In this issue of Blood, Johnsen et al have analyzed the frequency of coding sequence variants in the von Willebrand factor gene (VWF) and have identified 7 missense variants independently associated with levels of von Willebrand factor (VWF) or factor VIII (FVIII).1 Several rare missense variants have been previously identified, predominantly in European patients with von Willebrand disease (VWD),2,3 and some have now been reported to be common African American sequence variations.
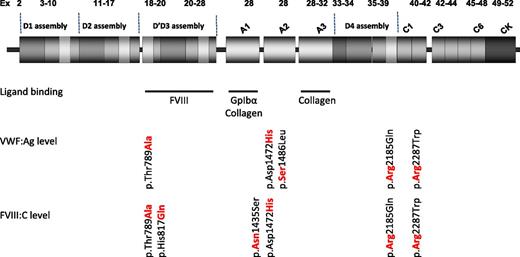
VWF alleles with an independent effect on VWF and FVIII levels. Independent effect on levels was evaluated by Johnsen et al by performing a multivariable regression model adjusted for age, sex, genetic ancestry, and the remaining missense variants.1 Updated structure of VWF (Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA. Sequence and structure relationships within VWF. Blood. 2012;120(2):449-458) showing ligand-binding regions for FVIII, GpIbα and collagen are indicated by blue bars. Amino acid residues shown in bold red text are associated with higher VWF or FVIII levels. Ex, exon.
VWF alleles with an independent effect on VWF and FVIII levels. Independent effect on levels was evaluated by Johnsen et al by performing a multivariable regression model adjusted for age, sex, genetic ancestry, and the remaining missense variants.1 Updated structure of VWF (Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA. Sequence and structure relationships within VWF. Blood. 2012;120(2):449-458) showing ligand-binding regions for FVIII, GpIbα and collagen are indicated by blue bars. Amino acid residues shown in bold red text are associated with higher VWF or FVIII levels. Ex, exon.
Differences in VWF levels between individuals of African origin and whites have been recognized for several years, and the basis for the difference has been investigated by the Zimmerman Program for the Molecular and Clinical Biology of VWD, which recently demonstrated that several previously reported VWF mutations were present in African Americans with normal VWF levels and no bleeding symptoms.4 A further recent study by Wang et al analyzed individuals from 14 different population groups and also highlighted that a small number of presumed VWF mutations that were rare in Europeans with type 1 and 2N VWD were common in African individuals.5 Both of these publications highlighted some of the same missense variants identified in the current study to be associated with alterations in VWF or FVIII level. Johnsen and colleagues have extended analysis of African American subjects to a panel of 4468 individuals, enabling the extent of effect on VWF and FVIII levels to be determined for each variant allele.1
Two of the seven variants identified by Johnsen et al that were independently associated with VWF level were particularly prevalent in African Americans: p.Thr789Ala (D' domain) and p.Asp1472His (A2 domain; see figure). p.Thr789Ala has been reported to have an association with risk of atherosclerosis and may be considered as a marker of endothelial dysfunction in elderly individuals.6 The current study demonstrated that each additional copy of the alanine allele contributes 6 to 8 IU/dL to levels of VWF antigen (VWF:Ag) and FVIII coagulant activity (FVIII:C), so that individuals homozygous for alanine may have average levels 15 IU/dL higher than those lacking it.
p.Asp1472His has been recognized recently to be associated with an elevated VWF:Ag level and with a reduced ratio of VWF:Ag to ristocetin cofactor activity (VWF:RCo/VWF:Ag) in individuals with 1 or 2 copies of the histidine allele, the majority of whom were African American.7 The effect is attributed to the histidine allele decreasing ristocetin-mediated interaction between VWF and GpIbα but likely has no physiological consequence in vivo. Johnsen and colleagues have confirmed the association with elevated VWF:Ag level in a much larger population that also results in an increment of 6 to 8 IU/dL VWF:Ag and FVIII:C per histidine allele.
The VWF p.His817Gln substitution had been previously associated with 2N VWD, and although it substantially reduced FVIII:C levels by 18 IU/dL per allele, it may not reduce levels sufficiently to often result in symptomatic 2N VWD. In contrast, for 2 missense variants that were rare among African Americans (p.Ser1486Leu and p.Arg2287Trp), each copy of the variant allele was associated with a 30- to 40-IU/dL alteration in VWF levels, and these variants were confirmed as likely to be pathogenic. The majority of the total of 30 missense variants identified by the study had no significant association with VWF or FVIII levels, suggesting that they are neutral variants.1
The VWF gene is relatively large, covering 178 kb of genomic DNA, with a 52-exon complementary DNA encoding the 2813 amino acids that comprise the large VWF monomer. Since the first sequence analysis of the gene in 1989, VWF has been recognized as having a high rate of sequence variation between individuals. Next-generation DNA sequence analysis (NGS) is a technique that enables many genes to be simultaneously sequenced. It was used in the current National Heart, Lung, and Blood Institute exome sequencing project to identify sequence variation in some of the study participants. The technique has been used in large-scale research studies and is beginning to be introduced for smaller-scale analysis of VWF along with other genes both in research and diagnostic settings.8 Until now, genetic analysis for many patients with VWD has been targeted to particular functional domains, especially those binding FVIII and GpIbα (see figure). Future use of NGS will facilitate rapid sequencing of the entire VWF coding region more rapidly and cost-effectively than current techniques allow. This will result in the identification of pathogenic and neutral sequence variations in addition to those that have a more minor effect on modifying VWF levels, such as p.Thr789Ala. Understanding the contribution of specific VWF sequence variants to phenotype is essential in discriminating those that influence VWF and FVIII level in patients with VWD from those that do not. There are currently 5856 variants documented within VWF on the single-nucleotide polymorphism database,9 averaging 1 every 30 nucleotides, and understanding their potential contribution to VWF level variation presents a significant challenge. For laboratories undertaking VWF sequence analysis, the additional information contained in the supplementary information accompanying this publication on allele frequency and prediction of pathogenicity of over 400 VWF sequence variants in African American and European American individuals presents a goldmine of information on the interpretation of sequence variation.
Analysis of VWF genotype in further large cohorts of individuals from additional population groups with known VWF and FVIII levels will extend our knowledge of the contribution of VWF sequence variation to phenotypic variability.
Conflict-of-interest disclosure: The author declares no competing financial interests.