Abstract
Epigenetic events including DNA methylation (ME) and histone modification (HistMod) regulate gene and protein expression levels. The HistMod code has mono, di and tri ME activation marks on histone 3 lysine 4 (H3K4me2, 3) and repressive ME n at H3K27 regulated by many methyl transferases (e.g ASH2L) and demethylases (LSD1). Likewise histone acetylation represses transcription regulated by several histone acetylases and deacetylases (HDAC1 to 6). Additional regulation comes from the Jumanji and Sirtulin families. To date the integrated status of the level of expression of these different regulators has not been studies at the protein level in AML.
To define the role of the HistMod proteins in AML we made a reverse phase protein array (RPPA) with protein from leukemia enriched cells from 511 new AML patients. Both bone marrow (n=387) and peripheral blood (n=283) samples were used, with 140 cases having both. The RPPA was probed with 231 strictly validated antibodies, including antibodies against 15 proteins known to be involved in HistMod: H3, H3K4Me2 and Me3, H3K27Me3, HDAC1,2,3, JumanjiD6, ASH2L, LSD1, SIRT1, BMI1 (unrepresses H2K119), Nucleolin (NCL, binds H1, causes chromatin decondensation), NPM1 (H3 chaperone) Trim24(H3K27 acetylator) and 4 with very strong correlation to these: CLPP, WTA,P Fli1 and hNRNPK. RPPA data was normalized as previously published. Expression was compared to that of normal bone marrow derived CD34+ cells.
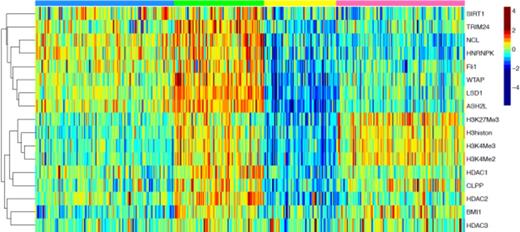
All 19 proteins had heterogeneous expression (Figure1). The optimal number of principal components based on protein clusters was determined to be 3 comprised of: 1) Histone 3 status (H3STAT. total, K4Me2, Me3 and K27 Me3) 2)Histone methylation regulator (MethReg, ASH2L, LSD1, WTAP, FLI1, Trim24, NCL, hNRNPK and Sirt 1 and 3) HDAC regulating (HDAC, HDAC1, 2, 3, along with BMI1, JMJD6,CLPP and NPM1. Another statistical approach determined that 4 patient groups was “optimal”. The 4 groups were 1)PanLow, characterized by below normal expression of all three proteins clusters (Yellow, n=70, 16.9%), 2) H3STAT-Only in which all members of the H3STAT cluster were highly expressed but the MethReg cluster had low expression and HDAC was weakly on (Blue n=134, 32.3%), 3) MethReg&HDAC, in which the MethReg group is highly expressed, HDAC is moderately on, but H3STAT is unmethylated (n=124, 29.9%), 4) PAN On in which all three clusters are highly expressed (n=87, 21%). In general the H3STAT, MethReg and HDAC principal components were expressed independently of each other. Cytogenetics were independent of HistMod patient group (p= 0.07) although 55% of favorable patients were in the Meth Reg. Patients with an antecedent hematological disorder wrere rarely in the PanLow group (p=0.008) and FLT3-ITD patients were most frequently in the PanOn group (p= 0.005). For patients with favorable prognosis cytogenetics those with active MethReg&HDAC did substantially better than patients in the other groups (only 2 relapses, 15/16 alive vs. 9/16 relapses and 7/16 alive , OS p = 0.002, RemDur p = 0.009). HistMod group was not prognostic in those with intermediate cytogenetics, even if stratified for FLT3 mutation status. Notably 55 patients (mostly older) were treated with histone demethylating or histone deacetylating agents alone or in combination. Among these patients, those in the PanOn cluster were much less likely to achieve remission 1 of 10 vs. 11/36) and had inferior overall survival (p= 0.001, median 14 weeks vs. 52).
Expression of HistMod members. Colors across top refer to groups defined above. Color scale shows expression on a Log-2 scale.
Expression of HistMod members. Colors across top refer to groups defined above. Color scale shows expression on a Log-2 scale.
HistMod proteins formed recurrent patterns of proteins expression with H3STAT and MethReg expression independent of each other. Having both motifs on was adverse, especially in favorable prognosis patients. Stem cell transplant is generally only used in favorable prognosis cytogenetics patients that relapse, but the HistMod group detected those likely to relapse and die, suggesting the ability to detect those that should go to stem cell transplant in first CR. Patients with high levels of proteins in all three groups also did poorly with demethylating and HDAC agents suggesting that such patients should receive alternate therapy. High ASH2L expression correlated strongly with functionally opposite LSD1, but seems to dominate as it was associated with higher H3K4methylation.
Ravandi:Sunesis: Consultancy, Honoraria, Research Funding; Celgene: Honoraria, Research Funding; Novartis: Honoraria, Research Funding; Bristol Myers Squibb: Honoraria, Research Funding; Teva: Consultancy, Honoraria; Pfizer: Honoraria; Merck: Research Funding; Bayer/Onyx: Consultancy, Honoraria; EMD Serono: Research Funding; Medimmune: Research Funding; Amgen: Consultancy, Honoraria; Seattle Genetics: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal