Key Points
Genetic loss of the transcriptional corepressor TRIM28 in adult mice results in deficient adult erythropoiesis in bone marrow, and anemia.
TRIM28 controls the mRNA levels of multiple erythroid transcription factors, heme biosynthetic enzymes, and the apoptotic apparatus.
Abstract
In previous mass spectrometry and coimmune precipitation studies, we identified tripartite motif-containing 28 (TRIM28; also known as transcriptional intermediary factor1β and Krüppel-associated box-associated protein-1) as a cofactor that specifically copurified with an NR2C1/NR2C2 (TR2/TR4) orphan nuclear receptor heterodimer that previous studies had implicated as an embryonic/fetal β-type globin gene repressor. TRIM28 has been characterized as a transcriptional corepressor that can associate with many different transcription factors and can play functional roles in multiple tissues and cell types. Here, we tested the contribution of TRIM28 to globin gene regulation and erythropoiesis using a conditional loss-of-function in vivo model. We discovered that Trim28 genetic loss in the adult mouse leads to defective immature erythropoiesis in the bone marrow and consequently to anemia. We further found that TRIM28 controls erythropoiesis in a cell-autonomous manner by inducibly deleting Trim28 exclusively in hematopoietic cells. Finally, in the absence of TRIM28, we observed increased apoptosis as well as diminished expression of multiple erythroid transcription factors and heme biosynthetic enzymes in immature erythroid cells. Thus, TRIM28 is essential for the cell-autonomous development of immature erythroblasts in the bone marrow.
Introduction
The supply of red blood cells (RBCs) is maintained by continuous production of erythroid cells in the bone marrow. Red and white blood cells are progeny of hematopoietic stem cells (HSCs) that reside in the bone marrow in adult animals. HSCs are endowed with classically defined properties of stem cells, containing both self-renewal capacity and multilineage differentiation potential. The first differentiation step of HSCs specifies multipotential progenitors (MPPs), which develop to common myeloid progenitors (CMPs) and lymphoid-primed MPPs. CMPs further develop into megakaryocyte-erythrocyte progenitors (MEPs) and granulocyte-macrophage progenitors (GMPs). The final differentiation commitment of MEPs exclusively to the erythroid lineage occurs in erythroblasts, which finally differentiate into enucleated reticulocytes in the bone marrow. Reticulocytes that are released from the bone marrow into the vascular network mature into RBCs while in circulation.1,2
During erythroid differentiation, initiation of globin gene transcription occurs at the erythroblast stages. The major form of hemoglobin, the essential vertebrate oxygen transporter, in the human fetal liver is fetal hemoglobin (α2γ2) and in the adult bone marrow is adult hemoglobin (α2β2). Robust induction of fetal hemoglobin in individuals bearing deleterious mutations in the adult β-globin gene (eg, in sickle cell anemia and β-thalassemia) ameliorates disease morbidity.3,4 To elucidate the molecular mechanism(s) that regulate fetal γ-globin gene repression in adult mammals, we previously reported the isolation and detailed characterization of the direct repeat erythroid-definitive complex as a candidate repressor of both the embryonic and fetal β-type globin genes5 and identified the DNA-binding orphan nuclear receptors NR2C1 (originally called TR2) and NR2C2 (also known as TR4) as the DNA-binding subunits of the repressor.6,7 We subsequently described a number of NR2C1/2-binding proteins that were hypothesized to function as possible corepressors through epigenetic modifying activities, including lysine-specific demethylase 1, DNA methyltransferase 1, and TRIM28.8
Tripartite motif-containing 28 (TRIM28; also known as transcriptional intermediary factor1β and Krüppel-associated box-associated protein-1) contains an N-terminal ring finger, 2 B-box zinc fingers, and a RING-B box-coiled-coil protein interaction domain as well as a C-terminal plant homeodomain/bromodomain transcriptional repressive sequence.9,10 TRIM28 recruits heterochromatin protein 1 (HP1) through the central HP1-binding domain11,12 and recruits the histone H3K9 methyltransferase SETDB1 through the homeodomain/bromodomain sequence.13 The ubiquitously expressed TRIM28 protein functions as a universal corepressor for Krüppel-associated box domain-containing zinc finger transcription factors by binding via its RING-B box-coiled-coil protein interaction domain.9,10
Genetically modified mice in which the Trim28 gene was inactivated die between embryonic days 5.5 and 8.8.14 TRIM28 function, among others, is required for the silencing of endogenous retroviruses in embryonic stem (ES) cells,15,16 for the pluripotency of ES cells,17,18 for proper DNA methylation in ES cells as well as for eliciting a timely transition from oocyte to embryo.19,20 TRIM28 mediates epigenetic repression in the forebrain and controls response to behavioral stress.21 In hematopoietic cells, TRIM28 has been shown to control the development and functions of B22 and T lymphoid cells.23-25 Although TRIM28 plays roles in multiple hematopoietic cells, its possible function(s) in myeloerythroid lineage cell development are unknown.
Because the TRIM28 protein was repeatedly recovered in immune complexes containing NR2C1/28 and is generally regarded as a corepressor, we hypothesized that TRIM28 might contribute functionally to the regulatory activity of the direct repeat erythroid-definitive complex, which represses embryonic and fetal β-type globin genes. Here, we tested the contribution of TRIM28 to β-type globin gene expression and erythropoiesis by ablating the Trim28 gene in all hematopoietic lineage cells of the mouse using the inducible Mx1Cre transgene.26 We conclude that TRIM28 is dispensable for embryonic/fetal globin gene silencing during definitive erythropoiesis but, rather, is required for the maturation of erythroid cells in the adult bone marrow.
Methods
Mice
The Trim28-floxed (Trim28L2, referred to here as Trim28flox),14 EpoR-Cre (EporCre),27 and Mx1Cre transgenic (TgMx1Cre)26 alleles have been described previously. Trim28flox/flox:EporCre/+ (TEC) mice were maintained in a C57BL/6:CD1 mixed background. Trim28flox/flox:TgMx1Cre (TMC) congenic mice were backcrossed for more than 7 generations with C57BL/6 mice. To induce Cre recombinase from the Mx1Cre transgene, 20 μg of poly(I:C) (GE Healthcare) was injected 5 times every other day. C57BL/6-Ly5B6 (CD45.2) mice and C57BL/6-Ly5SJL (CD45.1) mice were purchased from the Jackson Laboratory. Adoptive transfer experiments were performed as described previously.28 All animal experiments were approved and conducted according to the University Committee on Use and Care of Animals of the University of Michigan guidelines.
Cell preparation, flow cytometry, and cell staining
Cells were prepared and analyzed by flow cytometry as described previously.28,29 The following antibodies were also used: CD71(R17217), CD5(53-7.3), FcγR(93), CD45(30-F11), and Thy1.2(53-2.1). Peripheral blood samples were taken from the retro-orbital venous plexus using heparin-coated microtubes. Hematologic parameters were analyzed using the ADVIA 120 Hematology System (Siemens). Between 1 and 5 × 105 cells were used for cytospins followed by neutral benzidine staining30 or Wright-Giemsa staining (Sigma-Aldrich, WG16). Annexin V staining was performed as described previously.29
qPCR analysis of genomic DNA
Flow-sorted cells were lysed in Proteinase K plus sodium dodecyl sulfate buffer followed by phenol-chloroform extraction and isopropanol precipitation of genomic DNA. A Trim28 floxed allele-specific primer set was used to quantify gene deletion by quantitative polymerase chain reaction (qPCR) with SYBR green dye on an ABI Prism 7000 (Applied Biosystems). Primers used were 5’-GGTGGCGGCCGCTCTAGTAT-3’ and 5’-GCGGCCGCAGATCTCTATCTACT-3’. Total DNA content for normalization was analyzed using β-actin as the control.31 Quantification of Sry genomic DNA by qPCR was performed as described previously.31
qRT-PCR analyses
Total RNA was isolated from sorted cells using QIAGEN RNeasy Micro or Mini Kits with DNase treatment. Quantitative reverse transcription PCR (qRT-PCR) was performed as described previously.28 Expression levels were normalized to internal β-actin abundance. Primers used for the SYBR green assay were taken from the following citations: β-actin,32 εY-globin, βH1-globin, and adult β-globin.8 The adult β-globin primer set coamplifies both βmajor and βminor.
RNA-Seq analysis
Total RNA was isolated from sorted immature erythrocytes using QIAGEN RNeasy Mini Kit with DNase treatment. Intact total RNA showing an RNA integrity number of “10” (highest) on an Agilent Bioanalyzer was further processed using NuGEN Encore Complete RNA-Seq Library Systems at the University of Michigan Sequencing Core facility. The library was sequenced (50-50 bp paired-end) on an Illumina HiSeq2000. Raw reads were mapped to the mm10 mouse genome sequence using TopHat and further analyzed with Cuffdiff and CummeRbund.33 A default false discovery rate of 0.05 was used for the statistical significance analysis in Cuffdiff. The data are deposited as GEO: GSE49843.
Statistical analysis
Statistical significance was determined using the Student t test. Data were considered statistically significant when P < .05.
Results
β-globin transcription is unchanged in the absence of TRIM28
To test for possible contributions of the putative corepressor TRIM28 protein to globin gene regulation, the Trim28 gene was initially ablated using the previously well-characterized Trim28-floxed (Trim28flox)14 and EpoR-Cre knockin (EporCre)27 alleles; the latter is predominantly active in immature erythroid cells (supplemental Figure 1). CD71+TER119+ immature erythroid cells in the bone marrow isolated by flow sorting from TEC mutant adult mice retain 30% of (undeleted) floxed allele compared with Trim28flox/flox. The TEC mutant mouse exhibited normal β-type globin gene expression and erythropoiesis (supplemental Figures 2 and 3).
To achieve more efficient ablation of the conditional Trim28 alleles, we took advantage of the Mx1Cre transgene (TgMx1Cre) that we have effectively used for hematopoietic cell loxP-mediated deletion in the past.28,29 To induce Cre recombinase expression from TgMx1Cre, poly(I:C) was administered by intraperitoneal injection 5 times every other day. This treatment resulted in slightly diminished total bone marrow cellularity in TMC mutant mice both 2 and 4 weeks after poly(I:C) (Figure 1A). CD71+TER119+ cells in the bone marrow were isolated by flow sorting (Figure 1B), followed by quantification of the Trim28 floxed allele by qPCR. The remaining floxed (unaltered germ line) allele in this immature erythroid cell fraction in TMC mutant mice was 5.3% or 7.6% of control Trim28flox/flox mice at 2 and 4 weeks after poly(I:C), respectively (Figure 1C).
Erythropoiesis in mice inducibly ablated for Trim28. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected every 48 hours with poly(I:C) 5 times to transcriptionally activate the cre-expressing transgene.26 Uninjected Trim28flox/+ (flox/+) mice were used as controls. Mice were analyzed 2 weeks (2w) or 4 weeks (4w) after completion of poly(I:C) administration. (A) The absolute number of total bone marrow cells isolated from 2 femurs plus 2 tibias of individual animals (each circle) of various genotypes. The black bar represents the average. * indicates statistically significant, P < .05. NS: not significant, P > .05. (B) Flow cytometric analysis of erythropoiesis in total bone marrow cells from mutant and control mice. The numbers in each of the boxed areas indicate the mean frequency of cells in each hematopoietic population. (C) The abundance of Trim28 floxed allele in CD71+TER119+ immature erythroid cells was analyzed by qPCR. The CD71hiTER119- cells that aberrantly accumulated in the TMC mutant mice (panel B) were also analyzed. An average of the relative amount of the undeleted gene in the control Trim28flox/flox cell was set to 200, which is the copy number of the floxed allele in 100 cells. (D) The expression level of β-type globin genes normalized to β-actin in CD71+TER119+ immature erythroid cells was analyzed by qRT-PCR. The average of the relative amount in the control Trim28flox/flox cell was set to 1. (E) The absolute number of cells in each of the analyzed populations was calculated from the data shown in (A) and (B). Average with standard error of the mean (SEM). (F) Flow cytometric analysis of TER119+ cells in total bone marrow cells (separated by forward scatter and CD44 antigen according to Chen et al34 ). The numbers closest to the boxed areas indicate the mean percentage of cells in each population within parental TER119+ cells. (G) The absolute number of basophilic erythroblasts (r2), polychromatic erythroblasts (r3), orthochromatic erythroblasts (r4a), reticulocytes (r4b), and RBCs (r5) in the bone marrow was calculated from the data shown in panels (A) and (F). Error bars represent the SEM. (H) Hematologic parameters of peripheral blood cells. Data represent either a summary (A,C,D,E,G,H) or representative individuals (B,F) of more than 3 mice of each genotype from 2 to 4 independent experiments.
Erythropoiesis in mice inducibly ablated for Trim28. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected every 48 hours with poly(I:C) 5 times to transcriptionally activate the cre-expressing transgene.26 Uninjected Trim28flox/+ (flox/+) mice were used as controls. Mice were analyzed 2 weeks (2w) or 4 weeks (4w) after completion of poly(I:C) administration. (A) The absolute number of total bone marrow cells isolated from 2 femurs plus 2 tibias of individual animals (each circle) of various genotypes. The black bar represents the average. * indicates statistically significant, P < .05. NS: not significant, P > .05. (B) Flow cytometric analysis of erythropoiesis in total bone marrow cells from mutant and control mice. The numbers in each of the boxed areas indicate the mean frequency of cells in each hematopoietic population. (C) The abundance of Trim28 floxed allele in CD71+TER119+ immature erythroid cells was analyzed by qPCR. The CD71hiTER119- cells that aberrantly accumulated in the TMC mutant mice (panel B) were also analyzed. An average of the relative amount of the undeleted gene in the control Trim28flox/flox cell was set to 200, which is the copy number of the floxed allele in 100 cells. (D) The expression level of β-type globin genes normalized to β-actin in CD71+TER119+ immature erythroid cells was analyzed by qRT-PCR. The average of the relative amount in the control Trim28flox/flox cell was set to 1. (E) The absolute number of cells in each of the analyzed populations was calculated from the data shown in (A) and (B). Average with standard error of the mean (SEM). (F) Flow cytometric analysis of TER119+ cells in total bone marrow cells (separated by forward scatter and CD44 antigen according to Chen et al34 ). The numbers closest to the boxed areas indicate the mean percentage of cells in each population within parental TER119+ cells. (G) The absolute number of basophilic erythroblasts (r2), polychromatic erythroblasts (r3), orthochromatic erythroblasts (r4a), reticulocytes (r4b), and RBCs (r5) in the bone marrow was calculated from the data shown in panels (A) and (F). Error bars represent the SEM. (H) Hematologic parameters of peripheral blood cells. Data represent either a summary (A,C,D,E,G,H) or representative individuals (B,F) of more than 3 mice of each genotype from 2 to 4 independent experiments.
We hypothesized that TRIM28 might normally contribute to repression of the embryonic and/or fetal globin genes and, therefore, that the mouse εY- and/or βH1-globin genes (the structural homologues of human embryonic ε− and fetal γ-globin genes, respectively) would be derepressed in the absence of TRIM28. To test this hypothesis, we analyzed the expression of the β-type globin genes in the immature erythroid cell population by qRT-PCR. The level of εY-globin messenger RNA (mRNA) was unchanged. Although the level of βH1-globin mRNA increased slightly (not in a statistically significant manner), more than half of the animals analyzed had normal levels of these mRNAs (Figure 1D). A mild decrease in adult β-globin mRNA was observed (but again, not statistically different). These results clearly demonstrated that TRIM28 is dispensable for embryonic and fetal β-type globin gene repression in adult definitive erythropoiesis.
Trim28-deficient mice are anemic
Reduction in the absolute number of CD71+TER119+ immature erythroid cells was observed in the TMC mutant bone marrow (Figure 1B,E). Further detailed analysis of immature erythroid cell stages using an antibody recognizing CD4434 showed a reduction in the number of basophilic erythroblasts (r2) as well as later-stage erythroid cells in the TMC mutant bone marrow (Figure 1F-G). We also observed a significant increase in the number of CD71hiTER119- cells (Figure 1B,E, see below). In the peripheral blood of the TMC mutants, the RBC number as well as the hemoglobin and hematocrit parameters significantly decreased, whereas the mean corpuscular volume was unchanged (Figure 1H). This normocytic anemic phenotype is often observed in diseases displaying defects in bone marrow erythropoiesis.
Analyses of hematopoietic progenitor populations by flow cytometry revealed that the mutants bore an increased number of both CMPs and MEPs, whereas the HSC/MPP compartment (defined as Lin-Sca1+cKithi) and GMP populations were unaffected in the TMC mutant mice (Figure 2A-B). In accord with the observation that GMPs develop normally, the number of myeloid cells expressing Mac1 in the bone marrow was unaltered in the TMC mutant (Figure 2C). The number of monocytes and neutrophils in the peripheral blood of the TMC mutant animals at 2 or 4 weeks after poly(I:C) administration was not statistically altered when compared with control mice. The number of eosinophils was normal at 2 weeks and was reduced by 28% at 4 weeks. The number of basophils was transiently reduced to 35% of controls after 2 weeks, but recovered to normal levels by 4 weeks (data not shown). Thus, the number of myeloid lineage cells in the peripheral blood was essentially normal for monocytes and neutrophils.
Abnormal development of hematopoietic progenitor cells in Trim28 mutant mice. Adult TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected with poly(I:C). (A) Flow cytometric analysis of total bone marrow cells and division into individual early progenitor compartments (shown on the right) according to cell surface expression of cKit, Sca1, FcγR, CD34, and lineage (Lin) markers. Numbers in the boxed areas indicate the mean percentages of cells in the gated area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population (Frequency of Parent in FlowJo software). (B) The absolute number of HSC/MPP, CMP, MEP, and GMP was calculated from the data shown in panel (A) and total bone marrow cell number. Averages are indicated with SEM. * indicates statistically significant, P < .05. NS: not significant, P > .05. (C) The absolute number of Mac1+ cells in the bone marrow of mutant and control mice. (D) The absolute number of cKithi Lin- and cKithiLin+ cells in the bone marrow of control and conditionally mutant mice. (E) The absolute number of cKithiCD71hiTER119-, cKithiCD71-TER119-, and cKithiTER119+ cells in the bone marrow. The lineage mixture used for panels (A, B, and D) includes antibodies that recognize TER119, Mac1, Gr1, B220, CD19, CD5, CD4, and CD8a. Mice were analyzed for 2 weeks (A, B, D, E) or for 2 and 4 weeks (C) after completion of poly(I:C) administration. The data represent a summary (B-E) or are representative (A) of more than 3 mice of each genotype from 2 to 3 independent experiments. Each circle represents an individual mouse, and the black bars represent averages.
Abnormal development of hematopoietic progenitor cells in Trim28 mutant mice. Adult TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected with poly(I:C). (A) Flow cytometric analysis of total bone marrow cells and division into individual early progenitor compartments (shown on the right) according to cell surface expression of cKit, Sca1, FcγR, CD34, and lineage (Lin) markers. Numbers in the boxed areas indicate the mean percentages of cells in the gated area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population (Frequency of Parent in FlowJo software). (B) The absolute number of HSC/MPP, CMP, MEP, and GMP was calculated from the data shown in panel (A) and total bone marrow cell number. Averages are indicated with SEM. * indicates statistically significant, P < .05. NS: not significant, P > .05. (C) The absolute number of Mac1+ cells in the bone marrow of mutant and control mice. (D) The absolute number of cKithi Lin- and cKithiLin+ cells in the bone marrow of control and conditionally mutant mice. (E) The absolute number of cKithiCD71hiTER119-, cKithiCD71-TER119-, and cKithiTER119+ cells in the bone marrow. The lineage mixture used for panels (A, B, and D) includes antibodies that recognize TER119, Mac1, Gr1, B220, CD19, CD5, CD4, and CD8a. Mice were analyzed for 2 weeks (A, B, D, E) or for 2 and 4 weeks (C) after completion of poly(I:C) administration. The data represent a summary (B-E) or are representative (A) of more than 3 mice of each genotype from 2 to 3 independent experiments. Each circle represents an individual mouse, and the black bars represent averages.
We also observed a significant increase in the number of cKithiLin+ cells (Figure 2A,D). Those cKithiLin+ cells were CD71-negative (Figure 2E), and thus they differed from the CD71hiTER119- cells described previously (Figure 1B,E). The specific characteristics of the cKithiLin+ cells are not yet known. The accumulation of CMPs and MEPs in the TMC mutant suggests that TRIM28 is required for differentiation from either CMPs or MEPs into erythroblasts in the bone marrow.
Detailed characterization of the aberrant CD71hiTER119- cells that accumulate in the bone marrow of Trim28 mutant mice
Next, we wished to characterize the aberrant population of accumulated CD71hiTER119- cells in the TMC mutants in greater detail (Figure 1B,E). First, the residual undeleted Trim28 floxed allele in the CD71hiTER119- cells was determined to be 1.6% or 3.3% of control Trim28flox/flox mice at 2 and 4 weeks after poly(I:C) treatment, respectively (Figure 1C). We observed that when pelleted, the CD71hiTER119- cells were faint red (Figure 3A).
Detailed characterization of the aberrant CD71hiTER119-cell population that accumulates in Trim28 mutant bone marrow. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected with poly(I:C). Mice were analyzed 2 weeks after the final poly(I:C) administration. (A) Color of cells separated on the basis of CD71 and TER119 expression. Between 4 and 10 × 105 cells were flow sorted and suspended in PBS supplemented with 2% FBS in a 1.5-mL tube and centrifuged to form a pellet. Images were acquired using a Zeiss Stemi SV11 Apo stereomicroscope, AxioCam digital camera, and AxioVision LE software. (B) Flow cytometric analysis of forward scatter (FSC), CD45, cKit, Mac1, and B220 in the CD71hiTER119- bone marrow cells of control mice and Trim28 mutant mice. Numbers in the boxed areas indicate the mean percentages of cells in the boxed area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population. (C) The absolute number of different subpopulations in the CD71hiTER119- cells was calculated from panels (B) and normalized for total bone marrow cell number. Average with SEM. *indicates statistically significant, P < .05. NS: not significant, P > .05. (D) Neutral benzidine staining of individual cells recovered from flow-sorted populations of different genotype mice treated with poly(I:C). (E) Wright-giemsa staining. Images (D and E) were acquired using an Olympus BX-51 upright light microscope with the 100× oil immersion objective lens, DP-70 high-resolution digital camera, and DP Controller software. (F) Flow cytometric analysis of FSC and CD44 in CD71hiTER119- and CD71hiTER119+ cells. Numbers in or near the boxed areas indicate the mean percentage of cells in the boxed area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population. The data represent a summary (C) or representative individuals (A, B, D-F) of more than 3 mice of each genotype examined in 2 to 3 independent experiments.
Detailed characterization of the aberrant CD71hiTER119-cell population that accumulates in Trim28 mutant bone marrow. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected with poly(I:C). Mice were analyzed 2 weeks after the final poly(I:C) administration. (A) Color of cells separated on the basis of CD71 and TER119 expression. Between 4 and 10 × 105 cells were flow sorted and suspended in PBS supplemented with 2% FBS in a 1.5-mL tube and centrifuged to form a pellet. Images were acquired using a Zeiss Stemi SV11 Apo stereomicroscope, AxioCam digital camera, and AxioVision LE software. (B) Flow cytometric analysis of forward scatter (FSC), CD45, cKit, Mac1, and B220 in the CD71hiTER119- bone marrow cells of control mice and Trim28 mutant mice. Numbers in the boxed areas indicate the mean percentages of cells in the boxed area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population. (C) The absolute number of different subpopulations in the CD71hiTER119- cells was calculated from panels (B) and normalized for total bone marrow cell number. Average with SEM. *indicates statistically significant, P < .05. NS: not significant, P > .05. (D) Neutral benzidine staining of individual cells recovered from flow-sorted populations of different genotype mice treated with poly(I:C). (E) Wright-giemsa staining. Images (D and E) were acquired using an Olympus BX-51 upright light microscope with the 100× oil immersion objective lens, DP-70 high-resolution digital camera, and DP Controller software. (F) Flow cytometric analysis of FSC and CD44 in CD71hiTER119- and CD71hiTER119+ cells. Numbers in or near the boxed areas indicate the mean percentage of cells in the boxed area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population. The data represent a summary (C) or representative individuals (A, B, D-F) of more than 3 mice of each genotype examined in 2 to 3 independent experiments.
Next, we examined the expression of cell surface molecules in the accumulated CD71hiTER119- cells by flow cytometry. Although 95% of the CD71hiTER119- cells were CD45 positive in control mice, only 19% were positive in the analogous pool recovered from induced TMC mutant animals. CD45 is expressed in all hematopoietic lineage cells except for erythrocytes and platelets.35 The majority of CD45-negative cells in the induced TMC mutant were small in size (as indicated by flow forward scatter). More than 90% of the CD71hiTER119- cells were also negative for cKit, which is usually highly expressed in hematopoietic progenitor cells. Approximately 4% or 1% of the CD71hiTER119- cells were positive for macrophage marker Mac1 or B cell surface marker B220, respectively (Figure 3B-C). The CD71hiTER119- cells were positive when the cells were stained with benzidine, which detects heme molecules (Figure 3D). When the morphologic characteristics of these cells was examined by Giemsa staining, the CD71hiTER119- cells were found to be a mixture of round erythroblast-like cells and enucleated later-stage erythroid cells (Figure 3E). The CD44-FSC profile in the CD71hiTER119- cells was similar to that observed in the CD71hiTER119+ cells. The mutant cells were shifted toward slightly more immature stages (Figure 3F). On the basis of these observations, we finally concluded that the CD71hiTER119- cells that accumulate in the bone marrow of induced TMC mutant mice are r2-r4a erythroblast cells34 that fail to express TER119. Taken together with the previous analyses on Trim28 floxed allele deletion (Figure 1C), we concluded that TMC mutant mice generate 2 distinct types of immature erythroid cells: Δ, CD71hiTER119- r2-r4a-like cells with 1% to 4% of the Trim28 gene remaining undeleted, and δ, CD71hiTER119+ erythroblast cells with 5% to 8% undeleted (summarized in supplemental Figure 4).
TRIM28 is cell-autonomously required for immature erythropoiesis
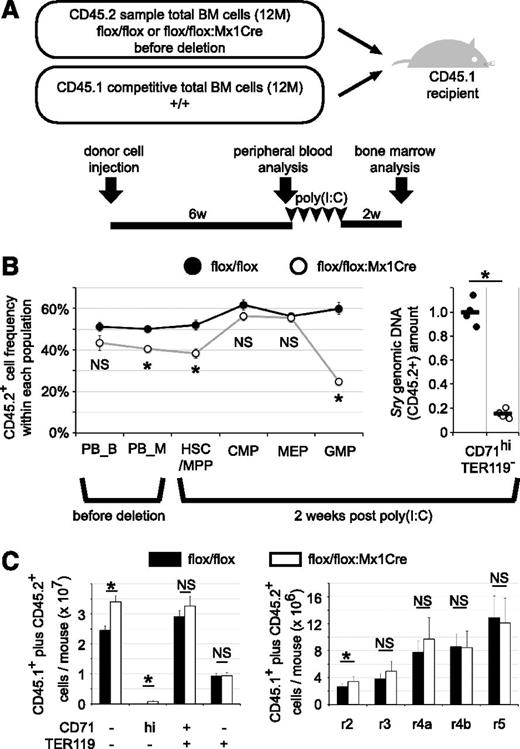
To test whether TRIM28 was intrinsically required for erythroid differentiation, mice were reconstituted with untreated TMC mutant hematopoietic cells by adoptive transfer. Total bone marrow donor cells isolated from Trim28flox/flox:TgMx1Cre before deletion or control Trim28flox/flox mice (1.2 × 107 CD45.2) were coinjected with wild-type competitive donor cells (1.2 × 107 CD45.1) into lethally irradiated wild-type recipient mice (CD45.1). CD45.1 wild-type donor cells were used in this experiment to protect mutant donor-alone injected animals from possible anemia. Although the same numbers of donor cells were injected, we unexpectedly observed a slightly lower contribution of TMC CD45.2+ cells compared with control (Trim28flox/flox) CD45.2+ cells in the peripheral blood of reconstituted animals (Figure 4B). We considered this difference to be within the range of experimental variation.
The contribution of TRIM28 to erythropoiesis is erythroid cell autonomous. (A) Experimental design. Total bone marrow donor cells isolated from Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre) or control Trim28flox/flox (flox/flox) mice (CD45.2) were coinjected with an approximately equal number of wild-type competitive donor cells (CD45.1) into lethally irradiated wild-type recipient mice (CD45.1). (B) CD45.2 and CD45.1 expression in each population in peripheral blood (before Cre induction) or in bone marrow (2 weeks post Cre activation) were analyzed by flow cytometry (left panel). Peripheral blood cells were analyzed 6 weeks after transplantation for CD45.2+ donor cell contribution in the Mac1+Gr1+ myeloid (PB_M) and B220+CD19+ B lymphoid (PB_B) populations. Total bone marrow cells were analyzed 2 weeks after completing poly(I:C) treatment for CD45.2+ donor cell contribution to the HSC/MPP, CMP, MEP, and GMP compartments. Each circle represents the average of control (black circle) or TMC mutant cells (open circle) with SEM (left panel). Data represent the summary of 8 recipient mice of each genotype from 2 independent experiments. Because erythroblasts and later-stage erythroid cells are negative for CD45,35 the contribution of CD45.2+ (male) cells to the immature erythroid cell fraction was analyzed by qPCR, quantifying the abundance of Sry genomic DNA (Y chromosome; right panel). The average of Trim28flox/flox mice was set to 1. Male CD45.2 and female CD45.1 cells were used in the first experiment. Each circle represents an individual mouse, and black bars represent the average (right panel). The summary of the first experiment with 4 recipients of each donor genotype is shown. In the second set of experiments, female CD45.2 and male CD45.1 cells were used; an (statistically insignificant) increased contribution of coinjected competitive CD45.1 cells was observed (data not shown). (C) The absolute number of different stages of erythroid cells in the bone marrow (2 femurs and 2 tibias). Mixtures of CD45.2 and CD45.1 cells are depicted. Average with SEM. * indicates statistically significant, P < .05. NS: not significant, P > .05.
The contribution of TRIM28 to erythropoiesis is erythroid cell autonomous. (A) Experimental design. Total bone marrow donor cells isolated from Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre) or control Trim28flox/flox (flox/flox) mice (CD45.2) were coinjected with an approximately equal number of wild-type competitive donor cells (CD45.1) into lethally irradiated wild-type recipient mice (CD45.1). (B) CD45.2 and CD45.1 expression in each population in peripheral blood (before Cre induction) or in bone marrow (2 weeks post Cre activation) were analyzed by flow cytometry (left panel). Peripheral blood cells were analyzed 6 weeks after transplantation for CD45.2+ donor cell contribution in the Mac1+Gr1+ myeloid (PB_M) and B220+CD19+ B lymphoid (PB_B) populations. Total bone marrow cells were analyzed 2 weeks after completing poly(I:C) treatment for CD45.2+ donor cell contribution to the HSC/MPP, CMP, MEP, and GMP compartments. Each circle represents the average of control (black circle) or TMC mutant cells (open circle) with SEM (left panel). Data represent the summary of 8 recipient mice of each genotype from 2 independent experiments. Because erythroblasts and later-stage erythroid cells are negative for CD45,35 the contribution of CD45.2+ (male) cells to the immature erythroid cell fraction was analyzed by qPCR, quantifying the abundance of Sry genomic DNA (Y chromosome; right panel). The average of Trim28flox/flox mice was set to 1. Male CD45.2 and female CD45.1 cells were used in the first experiment. Each circle represents an individual mouse, and black bars represent the average (right panel). The summary of the first experiment with 4 recipients of each donor genotype is shown. In the second set of experiments, female CD45.2 and male CD45.1 cells were used; an (statistically insignificant) increased contribution of coinjected competitive CD45.1 cells was observed (data not shown). (C) The absolute number of different stages of erythroid cells in the bone marrow (2 femurs and 2 tibias). Mixtures of CD45.2 and CD45.1 cells are depicted. Average with SEM. * indicates statistically significant, P < .05. NS: not significant, P > .05.
Two weeks after completion of poly(I:C) treatment, erythropoiesis in the recipient mice was analyzed. In contrast to the TMC mutant mice (Figure 2B), recipient mice reconstituted with hematopoietic TMC cells plus competitive wild-type cells exhibited only a slight increase in CD45.2+ frequency in CMP and MEP cells, whereas CD45.2+ frequency in LinKit+Sca-1+ cells was similar to the basal level (Figure 4B). Because immature erythroid cells are negative for CD45,35 we took advantage of gender difference. The amount of male-specific Sry genomic DNA allowed us to quantify the CD45.2 donor cell abundance in immature erythroid populations; to ensure that there was no gender bias in design, male CD45.2 animals were used for the first set and female CD45.2 animals were used as donors for the second set of experiments. However, this alternate approach is not applicable for later-stage (enucleated) mature erythrocytes. In keeping with the conclusions from the TMC mutant mice (Figure 1E), recipient mice reconstituted with hematopoietic TMC cells plus competitive wild-type cells exhibited a significant decrease in the CD45.2 donor cell contribution to CD71+TER119+ immature erythroid cells. These data demonstrate that differentiation of the mutant CMPs/MEPs into erythroblasts is significantly impaired when compared with the conversion of wild-type CMPs/MEPs into erythroblasts.
Although accumulation of CD71hiTER119- cells was also observed in the recipients reconstituted with hematopoietic TMC cells, the absolute number of aberrant cells per mouse was more than an order of magnitude lower than the number of aberrant cells that accumulated in the TMC mutant mice: 8.4 × 105 cells vs 93.7 × 105 cells (Figures 4C and 1E, respectively). These data demonstrate that differentiation of the mutant CMPs/MEPs into erythroblasts (r2-r4a-like cells) is defective when compared with the conversion of wild-type CMPs/MEPs into erythroblasts.
As expected (because of differentiation from the coinjected CD45.1 donor cells), 2 weeks after completion of poly(I:C) treatment, no anemic phenotype was observed in the peripheral blood (data not shown) or bone marrow (Figure 4C) of the reconstituted animals, in contrast to the TMC mutants (Figure 1E-H). These data demonstrate that deficient immature erythropoiesis resulting of the loss of TRIM28 is not a secondary response to systemic anemia or to the reduction in the number of erythroid lineage cells in the bone marrow, and that TRIM28 controls immature erythropoiesis in a hematopoietic cell–autonomous fashion.
Loss of TRIM28 alters erythroid lineage gene expression
To examine expression of the globin genes in greater detail and to address possible mechanisms that might be responsible for the observed defect in erythropoiesis in the TRIM28 mutant, we analyzed 2 immature erythroid populations in the TMC mouse (the δ and Δ cells shown in Figure 3F) 2 weeks after poly(I:C) treatment by RNA-Seq. The expression of TRIM28 mRNA in the TMC mutant is 26% in the δ (CD71hiTER119+) and 18% in the Δ (CD71hiTER119-) compared with CD71hiTER119+ cells isolated from control Trim28flox/flox bone marrow. Further analyses of TRIM28 mRNA by qRT-PCR (supplemental Figure 5) and RNA-Seq (supplemental Figure 6) revealed an almost complete absence of TRIM28 mRNA coding sequence.
First, and in contrast to our original expectations, in both the δ and Δ cells the level of εY-, βH1-, and βH2-globin mRNA was unchanged compared with that in control mice (Figure 5). Instead, we consistently observed an ∼50% (but not statistically meaningful) reduction in adult βmajor, βminor, and α-globin mRNAs in the Δ cells, but not in the δ cells (Figure 5). The level of α- and β-type globin mRNA was also analyzed and confirmed by qRT-PCR (supplemental Figure 5). In contrast to RNA-Seq, βH1-globin mRNA was approximately twofold (ranging from onefold to threefold) increased in both Δ and δ cells in the qRT-PCR assay. Although one might have anticipated that only the animals with the most complete deletion would express higher levels of βH1-globin mRNA, we detected no such inverse correlation of TRIM28 mRNA and the β-type globin mRNAs in the either δ or Δ cells (data not shown). The data obtained from RNA-Seq and qRT-PCR indicate that the contribution of TRIM28 to embryonic/fetal globin gene silencing in adult erythroblasts is very modest, if any. These data also underscore our previous conclusion that TRIM28 is dispensable for embryonic and fetal β-type globin gene repression in adult definitive erythropoiesis.
Expression profile of erythroid related genes in immature erythroid cells of Trim28 mutant mice. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox)] mice at ages 5 to 6 weeks were injected with poly(I:C) 5 times. Mice were analyzed 2 weeks after completing the poly(I:C) injections. CD71hiTER119+ immature erythroid cells (closed circle) were isolated from control Trim28flox/flox (flox/flox) mouse. CD71hiTER119+ (open circle) and CD71hiTER119- (open triangle) immature erythroid populations were isolated from mutant Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre) mouse. Cells were isolated from 3 animals of each genotype and were processed individually. RNA-Seq analysis was performed as described in “Methods.” Fragments per kilobase of exon per million mapped fragments for each animal are shown. Each circle represents an individual animal, whereas the black bars represent the averages. Gene names are shown at the top. Statistically significant changes in transcript expression analyzed in Cuffdiff analysis is depicted by a *. NS: not significant.
Expression profile of erythroid related genes in immature erythroid cells of Trim28 mutant mice. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox)] mice at ages 5 to 6 weeks were injected with poly(I:C) 5 times. Mice were analyzed 2 weeks after completing the poly(I:C) injections. CD71hiTER119+ immature erythroid cells (closed circle) were isolated from control Trim28flox/flox (flox/flox) mouse. CD71hiTER119+ (open circle) and CD71hiTER119- (open triangle) immature erythroid populations were isolated from mutant Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre) mouse. Cells were isolated from 3 animals of each genotype and were processed individually. RNA-Seq analysis was performed as described in “Methods.” Fragments per kilobase of exon per million mapped fragments for each animal are shown. Each circle represents an individual animal, whereas the black bars represent the averages. Gene names are shown at the top. Statistically significant changes in transcript expression analyzed in Cuffdiff analysis is depicted by a *. NS: not significant.
Next, we examined the expression of factors that might be responsible for repressing embryonic/fetal-globin genes3 by RNA-Seq (Figure 5) and qRT-PCR (supplemental Figure 5). The expression profiles suggested that reduced levels of SOX6, KLF1, and NR2C2 might be compensated by increased BCL11A to maintain embryonic/fetal-globin repression in the adult TMC mutant mouse. KDMA1A/lysine-specific demethylase 1 and DNA methyltransferase 1 gene levels were unchanged.
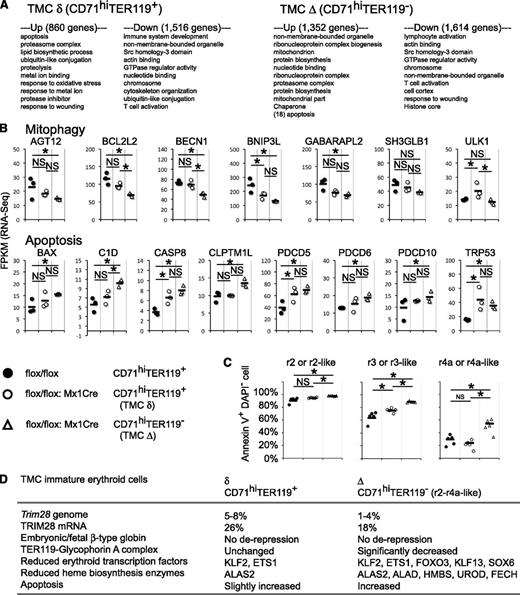
To delve into the molecular mechanisms that might cause deficient immature erythropoiesis in the TMC mutant, erythroid-related factors were evaluated from the RNA-Seq data (Figure 5) and by qRT-PCR (supplemental Figure 5). We found that many erythroid-affiliated transcription factors (KLF2, ETS1, FOXO3, KLF13, and SOX6) were affected in a statistically significant manner. The analysis also revealed a reduction in most of the enzymes required for heme biosynthesis (ALAS2, ALAD, HMBS, UROD, and FECH). One difference between Δ and δ immature erythrocytes is cell-surface TER119 expression, which is a molecule that associates with cell-surface glycophorin A (GYPA).36 The fact that there was a significant reduction of GYPA mRNA only in the Δ cells (Figure 5) suggests that the mutation should also have led to defective formation of the TER119-glycophorin A complex. Intriguingly, we found that many genes that are often used as internal standards showed unstable expression profiles in the TMC mutant cells (supplemental Figure 7). These data demonstrate that TRIM28 activates multiple transcription factors and heme biosynthetic enzymes in immature erythroid cells.
Cuffdiif statistical analyses revealed the presence of 1352 and 860 up-regulated genes, whereas 1614 and 1516 down-regulated genes were discovered in the Δ and δ immature erythroid cells, respectively. The gene lists were analyzed using the DAVID bioinformatics resource,37 and the top 10 categories for each are shown in Figure 6A. In agreement with a very recent report,38 mitochondrial-related genes differentially accumulated in the TMC mutant immature erythroid cells. We also confirmed diminished expression of mitophagy-related genes: AGT12, BCL2L2, BECN1, BNIP3L, and GABARAPL2 (Figure 6B). As novel observations, we found that several apoptosis-related mRNAs increased in the mutant, including BAX, C1D, CASP8, CLPTM1L, PDCD5, PDCD6, PDCD10, and TRP53 (Figure 6B; supplemental Figure 5).
Genomic transcriptional alterations in immature Trim28 mutant erythroid cells. (A) Gene ontology analysis. Genes that exhibit statistically significant differences between control mice and Trim28 mutant mice in the RNA-Seq experiment were analyzed by DAVID bioinformatics resources,37 and the top 10 categories for each list are shown. (B) Fragments per kilobase of exon per million mapped fragments from RNA-Seq for mitophagy and apoptosis-related genes. (C) Apoptosis analysis. Cells were isolated from bone marrow 2 weeks after poly(I:C) treatment, stained with antibodies and FITC-conjugated annexin V, and analyzed by flow cytometry. The frequency of annexin V+ DAPI- early apoptotic cells in each erythroid progenitor population is shown. CD71hiTER119+ or CD71hiTER119- cells were further subdivided by forward scatter and CD44 expression. Each circle represents an individual animal, whereas the black bars represent the averages. Data represent a summary from 2 independent experiments. * indicates statistically significant P < .05. NS: not significant, P > .05. (D) Summary of the phenotypes observed in the TMC mutant mouse.
Genomic transcriptional alterations in immature Trim28 mutant erythroid cells. (A) Gene ontology analysis. Genes that exhibit statistically significant differences between control mice and Trim28 mutant mice in the RNA-Seq experiment were analyzed by DAVID bioinformatics resources,37 and the top 10 categories for each list are shown. (B) Fragments per kilobase of exon per million mapped fragments from RNA-Seq for mitophagy and apoptosis-related genes. (C) Apoptosis analysis. Cells were isolated from bone marrow 2 weeks after poly(I:C) treatment, stained with antibodies and FITC-conjugated annexin V, and analyzed by flow cytometry. The frequency of annexin V+ DAPI- early apoptotic cells in each erythroid progenitor population is shown. CD71hiTER119+ or CD71hiTER119- cells were further subdivided by forward scatter and CD44 expression. Each circle represents an individual animal, whereas the black bars represent the averages. Data represent a summary from 2 independent experiments. * indicates statistically significant P < .05. NS: not significant, P > .05. (D) Summary of the phenotypes observed in the TMC mutant mouse.
To functionally test the hypothesis that TRIM28 normally represses apoptosis in immature erythroid cells, we analyzed the apoptotic status of TMC mutant mice by annexin V staining in combination with the DNA marker, 4,6 diamidino-2-phenylindole (DAPI). The data show that the frequency of early apoptotic cells (annexin V+ DAPI-) increased at all erythroblast stages in the Δ cells (Figure 6C). Less severe increase of early apoptotic cells was also observed in the r2 and r3 δ cells (Figure 6C). Thus, these data demonstrate that TRIM28 represses apoptosis in immature erythroid cells.
These data demonstrate that TMC mutant mice somehow generate 2 distinct types of immature erythroid cells, Δ and δ (Figures 3 and 6D; supplemental Figure 4). The majority of cells of both populations bear homozygous deletions of the Trim28 gene (Figure 1C), and both populations exhibited an almost complete absence of TRIM28 mRNA (supplemental Figures 5 and 6). Although these 2 populations were similar in morphologic appearance and CD44-FSC profile (Figure 3), the Δ cells showed more severe phenotype compared with the δ cells in the expression of TER119 (Figure 3F), GYPA, erythroid transcription factors, heme biosynthesis enzymes (Figure 5), mitophagy-related genes, and apoptosis-related genes (Figure 6B), as well as an increase in early apoptotic cells (Figure 6C). The frequency and absolute cell number of δ cells in bone marrow were similar to those of the Δ cells at 2 weeks, but were lower at 4 weeks after Trim28 inactivation (Figure 1B,E). These data indicate that TRIM28 is not required for the expression of TER119 for approximately half of the immature erythroid cells. Taken together, these data suggest that TRIM28 controls immature stages of erythropoiesis by regulating multiple erythroid transcription factors, multiple heme biosynthesis enzymes, and apoptosis.
Discussion
Erythroid-specific expression of the mammalian globin genes is controlled by multiple tissue-specific and tissue-restricted positive and negative transcriptional regulators. The nuclear receptors NR2C1 and NR2C2 were originally identified in erythroid cells as direct transcriptional repressors of the embryonic and fetal β-type globin genes.5-7 Multiple transcriptional cofactors, including TRIM28, were among numerous other NR2C1/NR2C2-associated proteins identified later.8 Here, we tested the requirement for TRIM28 on embryonic/fetal globin gene repression by inducibly ablating Trim28 in adult mice using the poly(I:C)-inducible Mx1Cre transgene, referred to as TMC mutants. Somewhat surprisingly, the expression of εY- and βH1-globin mRNAs was not altered after induced genetic loss of TRIM28; however, efficiently Trim28-deleted mice had severe normocytic anemia. Subsequently, defects in erythropoiesis in the bone marrow were observed at multiple erythroblast differentiation stages in the Trim28 mutants. CMP and MEP progenitors, but not HSC/MPP or GMP progenitors, abnormally accumulated in the TMC mutant animals. In competitive adoptive transfer experiments, a similar reduction of erythroblasts was observed when the mutants were assayed shortly after poly(I:C) administration/Trim28 ablation. In summary, these results demonstrated that TRIM28 is not required for the repression of embryonic/fetal globin genes during adult (definitive) erythropoiesis, but is required for the differentiation of erythroblasts from CMP and/or MEP progenitors in a cell-autonomous fashion.
In contrast to the erythroid defects, development of myeloid cells appeared to be quite normal in both the bone marrow (Mac1+) and peripheral blood (monocytes and neutrophils). Additionally, we recorded a statistically significant reduction in the number of lymphocytes in the peripheral blood of the TMC mutants: 42% of controls at 2 weeks and 36% at 4 weeks after Trim28 inactivation (data not shown). These data are consistent with recent findings showing that TRIM28 controls the development and functions of B and T lymphocytes as revealed by lymphocyte cell–specific ablation of TRIM28.22-25 These observations indicate that TRIM28 is essential for the development of multiple hematopoietic cell lineages. Because Mx1Cre is not active exclusively in erythroid cells,26 we could not exclude the possibility that loss of TRIM28 in these lymphoid cells or in other cells indirectly affects erythroblast differentiation. Our attempt to conditionally ablate Trim28 gene using Epor-Cre,27 which is predominantly active in immature erythroid cells, resulted in incomplete excision in CD71+TER119+ immature erythroid cells.
Multiple erythroid transcription factors and heme biosynthesis enzymes are down-regulated in the TMC mutant immature erythroid cells. We also confirmed decreased expression of mitophagy-related genes, consistent with another recent report.38 Furthermore, we documented induction of apoptosis-related genes in the TMC mutant immature erythroid cells, and experimentally confirmed the functional contribution of TRIM28 in normally repressing apoptosis in immature erythroid cells. We propose that diminished expression of multiple erythroid transcription factors, heme biosynthesis enzymes, and mitophagy-related proteins, as well as increased expression of apoptosis-related pathway enzymes, causes defective immature erythropoiesis in the bone marrow, thus leading to normocytic anemia in the Trim28 mutant mice.
Multiple transcription factors that have been shown to regulate embryonic and fetal globin genes were compromised in the TMC mutant mice. The balance between decreased (KLF1, SOX6, and NR2C2) and increased (BCL11A) factors seems to offset one another to maintain εY- and βH1-globin gene repression in the TMC mutant. Although these data demonstrate that TRIM28 is not required for the maintenance of embryonic/fetal β-type globin gene repression, it is nonetheless possible that TRIM28 may contribute to the initiation of their silencing.
RNA-Seq followed by Cuffdiif statistical analyses revealed 860 and 1352 up-regulated genes, and 1516 and 1614 down-regulated genes in the δ and Δ immature erythroid cells, respectively. This result was somewhat unexpected because TRIM28 has been shown to recruit HP1 and SETDB1, both of which function in transcriptional repression and was, in general, previously recognized solely as a corepressor.11-13 The RNA-seq results might additionally implicate TRIM28 as a coactivator, although not many examples have been reported to date.39 Alternatively, TRIM28 may function as a repressor of a repressor.
In conclusion, this work has revealed previously unknown functions for the transcriptional cofactor TRIM28 in erythropoiesis.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kevin Lane, Carmen Yu, and the Unit for Laboratory Animal Medicine at the University of Michigan for experimental support and excellent animal care. The authors also thank members of the Engel Laboratory for advice and discussion.
This work was supported by National Institutes of Health grants HL24415 and HL114368 (J.D.E.) and was supported in part by a fellowship from the Astellas Foundation for Research on Metabolic Disorders (T.H.).The research was also supported in part by the National Cancer Institute through the University of Michigan’s Cancer Center Support Grant (P30 CA046592) and the use of the following cores: Flow Cytometry, Morphology, Microscopy and Image Analysis Laboratory, Experimental Irradiation, and Sequencing.
Authorship
Contribution: T.H., O.T., and J.D.E. designed the study; R.L. generated an indispensable mouse reagent; T.H. and M.C. performed the experiments; T.H., M.C., and J.D.E. analyzed the data; and T.H. and J.D.E. wrote the paper.
Conflict-of-interest disclosure: The authors have no competing financial interests.
Correspondence: James Douglas Engel, 3071 BSRB, 109 Zina Pitcher Place, University of Michigan Medical School, Ann Arbor, MI 48109; e-mail: engel@umich.edu.

![Figure 1. Erythropoiesis in mice inducibly ablated for Trim28. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected every 48 hours with poly(I:C) 5 times to transcriptionally activate the cre-expressing transgene.26 Uninjected Trim28flox/+ (flox/+) mice were used as controls. Mice were analyzed 2 weeks (2w) or 4 weeks (4w) after completion of poly(I:C) administration. (A) The absolute number of total bone marrow cells isolated from 2 femurs plus 2 tibias of individual animals (each circle) of various genotypes. The black bar represents the average. * indicates statistically significant, P < .05. NS: not significant, P > .05. (B) Flow cytometric analysis of erythropoiesis in total bone marrow cells from mutant and control mice. The numbers in each of the boxed areas indicate the mean frequency of cells in each hematopoietic population. (C) The abundance of Trim28 floxed allele in CD71+TER119+ immature erythroid cells was analyzed by qPCR. The CD71hiTER119- cells that aberrantly accumulated in the TMC mutant mice (panel B) were also analyzed. An average of the relative amount of the undeleted gene in the control Trim28flox/flox cell was set to 200, which is the copy number of the floxed allele in 100 cells. (D) The expression level of β-type globin genes normalized to β-actin in CD71+TER119+ immature erythroid cells was analyzed by qRT-PCR. The average of the relative amount in the control Trim28flox/flox cell was set to 1. (E) The absolute number of cells in each of the analyzed populations was calculated from the data shown in (A) and (B). Average with standard error of the mean (SEM). (F) Flow cytometric analysis of TER119+ cells in total bone marrow cells (separated by forward scatter and CD44 antigen according to Chen et al34). The numbers closest to the boxed areas indicate the mean percentage of cells in each population within parental TER119+ cells. (G) The absolute number of basophilic erythroblasts (r2), polychromatic erythroblasts (r3), orthochromatic erythroblasts (r4a), reticulocytes (r4b), and RBCs (r5) in the bone marrow was calculated from the data shown in panels (A) and (F). Error bars represent the SEM. (H) Hematologic parameters of peripheral blood cells. Data represent either a summary (A,C,D,E,G,H) or representative individuals (B,F) of more than 3 mice of each genotype from 2 to 4 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/23/10.1182_blood-2013-04-496166/4/m_3798f1.jpeg?Expires=1769086387&Signature=16uG31IebjdnKObgUXo3dp0W4ed~3DDiso1yygZzoTIMi2lQDURh-FvzQ6dBHsM1QX1wW7LeH3Ov0V78smqeMVG9H5wwq43vCaAerZQN~oqeXFshat8~LEoYY3KkW9uw5tI7oDXPuAVABQ6SZi9wi~XNiyIw8JZnP5~irm6lWsBE7MkGkK8EGcKKklY2PkpWUVETJJX8gxPVdf1mhIOMtADbtqOIxVZp01SKAPoNxPNBp-TUUQ~vSNcVPL7YaXxQ~5C38xY0Fk5TqnvZC5V0qE6J6KNhW35tf2tx71AdqDXvqrCoZwNDdBy4pwSrhYlH4T7073l2gedOYz6UDrJABQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Abnormal development of hematopoietic progenitor cells in Trim28 mutant mice. Adult TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected with poly(I:C). (A) Flow cytometric analysis of total bone marrow cells and division into individual early progenitor compartments (shown on the right) according to cell surface expression of cKit, Sca1, FcγR, CD34, and lineage (Lin) markers. Numbers in the boxed areas indicate the mean percentages of cells in the gated area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population (Frequency of Parent in FlowJo software). (B) The absolute number of HSC/MPP, CMP, MEP, and GMP was calculated from the data shown in panel (A) and total bone marrow cell number. Averages are indicated with SEM. * indicates statistically significant, P < .05. NS: not significant, P > .05. (C) The absolute number of Mac1+ cells in the bone marrow of mutant and control mice. (D) The absolute number of cKithi Lin- and cKithiLin+ cells in the bone marrow of control and conditionally mutant mice. (E) The absolute number of cKithiCD71hiTER119-, cKithiCD71-TER119-, and cKithiTER119+ cells in the bone marrow. The lineage mixture used for panels (A, B, and D) includes antibodies that recognize TER119, Mac1, Gr1, B220, CD19, CD5, CD4, and CD8a. Mice were analyzed for 2 weeks (A, B, D, E) or for 2 and 4 weeks (C) after completion of poly(I:C) administration. The data represent a summary (B-E) or are representative (A) of more than 3 mice of each genotype from 2 to 3 independent experiments. Each circle represents an individual mouse, and the black bars represent averages.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/23/10.1182_blood-2013-04-496166/4/m_3798f2.jpeg?Expires=1769086387&Signature=EK4hD2Y5BWEaMgTW7LpQfTAJyptmFfQHnrHkzBjCglL38si3seSLsT~2mToFcAMe3jFt3ClUS4iIyoG59SE5-Uu~E~M~6ZjxYzdJw4DfWQF2JFuymxcbnshXyZlmHaIE9oeIuBwQeGJ5hQDC1grjPhah5UxzrTZn2iH0lzwVPub5R5XT~5cUe48ir92-SsYf4xxTUwB5yHltqcFALDGAUOVSoaKuGXEONWRsuKZXFrv8XfhnPmbOAcIi-awUdk-e6p09ICwYV3gNXrM8CdW34mZl13zfC0CUqL~mBGzY~SGjyd61qY9UFKbmQuQx423xKMvQ57Davz8kki78ftNQNA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Detailed characterization of the aberrant CD71hiTER119- cell population that accumulates in Trim28 mutant bone marrow. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox) and Trim28flox/+:TgMx1Cre (flox/+:Mx1Cre)] mice at ages 5 to 6 weeks were injected with poly(I:C). Mice were analyzed 2 weeks after the final poly(I:C) administration. (A) Color of cells separated on the basis of CD71 and TER119 expression. Between 4 and 10 × 105 cells were flow sorted and suspended in PBS supplemented with 2% FBS in a 1.5-mL tube and centrifuged to form a pellet. Images were acquired using a Zeiss Stemi SV11 Apo stereomicroscope, AxioCam digital camera, and AxioVision LE software. (B) Flow cytometric analysis of forward scatter (FSC), CD45, cKit, Mac1, and B220 in the CD71hiTER119- bone marrow cells of control mice and Trim28 mutant mice. Numbers in the boxed areas indicate the mean percentages of cells in the boxed area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population. (C) The absolute number of different subpopulations in the CD71hiTER119- cells was calculated from panels (B) and normalized for total bone marrow cell number. Average with SEM. *indicates statistically significant, P < .05. NS: not significant, P > .05. (D) Neutral benzidine staining of individual cells recovered from flow-sorted populations of different genotype mice treated with poly(I:C). (E) Wright-giemsa staining. Images (D and E) were acquired using an Olympus BX-51 upright light microscope with the 100× oil immersion objective lens, DP-70 high-resolution digital camera, and DP Controller software. (F) Flow cytometric analysis of FSC and CD44 in CD71hiTER119- and CD71hiTER119+ cells. Numbers in or near the boxed areas indicate the mean percentage of cells in the boxed area calculated by dividing the number of cells in that subpopulation by the number of cells in its immediate ancestor population. The data represent a summary (C) or representative individuals (A, B, D-F) of more than 3 mice of each genotype examined in 2 to 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/23/10.1182_blood-2013-04-496166/4/m_3798f3.jpeg?Expires=1769086387&Signature=Em46srR0RuUDWRI1PVsVXYM93DkjXXSflvh~rtslFPkq83kE-XYx~xKVkLg7Nd0ffbwezWqM-ifS0pe1TL7N8HQUg2wztz8Ack5PVwa1eTWAgPOaylkcot6mu6g3huLbFmaeW8ZtUBv5Fy8XDWhEjL2RxDBgmbj4F6VyFqKz9ZQcdGTgI6zWvmUOy1OzZ5MWoTC3S1qQu~L~vZxNQfnIhhdr~-6K2Uil8Ol2e29LLzAe0yhH-bDZUqisWNUx8gpWqju2DgZ3T4ryOJoPX6xHfEPaGie6d3L4WP7hzVNBUTBmzcqbkstcE2Qf18LR9Q-61-Ro-rHQy4CimwqP7jkZfw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Expression profile of erythroid related genes in immature erythroid cells of Trim28 mutant mice. TMC [Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre)] mutant or control [Trim28flox/flox (flox/flox)] mice at ages 5 to 6 weeks were injected with poly(I:C) 5 times. Mice were analyzed 2 weeks after completing the poly(I:C) injections. CD71hiTER119+ immature erythroid cells (closed circle) were isolated from control Trim28flox/flox (flox/flox) mouse. CD71hiTER119+ (open circle) and CD71hiTER119- (open triangle) immature erythroid populations were isolated from mutant Trim28flox/flox:TgMx1Cre (flox/flox:Mx1Cre) mouse. Cells were isolated from 3 animals of each genotype and were processed individually. RNA-Seq analysis was performed as described in “Methods.” Fragments per kilobase of exon per million mapped fragments for each animal are shown. Each circle represents an individual animal, whereas the black bars represent the averages. Gene names are shown at the top. Statistically significant changes in transcript expression analyzed in Cuffdiff analysis is depicted by a *. NS: not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/23/10.1182_blood-2013-04-496166/4/m_3798f5.jpeg?Expires=1769086387&Signature=zX7Qu0AwMBtwj1rC9dy-X~lKeaJsMiI4A8ei4OidAuzaE2bqRIfW7u00AHJtnP5X~NOiu4pfufgRv2VXMTEoiZdIMi3BFAqbhde9f5h2OETZ93qnTEMpA7fiGkwh8cLxMOHyLf7fvJvZ0uyGODvhp1psAwsWb-L7kYjMvkrHlimbx64YCCAJwJTMJHe5aWz6si1VDi-rPcOAaAx4j9d0QhnEiFP5YcrBMAZ-gKZ3ZlpXc9O7s8mc9jiPp377T6ZOj2~PheaK8Ev8NB0SGxFuDzMcrgC37fQ7AOU43g85kI7eEXKiYLCzBzpaBk~DblH68g2UeligKZVmHKc3fLaoIQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal