Abstract
In acute myeloid leukemia (AML), numerous genetic and epigenetic changes have been identified that result in loss of differentiation, apoptosis, and cell cycle arrest. In contrast, changes in protein expression are not as well characterized. To address these deficiencies, we performed RPPA analysis on 415 de novo AML patient samples and identified increased expression of heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a predictor of poor outcome. These elevated hnRNP K levels were most predictive in AML patients who also harbored mutant NPM1 and wild type FLT3. While NPM1MUT/FLT3WT status typically confers favorable prognoses, increased hnRNP K expression negated this effect, as greater than 90% of NPM1MUT/FLT3WTindividuals with elevated hnRNP K expression died within 12 months.
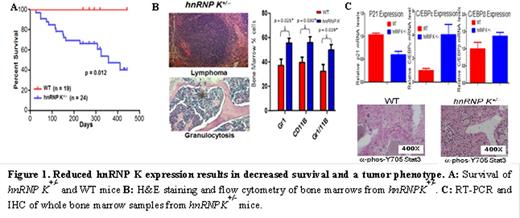
hnRNP K is a multifunctional protein that controls basic cellular functions through RNA, DNA, and protein-protein interactions (e.g.; p53) and whose expression is often altered in cancer. To examine the biological role of hnRNP K in vivo, we generated mouse models that either increase or decrease hnRNP K expression. Biallelic hnRNP K deletion results in embryonic lethality, while haploinsufficiency (hnRNP K+/-) results in a partial neonatal lethal phenotype. Surviving hnRNP K+/- mice have reduced survival and are more tumor prone than wild type mice (Fig. 1A and B). Analysis of hnRNP K+/- peripheral blood and bone marrow revealed significant hematologic neoplasms, including myeloid hyperplasias. The myeloid expansion appears to be a consequence of defects in proliferation (decreased p21) and differentiation (increased C/EBPβ and ε expression and activation of Stat3) pathways (Fig. 1C).
In vitro studies using hnRNP K+/- hematopoietic stem cells (HSC) and mouse embryo fibroblast (MEFs) likewise revealed defects in differentiation and proliferation potential. HSC were used in burst formation unit erythroid colony assays (BFU-E). In these experiments, we observed a significant increase in the number of hnRNP K+/- cells and immature cells as compared to wild type BFU-E (Fig. 2A and B). RT-PCR analysis of BFU-Es revealed deregulation of p53/p21 and TGFβ- pathway genes (Fig. 2C). Similarly, hnRNP K+/- MEFs failed to properly activate the p53/p21 pathway following exposure to ionizing radiation (Fig. 2D).
In contrast to diminished hnRNP K expression, overexpression results in activation of pro-growth and self-renewal pathway proteins in both humans and mice. RPPA analysis of AML patient samples that overexpress hnRNP K, as well as transient overexpression of hnRNP K in cell lines, results in increased expression of c-Myc. To directly examine the impact of hnRNP K overexpression in vivo, we generated transgenic mouse models. hnRNPK-transgenic mice express elevated levels of hnRNP K and are tumor prone.
While it is tempting to classify hnRNP K as either an oncogene or a tumor suppressor, our haploinsufficiency and overexpression data seem to indicate that abnormal expression in either direction has a significant impact on tumor predisposition. Mechanistically, hnRNP Kappears to be an influential regulator involved in proliferation, self-renewal, and differential programs. The functional consequences reduced and overexpression of hnRNP K is currently under investigation.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal