLs investigated, and an agreement between these methodologies and the capture/sequencing strategy was found in all but 2 instances (supplemental Table 4). In 1 case, a t(14;18) identified by G-banding was not detected by our methodology. A PCR-based assay that detects ∼80% of the IGH-BCL2 fusion8,9 was also negative for this DLBCL biopsy (supplemental Figure 1). A closer examination of the features of the capture sequencing strategy in this sample showed that it yielded ample reads (27,374,160 read-pairs) (#8789, supplemental Table 3), but essentially it had no signal above background on chromosome 18 and no consistent read-pairs between chr18 and chr14 to define the translocation. We did note that the “mapping percentage” for this sample was slightly lower than the average (78% vs 90%; supplemental Table 3), but we suggest that the large number of reads indicated above would be sufficient to offset this limitation. Presently, we cannot exclude the possibility that this tumor harbors a very uncommon breakpoint that maps to one of the few areas of low coverage in our capture library (supplemental Figure 1).
In the other discrepant sample, we identified a novel IGH fusion, as detailed below, despite the lack of 14q32 rearrangement as defined by conventional cytogenetics.
Novel IGH rearrangements in DLBCLs
We identified 3 hitherto unreported structural aberrations that bring the IGH locus to close proximity of genes relevant to B-cell biology (Figure 1): 2 DLBCLs displayed an IGH-IRF8 fusion, 1 had an IGH-EBF1 rearrangement, and in a fourth case, the IGH locus was juxtaposed to a gene-rich region on chromosome 17p13, which encompasses TNFSF13 (APRIL). All these novel fusions yield stable reciprocal rearrangements, targeting the switch region on der14, as confirmed by PCR and Sanger sequencing (Figure 1; supplemental Figure 4; supplemental Table 3). Unfortunately, no additional material was available from these cases to develop a FISH assay for detection of these fusions.
Diagrammatic representation of the novel IGH rearrangements identified with the capture/sequencing methodology. (A) IGH-IRF8 fusions were found in two DLCBL biopsies: the breakpoints on chromosome 16q24 mapped on a ∼40-kb range centromeric to the IRF8 locus, which is then translocated to the derivative chromosome 14. At the IGH locus, the breakpoints were located in the switch μ or γ regions, placing IRF8 under the control of the Eμ or Eα enhancers, respectively. (B) In the IGH-EBF1 rearrangement, the breakpoint on chromosome 5q34 mapped ∼1 kb telomeric to the EBF1 gene (transcribed from the telomere to the centromere) and was juxtaposed by a large segment of the IGH locus containing the Eμ enhancer. (C) In another DLBCL, a fusion was identified between the IGH locus and the intron 1-2 of the EIF4A1 gene, on chromosome 17p13. Three other genes, including the B-cell relevant TNFSF13 (APRIL), map immediately telomeric to the breakpoint and could also be influenced by the IGH regulatory elements; both derivative chromosomes are shown. PCR and Sanger sequencing in this case also suggested that this translocation is associated with an inversion within either chromosome 17p13 or 14q32. In each panel, the breakpoints are indicated by arrows, the genes and regulatory elements within the IGH locus are labeled, and the partner genes’ exons are numbered. Sequencing traces for each of the highlighted fusions are also shown.
Diagrammatic representation of the novel IGH rearrangements identified with the capture/sequencing methodology. (A) IGH-IRF8 fusions were found in two DLCBL biopsies: the breakpoints on chromosome 16q24 mapped on a ∼40-kb range centromeric to the IRF8 locus, which is then translocated to the derivative chromosome 14. At the IGH locus, the breakpoints were located in the switch μ or γ regions, placing IRF8 under the control of the Eμ or Eα enhancers, respectively. (B) In the IGH-EBF1 rearrangement, the breakpoint on chromosome 5q34 mapped ∼1 kb telomeric to the EBF1 gene (transcribed from the telomere to the centromere) and was juxtaposed by a large segment of the IGH locus containing the Eμ enhancer. (C) In another DLBCL, a fusion was identified between the IGH locus and the intron 1-2 of the EIF4A1 gene, on chromosome 17p13. Three other genes, including the B-cell relevant TNFSF13 (APRIL), map immediately telomeric to the breakpoint and could also be influenced by the IGH regulatory elements; both derivative chromosomes are shown. PCR and Sanger sequencing in this case also suggested that this translocation is associated with an inversion within either chromosome 17p13 or 14q32. In each panel, the breakpoints are indicated by arrows, the genes and regulatory elements within the IGH locus are labeled, and the partner genes’ exons are numbered. Sequencing traces for each of the highlighted fusions are also shown.
IGH-TNFSF13/EIF4A1 fusion
In this IGH-rearranged DLBCL biopsy, the breakpoint on derivative chromosome 17 (der17) was mapped to the first intron of EIF4A1, which encodes the oncogenic RNA helicase component of the 40S ribosomal subunit14 (Figure 1). Interestingly, TNFSF13, the gene encoding the B-cell proliferation-inducing ligand APRIL,15 maps 15 kb telomeric to the breakpoint, suggesting that it also could be deregulated by the IGH enhancers. To preliminarily investigate the potential contribution of EIF4A1 and TNFSF13 in this instance, we used real-time RT-PCR to quantify the expression of these genes in 20 nodal DLBCLs. The expression of EIF4A1 was not dissimilar among these lymphomas, whereas TNFSF13 expression was 2.8-fold higher in the DLBCL with the IGH-TNFSF13/EIF4A1 fusion than in the group of DLBCLs lacking this rearrangement (supplemental Figure 5).
IGH-IRF8 fusion
Two DLBCL cases in our series harbored an IGH-IRF8 fusion (Figure 1; supplemental Table 3). To start to examine the consequences of this chromosomal aberration, we quantified IRF8 expression by real-time RT-PCR in a collection of 21 nodal DLBCLs. The 2 tumors with the IGH-IRF8 fusion expressed IRF8 at levels markedly higher (6.4- and 4.5-fold, respectively) than the group of 19 DLBCLs without this rearrangement (Figure 2A). This finding was confirmed by immunohistochemistry in 1 rearranged case for which material was available. In brief, IRF8 staining was positive in >76% of neoplastic cells in the 3 DLBCL biopsies examined. Strong, uniform staining was observed in the IGH-IRF8–positive case, whereas a signal of weak to moderate intensity was observed in 2 cases lacking this rearrangement (Figure 2B).
IRF8 role in lymphomagenesis. (A) Real-time RT-PCR–based measurement of IRF8 expression in 21 nodal DLBCLs studied with the capture/sequence methodology demonstrates higher levels in the 2 tumors with the IGH-IRF8 fusion (ID #3271 and #6614). (B) Immunohistochemistry examination confirms the overexpression of IRF8 in a biopsy with an IGH-IRF8 fusion (#6614) compared with 2 DLBCLs lacking this rearrangement (600×). (C) Ectopic expression of IRF8 in 3 DLBCL cell lines led to the emergence of a lymphomagenic profile characterized by heightened expression of BCL6 and AID and suppression of PRMD1. (D) DLBCL cells ectopically expressing IRF8 became significantly resistant to apoptosis induced by H202 and serum deprivation (*P < .01, Student t test); ctrl (control) and 10% indicate the basal apoptosis rate in cells exposed to vehicle or grown in media supplemented with 10% fetal bovine serum, respectively. Bars labeled 2% or 0% correspond to cells grown in serum-deprived conditions. All data points were collected in triplicate at 24 hours, and the results were confirmed in 3 independent biological replicates; the results displayed represent the mean and standard deviation of a biological replicate.
IRF8 role in lymphomagenesis. (A) Real-time RT-PCR–based measurement of IRF8 expression in 21 nodal DLBCLs studied with the capture/sequence methodology demonstrates higher levels in the 2 tumors with the IGH-IRF8 fusion (ID #3271 and #6614). (B) Immunohistochemistry examination confirms the overexpression of IRF8 in a biopsy with an IGH-IRF8 fusion (#6614) compared with 2 DLBCLs lacking this rearrangement (600×). (C) Ectopic expression of IRF8 in 3 DLBCL cell lines led to the emergence of a lymphomagenic profile characterized by heightened expression of BCL6 and AID and suppression of PRMD1. (D) DLBCL cells ectopically expressing IRF8 became significantly resistant to apoptosis induced by H202 and serum deprivation (*P < .01, Student t test); ctrl (control) and 10% indicate the basal apoptosis rate in cells exposed to vehicle or grown in media supplemented with 10% fetal bovine serum, respectively. Bars labeled 2% or 0% correspond to cells grown in serum-deprived conditions. All data points were collected in triplicate at 24 hours, and the results were confirmed in 3 independent biological replicates; the results displayed represent the mean and standard deviation of a biological replicate.
Stable ectopic expression of IRF8 in DLBCLs
To investigate how constitutive expression of IRF8 could contribute to lymphomagenesis, we generated DLBCL cell lines ectopically expressing this transcription factor. In DLBCLs, stable IRF8 expression induced BCL6 and AID and suppressed PRDM1 expression (Figure 2C). Constitutive expression of IRF8 also rendered 3 independent DLBCL cell lines significantly more resistant to apoptosis induced by serum starvation and reactive oxygen species than their isogenic controls (Figure 2D).
IGH-EBF1 fusion
We used real-time RT-PCR to determine EBF1 expression within a cohort of primary DLBCLs. Unlike TNFSF13 and IRF8, EBF1 was not overexpressed in the IGH-EBF1–containing lymphoma (supplemental Figure 6A), suggesting that loss of normal regulatory control, not elevated levels, may play a dominant role in this instance.
Stable ectopic expression of EBF1 in DLBCL
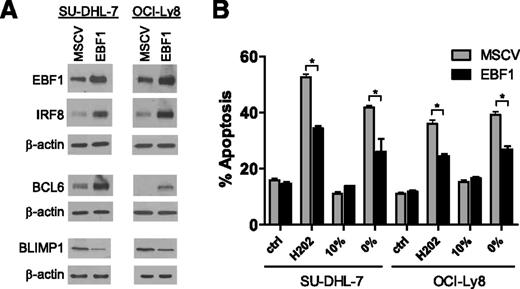
In murine B cells, EBF1 has been shown to induce the expression of PAX5 and IRF8.16,17 We generated a genetic model of EBF1 ectopic expression in DLBCL to define whether these events were relevant in human malignant mature B cells. In 2 independent DLCBL cell lines, EBF1 induced IRF8 but not PAX5 expression (Figure 3A; supplemental Figure 6B). The description that EBF1 occupies the BCL6 and PRDM1 promoters in murine and chicken B cells,18,19 respectively, prompted us to investigate these relationships in a DLBCL context. We showed that BCL6 is induced and PRMD1 suppressed in DLBCL cell lines ectopically expressing EBF1 (Figure 3A). Finally, we demonstrated that ectopic expression of EBF1 in DLBCL established an antiapoptotic profile (Figure 3B).
EBF1 role in lymphomagenesis. (A) Ectopic expression of EBF1 in 2 independent DLBCL cell lines induced IRF8 and BCL6 expression while decreasing PRMD1 levels. (B) DLBCL cells ectopically expressing EBF1 became significantly resistant to apoptosis induced by H2O2 and serum deprivation (*P < .01, Student t test); ctrl (control) and 10% indicate the basal apoptosis rate in cells exposed to vehicle or grown in media supplemented with 10% fetal bovine serum, respectively. Bars labeled 0% correspond to cells grown in serum-deprived conditions The H2O2 data were collected at 24 hours and the starvation data at 48 hours; these results were confirmed in 3 independent biological replicates, and the data displayed represent the mean and standard deviation of a biological replicate.
EBF1 role in lymphomagenesis. (A) Ectopic expression of EBF1 in 2 independent DLBCL cell lines induced IRF8 and BCL6 expression while decreasing PRMD1 levels. (B) DLBCL cells ectopically expressing EBF1 became significantly resistant to apoptosis induced by H2O2 and serum deprivation (*P < .01, Student t test); ctrl (control) and 10% indicate the basal apoptosis rate in cells exposed to vehicle or grown in media supplemented with 10% fetal bovine serum, respectively. Bars labeled 0% correspond to cells grown in serum-deprived conditions The H2O2 data were collected at 24 hours and the starvation data at 48 hours; these results were confirmed in 3 independent biological replicates, and the data displayed represent the mean and standard deviation of a biological replicate.
Discussion
We showed that targeted capture followed by next-generation sequencing is an effective strategy to diagnose known and discover new IGH rearrangements in B-cell lymphomas; this finding is in agreement with a recent report using a similar strategy in multiple myeloma.20 This methodology is implemented with DNA, thus abbreviating the need to obtain viable cells, a particularly important consideration in solid tumors, including lymphomas. Further, with this technical approach, all IGH fusions are immediately mapped at high resolution, allowing not only for the identification of the partner genes but also informing on the mechanism for the aberrant rearrangement (ie, variable diversity joining or class switch recombination). This nucleotide level output can also serve as a blueprint for the design of patient-tailored PCR-based approaches, which may be useful to monitor minimal residue disease. These are all important departures from current methodological standards used to identify IGH translocations.3,4
The potential for discovery of new IGH fusions further supports the relevance of this new strategy. Indeed, in this initial cohort of DLBCL, we identified 3 novel IGH rearrangements, all targeting B-lymphocyte relevant genes not previously shown to be disrupted by chromosomal translocation in mature B-cell malignancies. In all these cases, the breakpoints on der14 were mapped to the switch regions of the constant genes, suggesting the involvement of CSR in these aberrant processes.2
One of the new rearrangements brings the IGH regulatory elements to a gene-rich region on chromosome 17p13, suggesting that it may impact on transcription units on both derivative chromosomes. The breakpoint in this case maps to the first intron of EIF4A1, a putative oncogene in its own right,21 thus disrupting its reading frame that now would require an alternative ATG (in exon 2) for translation initiation. However, perhaps more relevant in this B-cell lymphoma context, TNFSF13 (APRIL), known for its potential pathogenic role in both autoimmunity and cancer,15 is located only 15 kb telomeric to breakpoint on chromosome 17p13. Although we cannot currently exclude the possibility that deregulation of genes on both derivative chromosomes play a role in this instance, as is the case for t(4;14)(p16.3;q32.3) found in myeloma,22 the relevance of APRIL to B-cell biology, the anatomy of the breakpoint in the EIF4A1 locus, and the significantly higher expression of TNFSF13 (but not EIF4A1) in this biopsy, all point to IGH-TNFSF13 as the main pathogenetic event in this rearrangement.
IRF8 is a transcription factor relevant for multiple hematopoietic lineages.23 It plays important roles throughout B-cell development, from the control of pre-B to B-cell transition to the germinal center reaction.16,23,24 It also often displays a reciprocal expression pattern with IRF4 that may be important in the transition of germinal center B cells to terminally differentiated plasma cells.24,25 Thus, deregulated expression of IRF8, which follows the acquisition of an IGH-IRF8 fusion, should be expected to contribute to lymhomagenesis. We were able to define some of steps in this process: we showed that in DLBCL models, IRF8 induces BCL6 expression, as has been reported in murine B cells,24 and suppresses PRMD1 expression, an event that may be secondary to BCL6 induction.26 Adding to this lymphomagenic profile, IRF8 also induced AID expression,24 which has the potential to generate further genomic stability and the emergence of aberrant somatic hypermutation or additional class switch recombination–mediated translocations. Certainly, because IRF8 is a transcription factor, the breadth of its dysfunction in mature B cells may go beyond the up-regulation of the oncogenic BCL6 and AID and down-regulation of the tumor suppressive protein PRDM1, although these event may be sufficient for lymphomagenesis. Indeed, our findings appear to suggest that the IGH-IRF8 fusion is a driver event in lymphoma biology. In agreement with this concept, in recent examinations of the DLBCL genome, 2 independent groups reported IRF8 as a target for somatic mutations.27,28 Albeit still functionally uncharacterized, these nucleotide changes displayed several features typical of oncogenic driver mutations29 : they were recurrent in a statistically significant manner, heterozygous and predominantly missense and clustered in the IRF8 DNA binding domain. Finally, we propose that the translocations targeting the IGH regulatory elements to the IRF8 locus may be more pervasive than it is immediately apparent. Using 24-color metaphase FISH, Bernicot et al12 identified 2 DLBCLs with a cryptic translocation, t(14;16)(q32;q2?2), which we propose reflects the IGH-IRF8 rearrangement that we are now reporting.
The potential involvement of EBF1 in the pathogenesis of DLBCLs closely recapitulates the PAX5 paradigm. Both are transcription factors with essential roles in early B-cell development, which function as tumor suppressor genes in B-acute lymphoblastic leukemia.16,30-32 Conversely, in B-cell lymphomas, PAX5 activation via IGH translocation yields a putative gain-of-function phenotype33,34 ; based on the findings reported here, we propose that EBF1 also acts as an oncogene in mature B cells. In addition, although the functional consequences for these mutations have not yet been established, both PAX5 and EBF1 are targeted by aberrant somatic hypermutation in DLBCLs.35,36 Interestingly, one of the main transcriptional targets of EBF1 in early B-cell development is PAX5. In our DLBCL model, constitutive expression of EBF1 did not influence PAX5 levels, in agreement with recent suggestions that the EBF1 regulatory network is markedly distinct at different levels of B-cell maturation.37 Instead, we found that the events of pathogenetic significance downstream to EBF1 include the engagement of an IRF8-BCL6-PRDM1 axis. In this model, EBF1 may directly induce BCL618 or modulate it via IRF8. Likewise, PRMD1 suppression could be a direct response to EBF1 activity19 or secondary to BCL6 up-regulation. Remarkably, irrespective of its direct or indirect transcriptional control of target genes, in our model, constitutive expression of EBF1 largely recapitulated the effects of IRF8 overexpression.
Here, we defined the value of a new methodology to identify IGH translocations in DLBCLs, discovered 3 new IGH fusion partners, and started to delineate the lymphomagenic properties of IRF8 and EBF1. Examination of additional cohorts of DLBCL and other mature B-cell malignancies will help define the actual incidence of these translocations and clarify whether they associate with any unique clinical, pathological, or molecular features found in these tumors. We postulate that the capture/sequencing strategy reported here can be expanded to other mature B-cell malignancies20 and adapted to the diagnosis and discovery of T-cell receptor translocations in T-cell malignancies. Although still computationally demanding, this approach has the potential to become a cost-effective clinical test that circumvents many of the limitations of FISH and conventional karyotyping in the diagnosis and clinical management of B-cell malignancies.
Note added in proof.
A paper has now appeared (Tinguely et al, Leuk Lymphoma, 2013, Jun 12) validating the observation that the t(14;16) (q32;q24) found in DLBCL results in an IGH-IRF8 fusion.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Patricia Dahia for productive discussions and suggestions throughout the execution of this project, Dhivya Arasappan and Benjamin Goetz for assistance in preparing figures and mapping statistics, and Kate Kim for technical help.
This work was supported by a grant from the Cancer Prevention Research Institute of Texas (CPRIT RP120372), Young Investigator Award from the Voelcker Fund (both to R.C.T.A.), and Cancer Center support grant P30 CA054174.
National Institutes of Health
Authorship
Contribution: H.B., S.A., A.-P.L., L.W., and D.J. conducted experiments; K.N.H. and M.C.K. provided well-characterized patient samples and performed immunohistochemistry analysis; S.H.-S. coordinated the next-generation sequencing and bioinformatics analysis; and R.C.T.A. conceived the project, designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ricardo Aguiar, Department of Medicine, University of Texas Health Science Center at San Antonio, 7703 Floyd Curl Dr, San Antonio, TX 78229; e-mail: aguiarr@uthscsa.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal