Key Points
We provide a functional DNA methylation map of human leukocyte subsets and identify cell-type–specific regulatory HMRs.
We illustrate use of this data by demonstrating a potential link between gene polymorphisms, DNA methylation, and immune-mediated disease.
Abstract
DNA methylation is an important mechanism by which gene transcription and hence cellular function are regulated. Here, we provide detailed functional genome-wide methylome maps of 5 primary peripheral blood leukocyte subsets including T cells, B cells, monocytes/macrophages, and neutrophils obtained from healthy individuals. A comparison of these methylomes revealed highly specific cell-lineage and cell-subset methylation profiles. DNA hypomethylation is known to be permissive for gene expression and we identified cell-subset–specific hypomethylated regions (HMRs) that strongly correlate with gene transcription levels suggesting these HMRs may regulate corresponding cell functions. Single-nucleotide polymorphisms associated with immune-mediated disease in genome-wide association studies preferentially localized to these cell-specific regulatory HMRs, offering insight into pathogenesis by highlighting cell subsets in which specific epigenetic changes may drive disease. Our data provide a valuable reference tool for researchers aiming to investigate the role of DNA methylation in regulating primary leukocyte function in health and immune-mediated disease.
Introduction
In complex organisms, different cell types display distinct gene expression profiles, which ultimately shape the function and phenotype of each tissue. Evidence is accumulating for a major role for epigenetic mechanisms in regulating tissue and cell-type–specific gene expression.1-3 The most extensively studied epigenetic mechanism is DNA methylation, which occurs at the 5′ position of the pyrimidine ring of cytosines, primarily, but not exclusively, in the context of the dinucleotide sequence CpG (5meC).4-7 Importantly, DNA methylation has been shown to regulate gene transcription and hence cellular function through its effect on DNA accessibility state.6,8 In principle, increased methylation of CpGs, particularly when located around transcription start sites (TSSs) that contain high densities of CpGs (ie, CpG islands [CGIs]), is associated with a condensed chromatin state leading to silencing of the respective gene. Hypomethylation has the opposite effect. However, recent studies have unraveled further, more complex mechanisms by which DNA methylation regulates gene expression, for example, the effect of intragenic methylation at alternative promoter regions.9
In addition to its role in fundamental biological processes such as x-chromosome inactivation, stem cell renewal, and genomic imprinting, aberrant DNA methylation is being increasingly implicated in causing disease.6 The fact that the epigenome is dynamic and can be modified in response to environmental factors has placed epigenetics in the focus of research into complex diseases including several immune-mediated conditions.10,11
To date our knowledge of the physiological role of DNA methylation in regulating gene expression/function in our immune system remains limited. This is primarily due to a lack of genome-wide DNA methylation data combined with gene expression analysis from purified primary human cell subsets.1,12-14 In this study, we set out to investigate the role of DNA methylation in regulating gene expression in primary purified subsets of leukocytes. We simultaneously extracted DNA and RNA from unseparated peripheral blood mononuclear cells (PBMCs) as well as purified CD4+ and CD8+ T cells, CD19+ B cells, CD14+ monocytes, and CD16+ neutrophils isolated from 6 healthy male individuals. For each sample, we generated detailed genome-wide methylome maps using the Infinium HumanMethylation450 BeadChip from Illumina15 and exon-level gene expression profiles using Affymetrix Human Gene 1.1 ST arrays. In addition, we generated methylation profiles using methylated DNA immunoprecipitation followed by second generation sequencing (MeDIP-seq) for all cell subsets and PBMCs from 1 individual.16,17 Our analyses reveal distinct lineage and cell-subset–specific DNA methylation profiles and identify cell-type–specific hypomethylated regions (HMRs) that correlate with gene transcription levels, suggesting a role for methylation in driving cell-type–specific gene expression. Our data provide a reference resource for future studies aiming to investigate the role of DNA methylation in health and immune-mediated diseases. The majority of these potential regulatory HMRs (rHMRs) are located in CGI shelves and shores, providing further evidence that regions outside CGIs play a major role in regulating gene expression. We demonstrate that single-nucleotide polymorphisms (SNPs) associated with a predisposition to several immune-mediated diseases (eg, Crohn's disease and type 1 diabetes[T1D]) are located within these cell-type–specific HMRs, suggesting a potential mechanism for the contribution of these SNPs to the predisposition of specific immune-mediated diseases.
Methods
Healthy volunteers
For this study, we recruited 6 white male volunteers aged 36 to 51 years. All individuals had no known current or previous medical conditions, were nonsmokers, and were not taking any regular medication (supplemental Table 1, available on the Blood Web site). Following informed consent in accordance with the Declaration of Helsinki, 100 mL of whole blood were collected from each individual and processed immediately. Venepuncture was performed at a similar time of day to minimize gene expression differences arising from circadian variation. Ethical approval for the study was obtained from the Cambridge Local Research Ethics Committee (08/H0308/176).
Cell separation
Cell separations of PBMCs and individual primary leukocyte cell subsets were performed as previously described.18 Full details are provided in the supplemental Methods.
DNA and RNA extraction
DNA and RNA were extracted simultaneously from the same sample using the AllPrep DNA/RNA mini kit (QIAGEN) following the manufacturer’s instructions. Following nucleic acid extraction, quality and quantity were assessed using an Agilent 2100 Bioanalyzer and a NanoDrop ND-1000 spectrophotometer, respectively.
Affymetrix Human Gene 1.1 ST array
Aliquots of total RNA (200 ng) were labeled using Affymetrix’s WT Sense Target labeling kit and hybridized to Human Gene 1.1 ST arrays (Affymetrix) following the manufacturer’s instructions. After washing, arrays were scanned using a Gene Titan instrument (Affymetrix).
DNA methylation analysis
Genome-wide DNA methylation analysis was performed on bisulphite-converted DNA using the Infinium HumanMethylation450 Bead-Chip according to the manufacturer’s instructions. In addition, MeDIP-seq was performed on a subset of samples. Genomic DNA (gDNA) (5 µg) was randomly sheared to a median fragment size of 200 base pairs (bp) and Illumina paired-end libraries prepared. MeDIP was performed using a Zymo Research IP kit with minor modifications. DNA library sequencing was performed on an Illumina Genome Analyzer II using 76 cycles. Full details are provided in the supplemental Methods. Infinium HumanMethylation450 Bead-Chip and MeDIP-seq data were validated using sequencing of bisulfite-converted DNA as described previously.19
Statistical analysis
Raw Illumina Infinium HumanMethylation450 BeadChip (K450) data were imported to the GenomeStudio (Version 2010.3) methylation module (1.8.2), which was used to extract the image intensities. The resulting data were imported into the R statistical programming language, and the lumi package from Bioconductor was used for normalization and further analysis.20
Differential methylation analysis (locus/probe level): methylation status for each probe was calculated using the M value, which is defined as the log2 ratio of methylated probe intensity and unmethylated probe intensity.21 Differential methylation at individual probe loci was assessed using the limma Bioconductor package22 to analyze the M-value data, setting a false discovery rate (FDR)-adjusted P value threshold of <.05 and a log fold-change cutoff >1.5.
For differentially methylated region analyses, the “gammaFitEM” function of the lumi package was used to fit a 2-component γ mixture model. The output of this function calls the methylation status of each probe as either “unmethylated,” “marginal,” or “methylated.” Subsequent analysis defined all marginal calls as “partially methylated.” HMRs were defined as regions spanning over ≥20 bp containing a minimum of 2 unmethylated or partially methylated CpG sites for 80% or more of the calls for each probe. Regions containing ranges of >500 bp without any Illumina probes were split into separate regions. Cell-type–specific differentially HMRs (dHMRs) were identified by comparison of each given cell subset to all remaining subsets.
Affymetrix expression array analysis was performed as described previously. Briefly, data were normalized using the variance-stabilizing normalization algorithm23 and the expression signal from probe sets representing exons summarized using robust multi-array analysis.24 These exon-level probe sets were mapped to known transcripts in the Ensembl database (www.ensembl.org). Transcripts were called as differentially expressed where 80% or more of their mapped exon probe set loci displayed differential expression in a comparison of the given cell subset to any of the remaining subsets (FDR-adjusted P < .05 and log fold-change >2).
Cell-subset–specific HMRs were described as having a positive correlation with gene transcript expression if they were located within 2000 bp of the TSS of a given transcript and a minimum of a 2-log–fold increase in transcript expression was observed.
Overlap of genome-wide association study (GWAS)–identified SNPs with differential HMRs was analyzed as follows. Linkage disequilibrium (LD) blocks were calculated for each SNP using the 1000 genomes phase 1 data to identify highly correlated nearby SNPs (r2 > 0.8). Significance of overlap of these LD blocks with HMRs, dHMRs, and rHMRs was determined by assessing the equivalent HMR overlap of 1000 randomly generated sets of GWAS SNPs, applying a normal approximation to generate the null distribution in each case.
MeDIP-Seq analysis was performed entirely using Bioconductor. The MEDIPS package (www.bioconductor.org) was used to generate absolute methylation status values, expressed as a normalized value between 0 and 1000, from the Burrows-Wheeler Alignment-mapped MeDIP-seq reads.25 Values below 200 were treated as fully unmethylated. Receiver operating characteristic (ROC) analysis was performed using the ROC Bioconductor package in R to compare methylation calls at each Illumina probe locus to that obtained using MEDIPS.
Results
Primary human leukocyte subsets have distinct DNA methylomes
We generated genome-wide methylation profiles for each of 5 purified cell subsets, including B cells (CD19+), T cells (CD4+ and CD8+), monocytes/macrophages (CD14+), neutrophils (CD16+) as well as PBMCs using the Infinium HumanMethylation450 BeadChip from Illumina (referred to as K450 array), which interrogates the methylation status of ∼480 000 CpG sites distributed across the human genome.15 CpG sites were classified as unmethylated, partially methylated, or methylated (see “Methods”). Although the total number of fully methylated CpG sites did not vary greatly between cell types, there were fewer unmethylated sites, and correspondingly, more sites showing partial methylation in cells of the lymphoid lineage (CD19+ B cells, and CD4+ and CD8+ T cells) than in cells of the myeloid lineage (CD14+ monocytes and CD16+ neutrophils) (myeloid:lymphoid ratio = 1.7, P = 7.2 × 10−6, Figure 1). Determination of the genomic context of the CpG sites revealed that unmethylated and partially methylated sites were more likely to be located upstream of or within TSSs (odds ratio = 3.8, Figure 1A) and be associated with CGIs or shores regardless of cell type (odds ratio = 8.6, Figure 1B). The methylated sites were more likely to be in the “open sea.”
Cell-subset–specific DNA methylation profiles of purified primary human leukocytes. (A-B) Genome-wide DNA methylation profiles of purified leukocyte cell subsets using the Illumina Infinium HumanMethylation450 BeadChip. The DNA methylation status of each CpG site was classified as “unmethylated,” “partially methylated,” or “methylated” and the number of probes of each status plotted per cell type. (A) Genomic location of individual sites in relation to TSSs. Location of probes referred to as within TSSs, 200 bp upstream or downstream from the TSSs, and far upstream and downstream (200-1500 bp from TSSs). (B) Distribution of sites with regard to their position relative to CGIs. CpG shores were defined as the 2-kb regions flanking CGIs, and the shelves as 2-kb regions extending from the shores. Probes located further away from CGIs are defined as being in open sea.
Cell-subset–specific DNA methylation profiles of purified primary human leukocytes. (A-B) Genome-wide DNA methylation profiles of purified leukocyte cell subsets using the Illumina Infinium HumanMethylation450 BeadChip. The DNA methylation status of each CpG site was classified as “unmethylated,” “partially methylated,” or “methylated” and the number of probes of each status plotted per cell type. (A) Genomic location of individual sites in relation to TSSs. Location of probes referred to as within TSSs, 200 bp upstream or downstream from the TSSs, and far upstream and downstream (200-1500 bp from TSSs). (B) Distribution of sites with regard to their position relative to CGIs. CpG shores were defined as the 2-kb regions flanking CGIs, and the shelves as 2-kb regions extending from the shores. Probes located further away from CGIs are defined as being in open sea.
To analyze the relationship between the different methylation profiles of each cell type and PBMCs, we performed principal component analysis (PCA) and hierarchical clustering. This revealed distinct clustering of individual cell subsets (Figure 2A, supplemental Figure 1), with the first principal component, which explains ∼52% of the variance, primarily separating cell subsets derived from the myeloid and lymphoid lineages. Although these data illustrate that each individual cell type has its unique methylome, the proximity of the CD4+ and CD8+ T-cell clusters suggests that functionally related cell types exhibit similar DNA methylation profiles (Figure 2A, supplemental Figure 1). Unseparated PBMC samples fall in the center of the PCA plot (Figure 2A) reflecting that this sample is composed of a mixture of 4 of the 5 cell subsets (CD16+ neutrophils are excluded by Ficoll centrifugation and not found in the PBMC fraction). This highlights the importance of cell separation prior to performing DNA methylation analyses, as has been shown for gene transcription analyses.26
PCA and differential methylation analysis of human leukocyte subsets. (A) PCA of leukocyte subsets and whole PBMC DNA methylation profiles. (B) Differential methylation analysis illustrating the number of differentially methylated loci between individual subsets. Analyses were performed on Illumina Infinium HumanMethylation450 BeadChip data for each cell type.
PCA and differential methylation analysis of human leukocyte subsets. (A) PCA of leukocyte subsets and whole PBMC DNA methylation profiles. (B) Differential methylation analysis illustrating the number of differentially methylated loci between individual subsets. Analyses were performed on Illumina Infinium HumanMethylation450 BeadChip data for each cell type.
We then compared DNA methylomes from the individual cell subsets with each other in order to identify both cell-type and lineage specific methylation differences. The number of differentially methylated loci varied greatly according to whether the comparison was between cell types of the same or different lineages (Figure 2B). For example, the comparison of CD8+ T cells and CD14+ monocytes identified 80 347 methylation variable positions while a comparison of CD14+ and CD16+ cells revealed only ∼14 948 methylation variable positions (Figure 2B).
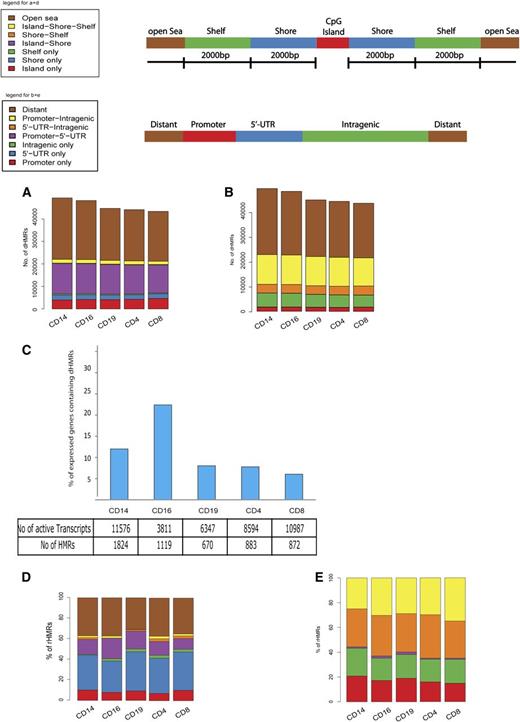
We also observed distinct clustering of hypomethylated and partially methylated CpG sites. The number of HMRs, total HMR length, and HMR length distribution for each cell type are illustrated in supplemental Figure 2. We defined these clusters as HMRs if they spread over >20 bp and contained a minimum of 2 unmethylated or partially methylated CpG sites (see “Methods”). We identified cell-subset–specific HMRs (ie, dHMRs) that were present in only 1 cell type, or, where there was a cell-subset–specific expansion (of >20 bp) to a HMR that was shared between several cell subsets (see “Methods”). Analysis of the distribution of dHMRs across the genome of the leukocyte subsets showed that dHMRs were more numerous in myeloid compared with lymphoid cell subsets (Figure 3A), supporting our initial findings at an individual CpG site level (Figure 1A). Analysis of the distribution of dHMRs across the genome revealed that a significant number of dHMRs were located in “open sea” regions, that is, not associated with CGIs (Figure 3A). dHMRs that were associated with CGIs were rarely found entirely within the CGI itself, but were usually located in CpG shores and shelves (Figure 3A). Moreover, cell-type–specific methylation differences seem to occur most frequently outside CGIs, in adjacent shelves and shores. dHMRs were most frequently found in areas distant from genes. Of those associated with genes, the majority overlap 5′ promoter and intragenic regions, with only a small fraction found exclusively within promoter regions (Figure 3B). Gene ontology analysis performed on genes associated with cell-type–specific dHMRs revealed enrichment for relevant cellular immune functions suggesting that at least a subset of these dHMRs is involved in regulating specific cellular phenotypes (supplemental Table 2). dHMRs found in “open sea” regions showed no association with genomic features such as centromere, telomere, or microRNA genes but were found to be preferentially associated with L1 long interspersed elements repeats compared with dHMRs associated with CGIs (data not shown).
Identification of cell-subset–specific HMRs and potential rHMRs. (A-B) Total number of cell-subset–specific identified HMRs are plotted. (A) Location of HMRs relative to CGIs. Shelves, shores, and open sea regions are defined as for Figure 1. (B) Distribution of HMRs relative to promoter regions, 5′ UTRs, intragenic and away from gene coding areas (ie, distant). (C) Total number and percentage of dHMRs correlating with differential gene transcript expression by cell type. (D-E) Proportion and distribution of cell-type–specific HMRs correlating with gene expression (ie, potential rHMRs) with regard to CGIs (D) and genomic location (E). UTR, untranslated region.
Identification of cell-subset–specific HMRs and potential rHMRs. (A-B) Total number of cell-subset–specific identified HMRs are plotted. (A) Location of HMRs relative to CGIs. Shelves, shores, and open sea regions are defined as for Figure 1. (B) Distribution of HMRs relative to promoter regions, 5′ UTRs, intragenic and away from gene coding areas (ie, distant). (C) Total number and percentage of dHMRs correlating with differential gene transcript expression by cell type. (D-E) Proportion and distribution of cell-type–specific HMRs correlating with gene expression (ie, potential rHMRs) with regard to CGIs (D) and genomic location (E). UTR, untranslated region.
Cell-type–specific HMRs are associated with cell-type–specific gene expression
To investigate the possible effect of cell-type–specific dHMRs on gene expression, messenger RNA levels were measured by microarray analysis and the location of the differentially expressed genes compared with the position of the dHMRs. Several hundred cell-specific dHMRs were found to be associated with differentially expressed genes (located in close proximity to the respective TSS, see “Methods”), suggesting that cell-type–specific transcription of these genes may be at least partly regulated by methylation (Figure 3C, supplemental Table 3). The number of such potentially regulatory dHMRs (rHMR) varied substantially between cell subsets, with the proportion of differentially expressed transcripts associated with at least 1 HMR being 12.4% in CD14+ monocytes and between 6% and 8% for the lymphoid subsets CD4+, CD8+, and CD19+. Strikingly, 22.4% of the relatively small numbers of differentially expressed transcripts in CD16+ neutrophils were associated with dHMRs (Figure 3C).
Increasing evidence indicates a major role for hypomethylation outside CGIs (ie, CpG shelves and shores) in the regulation of gene transcription.1,27-29 In support of these findings, our analyses reveal that the majority of potential rHMRs identified were located either partially or completely outside CGIs, in shores, shelves, or open sea (Figure 3D). Moreover, a significant proportion of rHMRs were located either partially or fully within gene bodies (Figure 3E), reflecting the importance of intragenic methylation in regulating gene transcription.9
Verification of K450 array data using MeDIP-seq
To verify the K450 array data, MeDIP-seq was performed on all cell subsets and PBMCs from 1 individual. While the K450 array interrogates DNA methylation of individual CpG sites based on bisulfite conversion, providing high-resolution (albeit not complete) coverage of the human genome, MeDIP-seq uses antibody-based affinity enrichment of methylated DNA followed by second-generation sequencing to generate a detailed map of methylated regions throughout the entire genome.16,17,30 Overall, we observed a strong correlation between the methylation status of CpG sites interrogated by the K450 array and corresponding MeDIP-seq data (supplemental Figures 3-4). Despite the inability of K450 arrays to distinguish 5-methyl cytosine from 5-hydroxymethylcytosine, between 39% and 52% of HMRs were predicted by both approaches. This is illustrated by analysis of the CD8A gene, differential expression of which defines the CD8+ T-cell subset (Figure 4A). Analysis of the K450 array data identified 3 cell-specific HMRs in the CD8+ T-cell subset located both upstream of and within the gene (indicated by the extended orange box in Figure 4B), with MeDIP-seq both confirming their existence and revealing an expansion in their size (black bar, Figure 4B). Thus, the K450 array HMRs identified using MeDIP-seq were frequently found to be larger, as might be expected in view of the method’s more extensive genome coverage (Figure 4B; supplemental Figure 5).
DNA methylation and gene expression of the CD8A gene as representative rHMR. Detailed illustration of CD8A as an example of a potentially rHMR. (A) Gene expression (ie, messenger RNA) levels of CD8A in leukocyte cell subsets. Gene expression is given as log-fold change of signal intensity. (B) DNA methylation profile of the CD8A gene in individual cell subsets. (Top panel) HMRs identified by the K450 array in the different cell subsets are displayed as a colored bar indicating extend of the HMR. (Middle panel) Raw MeDIP-seq analyses are plotted as number of mapped methylated DNA fragments. The extent of HMRs within CD8+ T-cells is indicated by a black bar. (Bottom panel) The location of CGIs (blue boxes), location of K450 array probes, and the position of the CD8A gene mapped to the reference genome. TSS and gene orientation are indicated by a black arrow.
DNA methylation and gene expression of the CD8A gene as representative rHMR. Detailed illustration of CD8A as an example of a potentially rHMR. (A) Gene expression (ie, messenger RNA) levels of CD8A in leukocyte cell subsets. Gene expression is given as log-fold change of signal intensity. (B) DNA methylation profile of the CD8A gene in individual cell subsets. (Top panel) HMRs identified by the K450 array in the different cell subsets are displayed as a colored bar indicating extend of the HMR. (Middle panel) Raw MeDIP-seq analyses are plotted as number of mapped methylated DNA fragments. The extent of HMRs within CD8+ T-cells is indicated by a black bar. (Bottom panel) The location of CGIs (blue boxes), location of K450 array probes, and the position of the CD8A gene mapped to the reference genome. TSS and gene orientation are indicated by a black arrow.
Location of immune-mediated GWAS SNPs within leukocyte-specific potential rHMRs
The genetic contributions made by common SNPs to complex immune-mediated diseases are being progressively revealed by data from large GWAS.31-33 The majority of these SNPs are noncoding and the mechanism(s) by which they increase an individual’s risk of developing disease remain largely unknown. We hypothesized that such SNPs may lead to altered transcription through DNA methylation changes and may fall within HMRs. We explored this hypothesis by looking for an overlap between GWAS-identified SNPs (and associated LD blocks, r2 > 0.8) and the HMRs, dHMRs, and potential rHMRs identified by the methylation array analysis. SNPs associated with susceptibility to the immune-mediated diseases Crohn's disease,34 ulcerative colitis,35 primary biliary cirrhosis,36 and T1D37 were found to be significantly enriched within HMRs and to a lesser extent dHMRs (Table 1).
Enrichment of SNPs predisposing to immune-mediated diseases within HMRs
| GWAS . | No. of associated SNPs . | HMR . | Cell-type–specific HMR* . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4 . | CD8 . | CD14 . | CD16 . | CD19 . | PBMC . | CD4 . | CD8 . | CD14 . | CD16 . | CD19 . | ||
| Crohn's disease | 70 | 30† | 30† | 33† | 32† | 29† | 28† | 12 (1)† | 9 (2)† | 10† | 11† | 12 (1)† |
| Primary biliary cirrhosis | 20 | 10† | 10† | 11† | 11† | 10† | 9 | 2† | 2† | 4† | 2 | 3 |
| T1D | 64 | 25† | 24† | 23† | 24† | 24† | 22† | 6 (2)† | 2 | 6 (1) | 6 (1) | 5 |
| Ulcerative colitis | 45 | 16 | 15 | 17 | 16 | 16 | 14 | 4 | 4 (1) | 3 | 5 (1) | 4 |
| GWAS . | No. of associated SNPs . | HMR . | Cell-type–specific HMR* . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4 . | CD8 . | CD14 . | CD16 . | CD19 . | PBMC . | CD4 . | CD8 . | CD14 . | CD16 . | CD19 . | ||
| Crohn's disease | 70 | 30† | 30† | 33† | 32† | 29† | 28† | 12 (1)† | 9 (2)† | 10† | 11† | 12 (1)† |
| Primary biliary cirrhosis | 20 | 10† | 10† | 11† | 11† | 10† | 9 | 2† | 2† | 4† | 2 | 3 |
| T1D | 64 | 25† | 24† | 23† | 24† | 24† | 22† | 6 (2)† | 2 | 6 (1) | 6 (1) | 5 |
| Ulcerative colitis | 45 | 16 | 15 | 17 | 16 | 16 | 14 | 4 | 4 (1) | 3 | 5 (1) | 4 |
HMRs that specifically occur in the given cell type (see “Methods”); values in parentheses indicate number of SNPs within the subset rHMRs (i.e., those associated with a change in gene expression).
P < .0001.
Depending on the disease and cell subset, between 31% and 55% of disease-associated SNPs overlapped HMRs. The most significant enrichment was found in SNPs associated with primary biliary cirrhosis, where depending on the cell type, 45% to 55% of the 20 SNPs analyzed were located in HMRs. As such regions are known to be functionally important,38 this observation strengthens the evidence that these noncoding, disease-associated SNPs may function by modulating gene expression. A significant number of the SNPs fell into cell-type–specific HMRs (Table 1). It is conceivable that in these cases, the SNPs function by modulating gene expression in a cell-type–specific manner. This suggestion is further reinforced by the finding that a small number of these SNPs overlap cell-type–specific HMRs that are associated with the cell-type–specific expression of an adjacent gene (Table 1, supplemental Table 4).
One example of this is SNP rs3087243 at the CTLA4 locus that is associated with susceptibility to T1D.37 This SNP overlaps a HMR that is only found in CD4 T cells and which correlates with the elevated expression of CTLA4 in this cell type. These SNPs may function by altering CpG content and thereby changing chromatin accessibility, and indeed a number of the SNPs mapping to these rHMRs potentially lead to the gain or loss of CpG sites (supplemental Table 4). Alternatively, they may alter transcription factor binding sites within these regions of open chromatin and therefore function as expression quantitative trait loci. More generally, our observations therefore provide a rational basis for selecting cell types in which to study the function of disease-associated SNPs in the absence of any other evidence.
Discussion
Changes in DNA methylation are established as one of the major epigenetic mechanisms in mammals. Several central biological mechanisms are known to be controlled at least in part through cytosine methylation, including x-chromosome inactivation, genomic imprinting, and tissue-specific gene expression.4 However, the exact mechanisms of how DNA methylation may be implicated in regulating different functional pathways remain poorly understood.
Here, we provide detailed methylomes for unseparated PBMCs and 5 purified primary leukocyte subsets obtained from 6 healthy male individuals. We demonstrate that each cell subset displays a unique methylation profile and observe substantial differences between cell subsets derived from either the myeloid or lymphoid lineage. PCA of these profiles pulls apart the individual cell types in a lineage-dependent manner, in a similar way to that seen using gene expression data.18 It also demonstrates that PBMC samples have a composite profile that is, therefore, likely to be sensitive to changes in cellular composition. For studies of the methylome in patients with inflammatory conditions, it is critical therefore that these are performed on separated cells to avoid observed differences merely being driven by alterations in cellular composition between individuals.
To identify regions that are likely to be involved in regulating gene expression through DNA methylation in a cell-subset–specific manner, we correlated DNA methylation with gene expression. Assuming that HMRs are likely to have a greater functional relevance than individual hypomethylated CpGs, we correlated cell-subset–specific HMRs with genes that were overexpressed in each cell type. A large number of HMRs correlated significantly with active gene expression. Importantly, a large proportion of these potential HMRs were confirmed using MeDIP-seq data. Not surprisingly, given the more extensive genomic coverage of this technique, many of these regions demonstrated even larger expansions than when observed by the K450 array, which only covers 480K of the 30 million CpGs present in the human genome. Analysis of the genomic distribution of these potential rHMRs revealed that the majority were found to be located outside of CGIs in shelves and shores or not associated with CGIs (ie, open sea), providing further evidence that these regions of the genome are crucial in regulating gene expression.1,28
Recent studies in both mice and humans have demonstrated the impact of DNA methylation on cellular differentiation during hematopoiesis, with myeloid commitment being associated with less global DNA methylation compared with lymphoid commitment.1,28 Our data extend this observation and suggest that maintenance of, as well as commitment to, the lymphoid lineage is associated with higher levels of DNA methylation. Given the well-documented impact of DNA methylation on gene expression, the extent of hypomethylation may reflect transcriptional plasticity. Fully differentiated cell types with stable transcriptional programs, such as neutrophils, show more extensive hypomethylation compared with lymphoid cells, such as CD8 T cells. This may well reflect the fact that lymphoid lineages contain more heterogeneous populations and comprise many naive cells that require an ongoing capacity for differentiation. By extension, one might expect to see a similar gradient within lymphoid populations with, for example, naive CD8 T cells having fewer HMRs than memory cells.
Over the past 5 to 8 years, large-scale GWAS have been conducted on many complex diseases and have led to the identification of genetic variants that predispose to these conditions.34-37 However, for the majority of these potential disease-predisposing SNPs, the mechanism by which changes in gene sequence contribute to disease pathogenesis remains unknown. The fact that a large proportion of these SNPs are located outside of coding regions suggests that they alter gene expression. Moreover, in addition to genetic predisposition, environmental factors undoubtedly play a major role in disease pathogenesis of complex diseases, providing another potential role for epigenetic mechanisms.
In this study, we provide a potential link between genetic predisposition and epigenetic mechanisms for complex immune-mediated diseases by demonstrating an enrichment of autoimmune GWAS SNPs within cell-subset–specific rHMRs. While the exact mechanism(s) involved remain to be identified, one may hypothesize that genetic modifications altering DNA methylation within potential rHMRs, may alter transcription factor binding and or the overall chromatin structure leading to changes in gene expression. A study by Ernst and colleagues also supports this hypothesis by demonstrating enrichment of top-scoring GWAS SNPs within enhancer elements, which were identified by mapping the chromatin state of several human cell types.3
Interestingly, our results reveal distinct differences in the number of GWAS SNPs overlapping HMRs between cell subsets as well as disease type, potentially providing further insight into pathogenesis. For example, at the CTLA4 locus the T1D-associated SNP rs3087243 overlaps a CD4 T-cell–specific HMR that correlates with expression of the gene in this cell type. Although further work is clearly required, this observation suggests that functional studies aimed at delineating the role of CTLA4 in diabetes pathogenesis should focus on its role in CD4 T cells, a cell type known to be important in T1D pathogenesis.
Taken together, in generating comprehensive functional methylome maps of PBMCs and 5 individual leukocyte subsets combined with gene expression data, our analyses support and further expand on recent findings with regard to the regulation of gene expression through DNA methylation. Importantly, our data provide a reference tool for researches aiming to investigate the role of DNA methylation in regulating primary leukocyte function in health and immune-mediated disease.
The raw data reported in this article are available from ArrayExpress (Affymetrix and Illumina arrays) and the European Genome-phenome Archive (methylated DNA immunoprecipitation sequencing [MeDIP-seq]), EGA study accession number: EGAS00001000490 (MeDIP-seq) and ArrayExpress accession number: E-ERAD-179 (K450 Illumina arrays), E-MTAB-2062 (Affymetrix arrays).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Panos Deloukas and Elin Grundberg for the generation of the Illumina Infinium HumanMethylation450 Beadchip data, Carol Scott for help with data analysis, Mike Quail and team for quantitation and dilution of samples prior to sequencing, John Burton and team for sequencing of MeDIP samples, Emma Gray and the Sample Management team for sample preparation, and the staff of the Wellcome Trust Sanger Institute for their invaluable contribution to the manuscript.
This work was supported by The Wellcome Trust (grant numbers 098051 and 094227/Z/10/Z); the Academy of Finland (grant number 251704 [A.P.]); the Academy of Finland, Centre of Excellence in Complex Disease Genetics (grant numbers 213506 and 129680 [A.P.]); the European Community’s Seventh Framework Programme (FP7/2007-2013); ENGAGE Consortium (grant agreement HEALTH-F4-2007-201413); EU/SYNSYS Synaptic Systems (grant number 242167 [A.P.]); the Sigrid Juselius Foundation, Finland (A.P.); BioSHaRE Consortium (FP7/2007-2013); Biobank Standardisation and Harmonisation for Research Excellence in the European Union, grant number 261433 (A.P.); gEUVADIS (grant agreement HEALTH-2010-261123); and a Wellcome Trust Strategic Award (079895 [Cambridge Institute for Medical Research]).
Authorship
Contribution: P.A.L., K.G.C.S., A.P., and M.Z. designed the experiments; M.Z., C.C., and A.J.C. performed most of the experiments; T.F.R., P.P., and C.J.J. performed data analysis; M.Z. and P.A.L. wrote the manuscript; and all authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul A. Lyons, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Rd, Cambridge, CB2 2XY, United Kingdom; e-mail: pal34@cam.ac.uk.
References
Author notes
M.Z. and T.F.R. contributed equally to the work.
P.A.L. and K.G.C.S. contributed equally to the work.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal