Key Points
The MDS1-EVI1 isoform of the MECOM locus is required for MLL-AF9–induced myeloid leukemia.
Within MDS1-EVI1, it is the PR domain that is essential.
Abstract
A subgroup of leukemogenic mixed-lineage leukemia (MLL) fusion proteins (MFPs) including MLL-AF9 activates the Mecom locus and exhibits extremely poor clinical prognosis. Mecom encodes EVI1 and MDS1-EVI1 (ME) proteins via alternative transcription start sites; these differ by the presence of a PRDI-BF1-RIZ1 (PR) domain with histone methyltransferase activity in the ME isoform. Using an ME-deficient mouse, we show that ME is required for MLL-AF9–induced transformation both in vitro and in vivo. And, although Nup98-HOXA9, MEIS1-HOXA9, and E2A-Hlf could transform ME-deficient cells, both MLL-AF9 and MLL-ENL were ineffective, indicating that the ME requirement is specific to MLL fusion leukemia. Further, we show that the PR domain is essential for MFP-induced transformation. These studies clearly indicate an essential role of PR-domain protein ME in MFP leukemia, suggesting that ME may be a novel target for therapeutic intervention for this group of leukemias.
Introduction
Acute myeloid leukemias bearing mixed-lineage leukemia (MLL) fusion proteins (MFPs) have a poor prognosis; chemotherapy is inadequate, indicating the need for more effective therapies.1-3 Insights into the molecular pathogenesis will greatly facilitate the development of targeted agents.
In a subset of MFP AMLs, specifically those lacking monocytic features,4 MFPs can bind to and activate transcription of MECOM,4-6 a highly conserved protooncogene encoding MDS1-EVI1 (ME), and EVI1 isoforms via distinct transcription sites (see Figure 1A). Relative to EVI1, ME possesses a PRDI-BF1-RIZ1 (PR) domain with histone methyltransferase (HMT) activity.7 In this study, using mouse alleles where ME is constitutively (MEm1) or conditionally (MEfl4) lost,8 we reveal the PR domain as being essential in MFP transformation in mice. These results strongly suggest that ME is a novel target for therapeutic intervention.
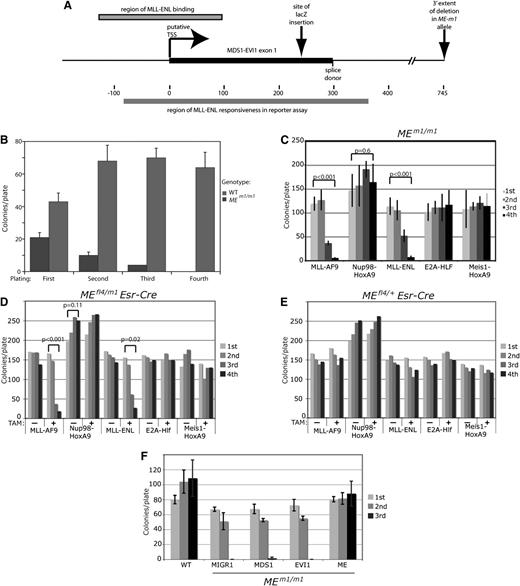
MEm1/m1bone marrow is resistant to MLL-AF9–induced transformation. (A) Diagram of the MEm1 allele, showing exon 1 of Mds1, with putative transcription start site and splice donor site, as well as site of lacZ insertion and the extent of the DNA of the first intron deleted. Also shown are the locations of MFP binding described by Arai et al,5 as well as the MFP-responsive region identified by this same group by luciferase reporter assays.5 (B) Quantitation of number of colonies formed in growth factor–supplemented methylcellulose at each of 4 replatings, for WT and MEm1/m1 bone marrow. Error bars represent standard deviation of platings done in triplicate, 1000 cells per plate. The experiment was repeated multiple times with the same result. (C-E) ME requirement is restricted to transformation by MLL fusion genes. (C) Quantitation of number of colonies formed in growth factor–supplemented methylcellulose at each of 4 replatings for MEm1/m1 LSK cells transduced with the virus indicated. Error bars are standard deviation; P values were determined by Student t test, comparing the first and fourth platings. (D-E) Serial replating assay of bone marrow from mice of the genotype indicated, transduced with the leukemogenic oncogene indicated, and treated or not with 4-OH TAM (1 µM) as indicated. Error bars are not shown but are within 10% of the value of each bar; P values were calculated by Student t test, comparing first and fourth platings. (F) ME but not EVI1 or MDS1 can rescue the transformation deficiency of MEm1/m1 bone marrow. Serial replating transformation assay of LSK cells isolated from either WT or MEm1/m1 bone marrow, transduced with MLL-AF9, as well as retroviral expression construct indicated: MIGR1 (empty vector), MDS1, EVI1, or ME. Error bars denote standard deviation. Third replating yielded no colonies for MIGR1- and EVI1-transduced cells.
MEm1/m1bone marrow is resistant to MLL-AF9–induced transformation. (A) Diagram of the MEm1 allele, showing exon 1 of Mds1, with putative transcription start site and splice donor site, as well as site of lacZ insertion and the extent of the DNA of the first intron deleted. Also shown are the locations of MFP binding described by Arai et al,5 as well as the MFP-responsive region identified by this same group by luciferase reporter assays.5 (B) Quantitation of number of colonies formed in growth factor–supplemented methylcellulose at each of 4 replatings, for WT and MEm1/m1 bone marrow. Error bars represent standard deviation of platings done in triplicate, 1000 cells per plate. The experiment was repeated multiple times with the same result. (C-E) ME requirement is restricted to transformation by MLL fusion genes. (C) Quantitation of number of colonies formed in growth factor–supplemented methylcellulose at each of 4 replatings for MEm1/m1 LSK cells transduced with the virus indicated. Error bars are standard deviation; P values were determined by Student t test, comparing the first and fourth platings. (D-E) Serial replating assay of bone marrow from mice of the genotype indicated, transduced with the leukemogenic oncogene indicated, and treated or not with 4-OH TAM (1 µM) as indicated. Error bars are not shown but are within 10% of the value of each bar; P values were calculated by Student t test, comparing first and fourth platings. (F) ME but not EVI1 or MDS1 can rescue the transformation deficiency of MEm1/m1 bone marrow. Serial replating transformation assay of LSK cells isolated from either WT or MEm1/m1 bone marrow, transduced with MLL-AF9, as well as retroviral expression construct indicated: MIGR1 (empty vector), MDS1, EVI1, or ME. Error bars denote standard deviation. Third replating yielded no colonies for MIGR1- and EVI1-transduced cells.
Study design
Mice
Retroviral constructs
Serial replating assay
Lineage-negative/Sca-1+/c-kit+ (LSK) cells were isolated (t = 0), infected8 with retrovirus (t = 15 hours), sorted (t = 63 hours), and plated in M3434 (StemCell). For Figure 1F, add-back infections were performed following MLL-AF9 sort (t = 64 hours), sorted for green fluorescent protein (GFP) (t = 90 hours), and plated in M3434.13
In vivo leukemogenesis assay
Primary leukemias were harvested and explanted to culture, +/− 4-OH tamoxifen (TAM); 106 spleen cells per mouse were transplanted into sublethally irradiated secondary recipient mice via tail vein injection. Leukemia development in transplanted mice was monitored by assessing complete blood counts (CBCs) every week.
Results and discussion
Knockout of Mds1/ME isoforms abrogates the ability of MLL-AF9 to transform bone marrow progenitors; add-back assay shows primary role for PR domain of ME isoform
To test the role of ME in MLL-AF9 leukemogenesis, we infected bone marrow cells from MEm1/m1 mice,8 which bear a lacZ insertion in exon 1 of Mds1 (Figure 1A) and lack MDS1 and ME but express normal levels of EVI1, and performed a serial replating assay.13-16 Although MLL-AF9 induced wild-type (WT) cells to form colonies at each cycle, it was unable to transform MEm1/m1 cells (Figure 1B). These findings indicate a dependency of MLL-AF9 transformation on functional Mds1 and/or ME.
We then tested if the transformation block was specific for MLL-AF9: MEm1/m1 cells were transduced with MLL-AF9, Nup98-HoxA9, MLL-ENL, E2A-Hlf, or Meis1-HoxA9 and were assayed for transformation. This revealed that cells lacking ME were resistant to transformation only by MLL-AF9 or MLL-ENL (Figure 1C). Thus, the block to transformation is oncogene specific. To further test this finding, MEfl4/m1/Esr-Cre (Figure 1D) and MEfl4/+/Esr-Cre (Figure 1E) LSK cells were transduced with the same oncogenes, split into 2 treatment groups (vehicle or 4-OH TAM), and assayed for transformation. All vehicle-treated cell samples, regardless of genotype, were fully capable of being transformed with all oncogenes (Figure 1D-E), whereas 4-OH TAM–treated MEfl4/m1/Esr-Cre cells displayed the same selective resistance to transformation; 4-OH TAM–treated MEfl4/+/Esr-Cre cells were susceptible to transformation by all oncogenes. Together, these data indicate a selective requirement for a functional ME allele for transformation by MFP oncogenes.
To test which isoform of Mecom is required for MLL-AF9 transformation, we retrovirally transduced LSK cells from MEm1/m1 mice with MIGR1, or with constructs for MDS1, EVI1, or ME, and then infected them with MLL-AF9 retrovirus; transduced cells were assayed for transformation. Although neither MDS1 nor EVI1 was able to rescue the deficiency, ME was able to (Figure 1F), confirming that in the context of the MEm1/m1 genotype, ME is the essential isoform that is lacking. Because the major difference between ME and EVI1 is the PR domain, it is clear that this domain is critical for ME activity in the setting of MLL-AF9–induced leukemogenesis.
Knockout of ME results in suppression of MLL-AF9 leukemia development in transplanted mice
To test if ME is required for transformed MLL-AF9 AML cells to survive in vivo, we established transplantable leukemias and then deleted the gene and assayed for leukemogenesis by transplantation (Figure 2A). MEfl4/m1/Esr-Cre or MEfl4/+/Esr-Cre LSK cells were transduced with MLL-AF9 and transplanted into irradiated recipients. After 3 months, primary AMLs developed, and leukemic spleen cells were harvested, induced with 4-OH TAM to delete ME (yielding genotypes MEko4/m1 and MEko4/+, respectively), and injected into irradiated recipients. Weekly CBCs noted an increasing leukocyte count and persistently low hematocrit and platelet count in mice receiving MEfl4/m1/Esr-Cre cells not treated with 4-OH TAM (Figure 2B-D), indicating infiltration of leukemic cells into the blood and bone marrow; in mice receiving 4-OH TAM–treated MEfl4/m1/Esr-Cre cells, a normal leukocyte level was maintained, and platelets and hematocrit recovered to normal levels. The control group mice receiving MEfl4/+/Esr-Cre cells with and without 4-OH TAM treatment all develop leukemia as expected (not shown).
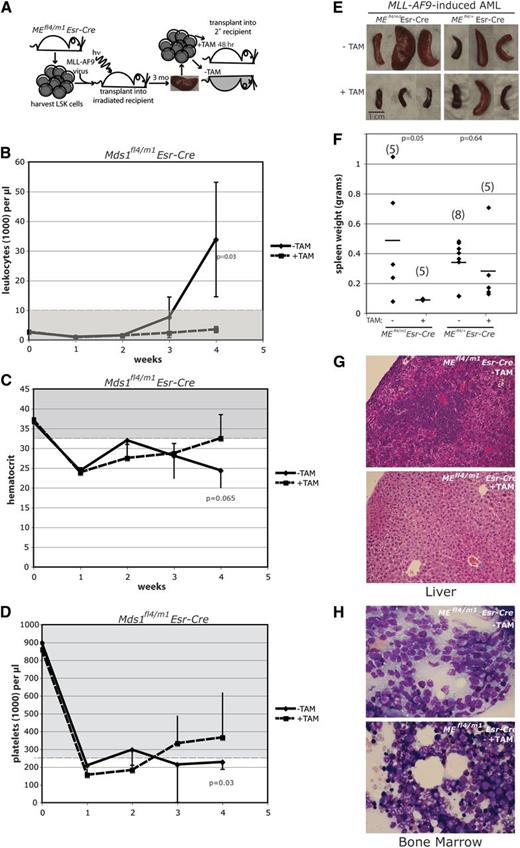
Deletion of ME results in failure of MLL-AF9 leukemic cells to transplant into syngeneic sublethally irradiated recipient mice. (A) Diagram depicts experimental procedures. (B-D) Shown are CBC data from weeks 0 to 4 posttransplant for recipients of MEfl4/m1 cells, with and without pretreatment with 4-OH TAM; extent of normal values indicated by grayed zone. Mice receiving cells with no 4-OH TAM pretreatment became frankly leukemic, anemic, and thrombocytopenic over the 4 weeks of monitoring. At week 4, most recipients of the untreated cells were moribund, and the mice were necropsied. Error bars depict standard deviation. Statistical significance (Student t test) was observed at week 4 for leukocytes and platelets (P < .05). (E-H) Deletion of ME results in failure of leukemic cells to significantly infiltrate organs of irradiated recipients. Mice receiving 4-OH TAM–pretreated MLL-AF9 leukemia cells maintained normal spleen weights (E-F), as well as livers and bone marrow essentially devoid of infiltrating leukemia; non-pretreated cells infiltrated spleen, liver, and bone marrow (G-H). (E) Gross photographs of spleens from mice injected with untreated and 4-OH TAM–pretreated cells. (F) Scattergrams of spleen weights of the 4 experimental groups, as indicated; number of spleens in cohort in parentheses. Average is demarcated by horizontal bar; P value determined by Student t test. (G-H) Photomicrographs of liver (G) and bone marrow (H), showing extensive infiltration by leukemic cells (original magnification ×200 magnification; hematoxylin and eosin staining).
Deletion of ME results in failure of MLL-AF9 leukemic cells to transplant into syngeneic sublethally irradiated recipient mice. (A) Diagram depicts experimental procedures. (B-D) Shown are CBC data from weeks 0 to 4 posttransplant for recipients of MEfl4/m1 cells, with and without pretreatment with 4-OH TAM; extent of normal values indicated by grayed zone. Mice receiving cells with no 4-OH TAM pretreatment became frankly leukemic, anemic, and thrombocytopenic over the 4 weeks of monitoring. At week 4, most recipients of the untreated cells were moribund, and the mice were necropsied. Error bars depict standard deviation. Statistical significance (Student t test) was observed at week 4 for leukocytes and platelets (P < .05). (E-H) Deletion of ME results in failure of leukemic cells to significantly infiltrate organs of irradiated recipients. Mice receiving 4-OH TAM–pretreated MLL-AF9 leukemia cells maintained normal spleen weights (E-F), as well as livers and bone marrow essentially devoid of infiltrating leukemia; non-pretreated cells infiltrated spleen, liver, and bone marrow (G-H). (E) Gross photographs of spleens from mice injected with untreated and 4-OH TAM–pretreated cells. (F) Scattergrams of spleen weights of the 4 experimental groups, as indicated; number of spleens in cohort in parentheses. Average is demarcated by horizontal bar; P value determined by Student t test. (G-H) Photomicrographs of liver (G) and bone marrow (H), showing extensive infiltration by leukemic cells (original magnification ×200 magnification; hematoxylin and eosin staining).
Four weeks posttransplant, mice receiving MEfl4/+/Esr-Cre cells with and without 4-OH TAM treatment, and mice receiving MEfl4/m1/Esr-Cre cells untreated with 4-OH TAM became moribund; in contrast, the cohort receiving MEfl4/m1/Esr-Cre cells treated with 4-OH TAM remained healthy. Necropsy confirmed that the moribund mice had widely disseminated disease, with enlarged spleens (Figure 2E-F), and, on histopathology, leukemic cell infiltration of liver (Figure 2G), bone marrow (Figure 2H), and spleen (not shown). In contrast, mice receiving 4-OH TAM–treated MEfl4/m1/Esr-Cre leukemic cells remained healthy and had normal-sized spleens at necropsy (Figure 2E-F), with no or minimal AML infiltration into organs, as assessed by histopathology (Figure 2G-H).
Bindels et al showed overexpression of EVI1 in a subset of MFP leukemias; those expressing EVI1 were phenotypically distinct in that they rarely showed monoblastic phenotype.4 Thus, it appears that not all cases of MFP-induced leukemias express the MECOM locus and that there is a distinct phenotype (monoblastic) when MECOM is not activated. It is likely that MECOM nonexpressing MFP-induced leukemias arise via transformation of a cell that is beyond the hematopoietic stem cell/common myeloid progenitor (HSC/CMP) stage, at which the MECOM locus is normally silenced.17 Arai et al established that MFPs can upregulate transcription of both EVI1 and ME.5 Although our results reveal an essential role for ME, they do not preclude that EVI1 isoforms are also essential. Nor do they exclude the possibility that it is the ratio of ME to EVI1 that is critical.
Currently, there are no clinically available targeted therapies for MFP leukemias. Published reports suggest several targeted therapies for these leukemias: inhibitors of glycerol synthase kinase,18 DOT1L,19 and the MFP–multiple endocrine neoplasia 1 interaction.20 However, none of these has yet made it to the clinic, and furthermore, each is likely toxic, based on interpolation from genetic studies.21-23 Thus, despite these reports, there is still a need for additional avenues of therapeutic intervention. This study reveals the novel discovery we have made that ME is necessary for MFP transformation.
The fact that ME but not EVI1 can rescue the mutant phenotype centers attention on the PR domain as being essential for ME function in the setting of MFP leukemogenesis. Recent studies have shown that this domain harbors H3K9 monomethyltransferase activity,7 which was expected based on the finding of HMT activity in closely related proteins.24 This may open an avenue for novel therapeutic intervention in the treatment of MFP leukemias. The fact that ME is a nonessential gene for cellular and organismal survival8 suggests that therapies that inhibit its function are likely to be nontoxic and well tolerated. Future studies will be focused on further characterization of the PR domain and inhibition of its HMT activity.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Johan Jansson and Katie Brock for technical help, and Hongbo Yu for histology.
This work was supported by grants from the National Institutes of Health, National Cancer Institute (R01CA120313) (A.S.P.) and New York State Stem Cell Science (C026423) (Y.Z.) (C026406) (A.S.P).
Authorship
Contribution: Y.Z., F.C., and A.S.P. designed the study; Y.Z., K.O., K.K., L.H., and F.C. performed the study; and Y.Z., C.H.G., and A.S.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Archibald S. Perkins, Department of Pathology and Laboratory Medicine, University of Rochester, Box 626, 601 Elmwood Ave, Rochester, NY 1464; e-mail: archibald_perkins@URMC.rochester.edu; and Yi Zhang, Department of Pathology and Laboratory Medicine, University of Rochester, Box 626, 601 Elmwood Ave, Rochester, NY 14642; e-mail: yi_zhang@urmc.rochester.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal