Key Points
Hsp90 oncoproteome analysis identifies relevant pathways in KSHV-associated primary effusion lymphoma that can inform novel combinatorial therapies.
The Hsp90 inhibitor PU-H71 affects chaperoning of KSHV viral proteins, blocking latent and lytic viral functions.
Abstract
PU-H71 is a purine-scaffold Hsp90 inhibitor that, in contrast to other Hsp90 inhibitors, displays unique selectivity for binding the fraction of Hsp90 that is preferentially associated with oncogenic client proteins and enriched in tumor cells (teHsp90). This property allows PU-H71 to potently suppress teHsp90 without inducing toxicity in normal cells. We found that lymphoma cells infected by Epstein-Barr virus or Kaposi sarcoma-associated herpes virus (KSHV) are exquisitely sensitive to this compound. Using PU-H71 affinity capture and proteomics, an unbiased approach to reveal oncogenic networks, we identified the teHsp90 interactome in KSHV+ primary effusion lymphoma cells. Viral and cellular proteins were identified, including many involved in nuclear factor (NF)-κB signaling, apoptosis, and autophagy. KSHV vFLIP is a viral oncoprotein homologous to cFLIPs, with NF-κB–activating and antiapoptotic activities. We show that teHsp90 binds vFLIP but not cFLIPs. Treatment with PU-H71 induced degradation of vFLIP and IKKγ, NF-κB downregulation, apoptosis and autophagy in vitro, and more importantly, tumor responses in mice. Analysis of the interactome revealed apoptosis as a central pathway; therefore, we tested a BCL2 family inhibitor in primary effusion lymphoma cells. We found strong activity and synergy with PU-H71. Our findings demonstrate PU-H71 affinity capture identifies actionable networks that may help design rational combinations of effective therapies.
Introduction
Hsp90 is a molecular chaperone that plays a crucial role in the proper folding and function of a number of proteins with critical roles in cell survival signal transduction. Hsp90 is considered a promising target for cancer therapy because it is highly expressed in a number of malignancies and acts as a chaperone for oncogenic signaling complexes.1 A role for Hsp90 in virus-associated lymphomas has been proposed, including those caused by infection with Epstein-Barr virus (EBV/human herpesvirus-4) and Kaposi sarcoma (KS) herpesvirus (KSHV/human herpesvirus-8).2,3
KSHV is a γ-herpesvirus and the etiologic agent of KS, multicentric Castleman disease, and primary effusion lymphoma (PEL).4-6 KS is a tumor of endothelial origin, whereas PEL is a rare non-Hodgkin B-cell lymphoma commonly manifesting as lymphomatous effusions. Combined antiretroviral therapy is effective in some patients with KS, and other noncurative approaches such as radiation, surgery, and chemotherapy are helpful. However, a large percentage of patients are refractory to these treatments and there is a pressing need for specific drug development.
Several studies have pointed to an essential role for Hsp90 in the survival of KSHV-associated malignancies. Field et al showed that the viral oncoprotein vFLIP is in complexes containing IKKγ and Hsp90; inhibition of Hsp90 by geldanamycin decreased nuclear factor (NF)-κB activity induced by vFLIP and killed PEL cells.2 Inhibition of extracellular Hsp90 blocked viral gene expression during de novo infection by KSHV.7 Hsp90β was uncovered as a binding partner of the KSHV K1 protein,8 a viral protein that immortalizes endothelial cells in vitro.9 Furthermore, Hsp90 inhibition led to the degradation of LANA, an important viral protein expressed during latency, in KSHV-infected endothelial cells.10 Hsp90 also serves crucial functions in the transformation and maintenance of malignant cells by EBV. Hsp90 inhibition blocked EBV-induced transformation of B cells3 and was toxic to EBV-infected lymphoblastoid cell lines, in part by directly repressing the transcription of EBV EBNA1, the functional equivalent of KSHV LANA in terms of viral episome maintenance.11 Additionally, Hsp90 inhibition by geldanamycin or 17-AAG induced apoptosis of EBV-associated NK/T-cell lymphoma cell lines.12
As an alternative to classical toxic quinone inhibitors of Hsp90,13 Chiosis et al developed a new class of purine scaffold nonquinone Hsp90 inhibitors that inhibit Hsp90 function by competing for the adenosine triphosphate (ATP)-binding pocket,14 preventing the completion of the chaperone cycle. Among these is PU-H71, which has been well characterized in multiple models of solid and lymphoid malignancy with low toxicity.15 Significantly, PU-H71 accumulates at levels up to 25× higher in malignant cells compared with primary cells.16 Hsp90 inhibition by PU-H71 leads specifically to the disruption of oncoprotein-containing complexes over normal signaling complexes containing Hsp90.17 This observation was demonstrated in cancer cell lines such as chronic myelogenous leukemia and acute myelogenous leukemia, where PU-H71 pull-down revealed preferential association with BCR-ABL, rather than the normal ABL protein.18 Affinity capture thus demonstrated that this compound specifically binds the fraction of Hsp90 that is associated with oncogenic proteins and enriched in tumor cells, designated tumor-enriched Hsp90 (teHsp90). A possible mechanism for this preferential association to proteins of particular importance to tumor cells is that these proteins require active chaperoning and are preferentially bound to Hsp90 in complex with cochaperones, which in turn are preferentially identified by PU-H71.18
Based on the reported importance of Hsp90 in viral lymphomagenesis, we evaluated the effect of teHsp90 inhibition using PU-H71 in virus-infected lymphoma cell lines and performed a global analysis of the teHsp90 interactome using PEL as a model. We found that PU-H71 was highly effective against EBV- and KSHV-infected lymphoma cells. Treatment caused viral onco-signaling disruption that led to cell death, and these results were translatable to a mouse xenograft model of PEL. Based on the teHsp90 interactome, we validated the hypothesis that inhibition of antiapoptotic proteins would lead to PEL cell death and synergize with PU-H71. These results indicate that characterization of the teHsp90 interactome can identify vulnerable pathways in tumor cells, and specific inhibitor combinations that merit consideration for clinical development.
Materials and methods
Cell culture, viability, and apoptosis assays
Cell were cultured in RPMI-1640 medium (Invitrogen) supplemented with 50 μg/mL. Gentamicin (Sigma-Aldrich), and 10% or 20% heat-inactivated fetal bovine serum. Cell viability was determined using the Cell Proliferation Kit (3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide [MTT]) (Roche, Indianapolis, IN). Apoptosis was evaluated by flow cytometry for Annexin V. Further details are provided in the supplemental Materials section on the Blood website.
Autophagy detection: western blotting for LC3B
Rabbit Anti-LC3B monoclonal antibody (Cell Signaling Technology Inc., Danvers, MA) was used for detection of cleavage of LC3 in BC3 cells after PU-H71 treatment. The western blotting was conducted as described previously.19
Immunoblotting and co-immunoprecipitation
PU-H71 affinity capture and proteomics
Affinity capture and proteomics analysis were performed as described previously.18 Experimental details are provided in the supplemental Materials section. Data were analyzed through the use of Ingenuity Pathway analysis (Ingenuity Systems, www.ingenuity.com).
In vivo xenograft model
The in vivo xenograft model was set up as previously described.20 Details are provided in the supplemental Materials section. Essentially, nonobese diabetic–severe combined immunodeficiency mice (Jackson Labs) were injected intraperitoneally (IP) with 1 × 107 BC3-NF-κB-luc cells and followed by in vivo imaging. For assessment of disease-free survival and tumor burden, Kaplan-Meier analysis was performed using GraphPad Prism software, and P values determined by 2-tailed analysis with log-rank tests.
Combinatorial cell viability assays
We determined cell viability by using a luminescent ATP-content method (CellTiter-Glo, Promega, Madison, WI) and Trypan blue dye-exclusion. Details are provided in the supplemental Materials section.
Ethics statement
These studies were approved by the Weill Cornell Medical College Institutional Animal Care and Use Committee.
Results
KSHV-positive PEL cell lines are highly sensitive to teHsp90 inhibition by PU-H71
Previous studies had shown that KSHV-infected PEL cell lines are sensitive to several Hsp90 inhibitors, including geldanamycin, 17-AAG, and 17-DMAG.8 We extended these studies by testing the efficacy of the 2 major classes of Hsp90 inhibitors (quinone and protein scaffold) in a wide range of lymphoma cell lines, including 3 KSHV+ PEL, 3 KSHV+/EBV+ PEL, 1 EBV+ lymphoblastoid cell line, 1 EBV+ immunoblastic lymphoma line, 1 EBV+ Hodgkin lymphoma cell line, and 3 virus-negative diffuse large B-cell lymphoma cell lines. We performed MTT assays and determined concentration of compound that inhibited viability of the cell lines by 50% (growth inhibition, GI50), confirming that KSHV- and EBV-infected lymphoma cell lines were sensitive to Hsp90 inhibition by 17-AAG (Table 1). We found that the infected cells were exquisitely sensitive to PU-H71. GI50s ranged from 27 to 337 nM, with no obvious explanation for the variability within this range. For all virus-positive cell lines, the GI50 was lower with PU-H71 than with 17-AAG.
GI50s of PU-H71 and 17-AAG in KSHV- and EBV-positive cell lines
| Cell lines . | PU-H71 . | 17-AAG . | Lymphoma type . | Viral status . |
|---|---|---|---|---|
| GI50 (nM) . | GI50 (nM) . | |||
| BC-1 | 36.9 | 50.9 | PEL | KSHV+/EBV+ |
| BC-2 | 27.1 | 78.6 | PEL | KSHV+/EBV+ |
| BC-5 | 122.7 | 650 | PEL | KSHV+/EBV+ |
| BC-3 | 63.4 | 605.4 | PEL | KSHV+ |
| BCBL-1 | 217.0 | 1009.2 | PEL | KSHV+ |
| VG-1 | 281.5 | 608.9 | PEL | KSHV+ |
| L591 | 337.0 | 952.7 | HL | EBV+ |
| BCKN-1 | 70.9 | 548 | Effusion DLBCL | EBV+ |
| IBL-1 | 87.0 | 833.4 | DLBCL | EBV+ |
| Ly18 | 308.3 | 308.4 | DLBCL | Negative |
| Ly3 | 376.7 | ND | DLBCL | Negative |
| Ly10 | 256.8 | ND | DLBCL | Negative |
| TIVE | 20 561 | ND | Endothelial | Negative |
| TIVE-LTC | 1423 | ND | Endothelial | KSHV+ |
| Cell lines . | PU-H71 . | 17-AAG . | Lymphoma type . | Viral status . |
|---|---|---|---|---|
| GI50 (nM) . | GI50 (nM) . | |||
| BC-1 | 36.9 | 50.9 | PEL | KSHV+/EBV+ |
| BC-2 | 27.1 | 78.6 | PEL | KSHV+/EBV+ |
| BC-5 | 122.7 | 650 | PEL | KSHV+/EBV+ |
| BC-3 | 63.4 | 605.4 | PEL | KSHV+ |
| BCBL-1 | 217.0 | 1009.2 | PEL | KSHV+ |
| VG-1 | 281.5 | 608.9 | PEL | KSHV+ |
| L591 | 337.0 | 952.7 | HL | EBV+ |
| BCKN-1 | 70.9 | 548 | Effusion DLBCL | EBV+ |
| IBL-1 | 87.0 | 833.4 | DLBCL | EBV+ |
| Ly18 | 308.3 | 308.4 | DLBCL | Negative |
| Ly3 | 376.7 | ND | DLBCL | Negative |
| Ly10 | 256.8 | ND | DLBCL | Negative |
| TIVE | 20 561 | ND | Endothelial | Negative |
| TIVE-LTC | 1423 | ND | Endothelial | KSHV+ |
DLBCL, diffuse large B-cell lymphoma; HL, Hodgkin lymphoma; ND, not done.
Identification of the Hsp90 interactome in PEL cells
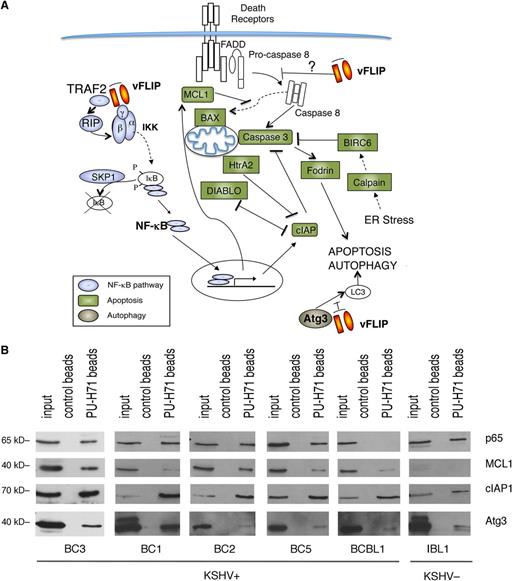
We performed unbiased affinity-capture using PU-H71-conjugated agarose beads to determine the teHsp90 interacting proteins in the KSHV-infected BC3 PEL cell line. The entire cargo isolated by the beads was subjected to proteomic analysis by mass spectrometry. A total of 1756 proteins were identified (supplemental Table 1). One of the top signaling networks identified by Ingenuity Pathway analysis was “NF-κB activation by viruses” (supplemental Figure 1). We found a remarkable number of proteins in the NF-κB pathway, including TRADD, TRAF2, IKKα, IKKβ, IKKγ, and 4 REL/NF-κB proteins (p65/RELA, RELB, p50, and p52) (Figure 1A). Additionally, there was a particular enrichment of molecules involved in apoptosis and autophagy, a feature not previously seen in other tumor types.18 To validate these findings, we used a targeted approach, consisting of pull-down by PU-H71–conjugated beads followed by immunoblot analysis. We focused on representative proteins in the apoptosis (MCL1 and cIAP1), autophagy (ATG3), and NF-κB pathways (p65), and tested additional PEL cell lines (BC3, BC1, BC2, BC5, and BCBL1) as well as 1 EBV+ immunoblastic lymphoma cell line (IBL1) (Figure 1B). We found that in almost all PEL cells and in IBL1, teHsp90 binds these representative proteins, validating the unbiased proteomic approach. The only exceptions were an absence of p65 in BCBL1 cells and low levels of MCL1 in IBL1 cells.
PU-H71 association-based identification of proteins involved in NF-κB activation, apoptosis, and autophagy in PEL cells. (A) PU-H71 affinity capture of BC3 cell extracts was followed by proteomic analysis. Proteins identified in these pull-downs are as follows: those involved in NF-κB signaling are blue; those involved in apoptosis are green; and ATG3, involved in autophagy, is brown. vFLIP, a key nodal protein, is shown in orange. Results shown are from 1 of 2 independent experiments. (B) Immunoblot from PU-H71 pull-downs, but not control bead pull-downs, using extracts from the indicated cell lines, confirmed that Hsp90 interacts with selected proteins in key pathways, including NF-κB (p65), apoptosis (MCL1, cIAP1), and autophagy (ATG3).
PU-H71 association-based identification of proteins involved in NF-κB activation, apoptosis, and autophagy in PEL cells. (A) PU-H71 affinity capture of BC3 cell extracts was followed by proteomic analysis. Proteins identified in these pull-downs are as follows: those involved in NF-κB signaling are blue; those involved in apoptosis are green; and ATG3, involved in autophagy, is brown. vFLIP, a key nodal protein, is shown in orange. Results shown are from 1 of 2 independent experiments. (B) Immunoblot from PU-H71 pull-downs, but not control bead pull-downs, using extracts from the indicated cell lines, confirmed that Hsp90 interacts with selected proteins in key pathways, including NF-κB (p65), apoptosis (MCL1, cIAP1), and autophagy (ATG3).
Other top networks identified have been shown to be functionally significant in PEL cells, including protein kinase B (AKT)/phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin signaling (supplemental Figure 2), and interleukin-6 signaling (supplemental Figure 3). Interestingly, among the top pathways were those associated with angiogenesis, such as Ras-related C3 botulinum toxin substrate (supplemental Figure 4) and vascular endothelial growth factor signaling (supplemental Figure 5).
Several KSHV viral proteins were also pulled down (Table 2). With the exception of vCyclin and viral interleukin (vIL)-6, the viral proteins identified as bound to PU-H71 are known to be only expressed or upregulated during lytic replication. Although most PEL cells have latent KSHV in culture, a small fraction express lytic viral proteins, so it is likely that the lytic proteins detected derive from these scarce lytic cells because they express lytic genes at much higher levels.
Viral proteins identified by proteomic analysis of PU-H71 pull-downs
| Identified proteins . | Molecular weight . | Classification . | Known function(s) . |
|---|---|---|---|
| ORF20 | 28 kDa | Late | UL24 family: induces cell-cycle arrest and apoptosis |
| ORF18 | 29 kDa | Late | Function not known for KSHV: involved in late gene transcription in other herpesviruses |
| ORF64 | 290 kDa | Early | Tegument protein with deubiqutinase activity: suppresses RIG-I–mediated interferon signaling by reducing the ubiquitination of RIG-I |
| ORF K2 | 23 kDa | Early/latent* | vIL-6: viral homolog of interleukin-6 |
| ORF57 | 51 kDa | Early | Messenger RNA biogenesis |
| ORF22 | 81 kDa | Late | Virion envelope glycoprotein |
| ORF72 | 28 kDa | Latent | vCyclin: viral homolog of cyclin D |
| ORF25 | 153 kDa | Late | Major capsid protein |
| ORK9 | 113 kDa | Early | DNA polymerase |
| ORF27 | 32 kDa | Late | Membrane glycoprotein that contributes to intercellular viral spread |
| ORF63 | 104 kDa | Early | Viral homolog of human NLRP1 (blocks innate immune responses/inflammasome) |
| ORF6 | 125 kDa | Early | Multifunctional DNA-binding proteins with roles central to viral DNA replication |
| ORF56 | 96 kDa | Early | Primase |
| ORF4 | 61 kDa | Late | Complement inhibitor: KCP for complement control protein |
| ORF35 | 17 kDa | Early | Induced by RTA, required for in vitro infection |
| ORF21 | 65 kDa | Early | Thymidine kinase |
| ORF19 | 61 kDa | Early | Tegument protein |
| ORF K4.1 | 13 kDa | Early | vMIP-III/vCCL3-CCR4 agonist |
| ORF17.5 | 31 kDa | Early | Assembly/scaffolding protein |
| ORF62 | 36 kDa | Late | Triplex-1 structural protein |
| ORF2 | 23 kDa | Early | vDHFR: viral homolog of dihydrofolate reductase |
| Identified proteins . | Molecular weight . | Classification . | Known function(s) . |
|---|---|---|---|
| ORF20 | 28 kDa | Late | UL24 family: induces cell-cycle arrest and apoptosis |
| ORF18 | 29 kDa | Late | Function not known for KSHV: involved in late gene transcription in other herpesviruses |
| ORF64 | 290 kDa | Early | Tegument protein with deubiqutinase activity: suppresses RIG-I–mediated interferon signaling by reducing the ubiquitination of RIG-I |
| ORF K2 | 23 kDa | Early/latent* | vIL-6: viral homolog of interleukin-6 |
| ORF57 | 51 kDa | Early | Messenger RNA biogenesis |
| ORF22 | 81 kDa | Late | Virion envelope glycoprotein |
| ORF72 | 28 kDa | Latent | vCyclin: viral homolog of cyclin D |
| ORF25 | 153 kDa | Late | Major capsid protein |
| ORK9 | 113 kDa | Early | DNA polymerase |
| ORF27 | 32 kDa | Late | Membrane glycoprotein that contributes to intercellular viral spread |
| ORF63 | 104 kDa | Early | Viral homolog of human NLRP1 (blocks innate immune responses/inflammasome) |
| ORF6 | 125 kDa | Early | Multifunctional DNA-binding proteins with roles central to viral DNA replication |
| ORF56 | 96 kDa | Early | Primase |
| ORF4 | 61 kDa | Late | Complement inhibitor: KCP for complement control protein |
| ORF35 | 17 kDa | Early | Induced by RTA, required for in vitro infection |
| ORF21 | 65 kDa | Early | Thymidine kinase |
| ORF19 | 61 kDa | Early | Tegument protein |
| ORF K4.1 | 13 kDa | Early | vMIP-III/vCCL3-CCR4 agonist |
| ORF17.5 | 31 kDa | Early | Assembly/scaffolding protein |
| ORF62 | 36 kDa | Late | Triplex-1 structural protein |
| ORF2 | 23 kDa | Early | vDHFR: viral homolog of dihydrofolate reductase |
vIL-6 is increased upon lytic reactivation, but is expressed in latently infected PEL.40
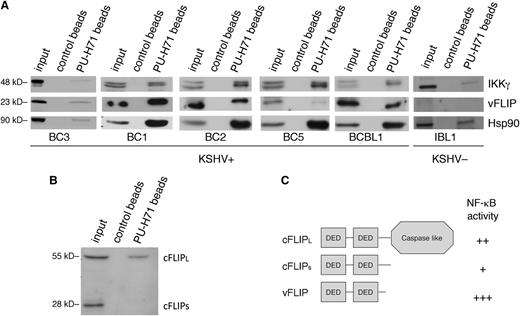
An examination of the major pathways enriched in the pull-down analysis (ie, NF-κB activation, apoptosis, and autophagy) suggested KSHV vFLIP, the viral homolog of cellular FLIP (cFLIP), as a common viral link because this protein has previously been shown to be essential for the enhanced survival and NF-κB–induced antiapoptotic signaling in PEL cells21 and to inhibit autophagy.22 Thus, we validated our findings by directly examining the binding and effect of PU-H71 on vFLIP. In PEL cells, vFLIP has been found to exist primarily as part of the IKK signalosome, comprising IKKs α, β, γ, and Hsp90. Although vFLIP was not directly identified by targeted pull-down proteomic analysis, this method favors proteins expressed abundantly and may fail to identify relevant interactions. Therefore, we assessed the presence of vFLIP in complex with teHsp90 by performing pull-down analysis in 4 different PEL cell lines using PU-H71–conjugated beads. Both vFLIP and IKKγ were identified, implying their presence in complexes with teHsp90 (Figure 2A).
PU-H71 recognizes Hsp90 bound to the vFLIP signalosome and shows preferential binding to the long form of cFLIP. (A) Immunoblot from PU-H71 pull-downs, but not control bead pull-downs, using extracts from 5 different KSHV+ PEL cell lines (BC3, BC1, BC2, BC5, and BCBL1), recognized vFLIP and IKKγ. Control extracts from a B-cell lymphoma cell line lacking KSHV and thereby vFLIP (IBL1) showed binding of Hsp90 to IKKγ, but no bands were seen with the vFLIP antibodies, indicating specificity of the vFLIP band. Results shown are representative of 2 independent experiments. (B) Immunoblots from PU-H71 pull-downs were evaluated for Hsp90 binding to cFLIP, the cellular homolog of vFLIP. Although vFLIP is more closely structurally related to the short form (cFLIPS), PU-H71 failed to recognize this protein, and only the long form (cFLIPL) was identified. (C) Schematic representation of cFLIP long and short forms is shown; the most active of these proteins in terms of ability to activate NF-κB is vFLIP, followed by cFLIPL and the by cFLIPS.21,23
PU-H71 recognizes Hsp90 bound to the vFLIP signalosome and shows preferential binding to the long form of cFLIP. (A) Immunoblot from PU-H71 pull-downs, but not control bead pull-downs, using extracts from 5 different KSHV+ PEL cell lines (BC3, BC1, BC2, BC5, and BCBL1), recognized vFLIP and IKKγ. Control extracts from a B-cell lymphoma cell line lacking KSHV and thereby vFLIP (IBL1) showed binding of Hsp90 to IKKγ, but no bands were seen with the vFLIP antibodies, indicating specificity of the vFLIP band. Results shown are representative of 2 independent experiments. (B) Immunoblots from PU-H71 pull-downs were evaluated for Hsp90 binding to cFLIP, the cellular homolog of vFLIP. Although vFLIP is more closely structurally related to the short form (cFLIPS), PU-H71 failed to recognize this protein, and only the long form (cFLIPL) was identified. (C) Schematic representation of cFLIP long and short forms is shown; the most active of these proteins in terms of ability to activate NF-κB is vFLIP, followed by cFLIPL and the by cFLIPS.21,23
To determine whether cFLIP also binds teHsp90 (directly or indirectly), we performed pull-down analysis of BC3 cell extracts followed by immunoblotting with an antibody to cFLIP, which recognizes the long and short form in PEL cells. We identified only cFLIPL (long isoform) and not cFLIPS (short isoform) in association with PU-H71 (Figure 2B), in spite of vFLIP being most closely related structurally to cFLIPS. This confirms a selective targeting of oncocomplexes because vFLIP and cFLIPL to a lesser extent are both active in their ability to induce NF-κB signaling (Figure 2C), leading to downstream antiapoptotic and pro-proliferative signaling in PEL cells.21 In contrast, the ability of cFLIPS to activate NF-κB is minor.23
PU-H71 induces PEL cell death by apoptosis and autophagy
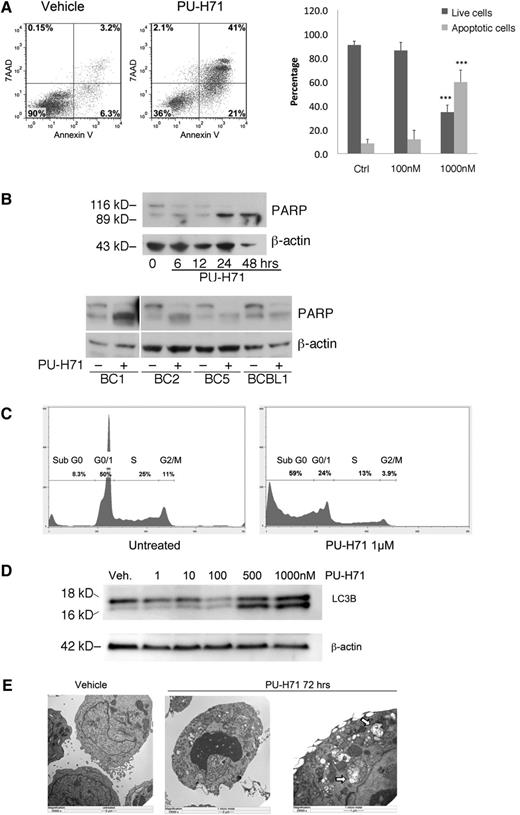
We used Annexin V and 7-AAD staining of BC3 cells to assess the mechanisms involved in cellular cytotoxicity of PU-H71 and found extensive apoptosis, with expansion of both early and late apoptotic compartments, by 72 hours at 1 μM PU-H71 (Figure 3A). Treatment with 1 μM PU-H71 led to a 6-fold increase in apoptosis. Interestingly, 100 nM PU-H71 did not lead to significant apoptosis, although this dose is higher than the GI50 (63.4 nM, Table 1). This discrepancy indicates that the metabolic inhibition as determined by MTT assay (which measures mitochondrial respiratory chain activity) does not accurately reflect cell death, although it is a standard method for determining drug sensitivity. Of note, the GI50 obtained for BC3 cells using a luminescent ATP content method was 290 nM (described in the following section), indicating that this method may be a better indicator of cell viability. Poly (ADP-ribose) polymerase (PARP) cleavage was used as a second method for testing apoptosis, and it was observed by 24 hours posttreatment in BC3 cell lines as well as in all 4 additional PEL cell lines examined at 24 hours posttreatment with PU-H71 (Figure 3B). Additionally, a large increase in the sub-G1 population, which is indicative of DNA fractionation accompanying apoptosis, was also found using DNA content cell cycle analysis (Figure 3C). Unlike 17-AAG, which has been shown to induce cell-cycle arrest in PEL cells,8 PU-H71–treated cells did not reveal such arrest.
PU-H71 induces PEL cell death by apoptosis and autophagy. (A) BC3 cells treated with 1 µM PU-H71 for 72 hours were examined by flow cytometry after staining for 7-AAD and Annexin V. Results were quantified into percentages of live and apoptotic cells (Annexin V positive). ***Statistical significance of P < .001 as compared with the control (untreated) group. (B) Top: Immunoblot analysis using extracts from BC3 cells treated at the indicated time points with 2 µM PU-H71 for the indicated time points shows PARP cleavage by 24 hours as indication of apoptosis. Lower: PARP cleavage is apparent after PU-H71 treatment (2 μM for 24 hours) in additional PEL cell lines. (C) BC3 cells treated with 1 µM PU-H71 for 72 hours were examined by flow cytometry for DNA content cell-cycle analysis. Only the Sub-G0 fraction, indicative of apoptosis, was found to increase in treated cells. (D) Autophagy induction was examined by immunoblot of PU-H71–treated cells. Appearance of the cleavage product of LC3B is indicative of autophagy. (E) BC3 cells treated with 1 µM PU-H71 were examined for the appearance of autophagosomes containing degraded cellular organelles using electron microscopy. Autophagosomes are clearly visible in the right panel at higher magnification (arrows).
PU-H71 induces PEL cell death by apoptosis and autophagy. (A) BC3 cells treated with 1 µM PU-H71 for 72 hours were examined by flow cytometry after staining for 7-AAD and Annexin V. Results were quantified into percentages of live and apoptotic cells (Annexin V positive). ***Statistical significance of P < .001 as compared with the control (untreated) group. (B) Top: Immunoblot analysis using extracts from BC3 cells treated at the indicated time points with 2 µM PU-H71 for the indicated time points shows PARP cleavage by 24 hours as indication of apoptosis. Lower: PARP cleavage is apparent after PU-H71 treatment (2 μM for 24 hours) in additional PEL cell lines. (C) BC3 cells treated with 1 µM PU-H71 for 72 hours were examined by flow cytometry for DNA content cell-cycle analysis. Only the Sub-G0 fraction, indicative of apoptosis, was found to increase in treated cells. (D) Autophagy induction was examined by immunoblot of PU-H71–treated cells. Appearance of the cleavage product of LC3B is indicative of autophagy. (E) BC3 cells treated with 1 µM PU-H71 were examined for the appearance of autophagosomes containing degraded cellular organelles using electron microscopy. Autophagosomes are clearly visible in the right panel at higher magnification (arrows).
KSHV vFLIP has been shown to prevent autophagosome biogenesis by competing with the ubiquitin-like LC3 for binding to the E2-like ATG3 protein.22 Commonly used markers of autophagy are the appearance of the LC3-IIb cleavage product as well as appearance of autophagosomes, both of which were observed 48 hours posttreatment (Figure 3D-E).
PU-H71 leads to the elimination of the vFLIP-IKKγ signaling complex and inhibition of NF-κB signaling in PEL cells
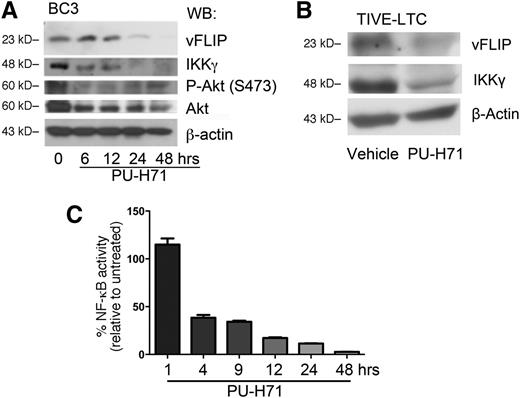
Classical Hsp90-regulated clients depend on Hsp90 for their stability, and their steady-state concentration decreases upon Hsp90 inhibition.18,24,25 Thus, we speculated that this chaperone might play a role in the stabilization of the vFLIP signalosome. Immunoblotting showed that inhibition of teHsp90 led to the decrease of IKKγ as well as the known Hsp90 target AKT (both total and phosphorylated forms) by 6 hours (Figure 4A). In addition, vFLIP protein levels were also reduced by 24 hours. Co-immunoprecipitation using anti-IKKγ to pull down associated vFLIP and Hsp90 revealed a decrease of signalosome components (supplemental Figure 6).
PU-H71 leads to the loss of the vFLIP-IKKγ signaling complex. (A) BC3 cells were treated with 2 µM PU-H71 for the time course indicated and subjected to immunoblot analysis. Treatment resulted in decrease of vFLIP, IKKγ, phospho-Akt, and total Akt. This result is representative of at least 4 independent experiments. (B) TIVE-LTC (KSHV+) cells were treated with 2 µM PU-H71 for 48 hours, followed by immunoblot with the indicated antibodies. Results are representative of three independent experiments. (C) BC3-NF-κB-luc reporter cells were treated with 2 µM PU-H71, and luciferase assays performed at the indicated time points. Results shown are the average of 3 independent experiments.
PU-H71 leads to the loss of the vFLIP-IKKγ signaling complex. (A) BC3 cells were treated with 2 µM PU-H71 for the time course indicated and subjected to immunoblot analysis. Treatment resulted in decrease of vFLIP, IKKγ, phospho-Akt, and total Akt. This result is representative of at least 4 independent experiments. (B) TIVE-LTC (KSHV+) cells were treated with 2 µM PU-H71 for 48 hours, followed by immunoblot with the indicated antibodies. Results are representative of three independent experiments. (C) BC3-NF-κB-luc reporter cells were treated with 2 µM PU-H71, and luciferase assays performed at the indicated time points. Results shown are the average of 3 independent experiments.
Because KS is a tumor of endothelial cell origin, we evaluated whether PU-H71 is also cytotoxic to KSHV-infected endothelial cells. We used a long-term telomerase-immortalized endothelial cell line as an in vitro model for KS.26 The infected cell line was approximately 14-fold more sensitive than the uninfected control (Table 1). In these cells, PU-H71 also led to a decrease in both IKKγ and vFLIP protein levels (Figure 4B).
Because the vFLIP-IKKγ-Hsp90 signaling complex is directly upstream of NF-κB activation, the effect of teHsp90 inhibition by PU-H71 on NF-κB signaling was studied in an NF-κB reporter luciferase BC3 cell line (BC3-NF-κB-luc).20 A time course of treatment revealed decrease of NF-κB reporter luciferase activity within 4 hours (Figure 4C).
PU-H71 does not reduce the levels of other latent viral proteins and inhibits lytic reactivation in PEL cells
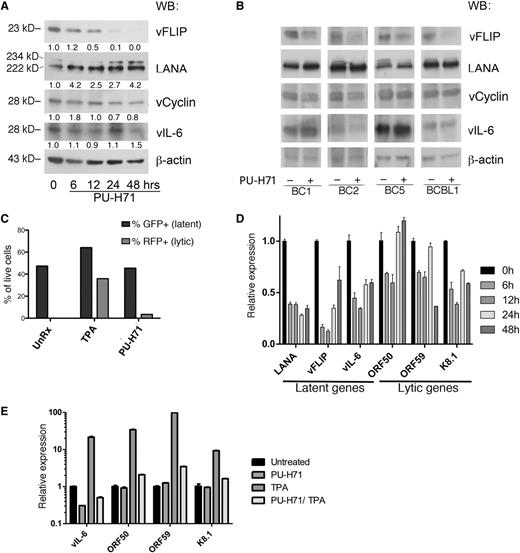
We evaluated the effect of PU-H71 on the other KSHV latent proteins—LANA, vCyclin, and vIL-6—and found a partial decrease in vCyclin and no consistent changes in LANA and vIL-6 upon treatment (Figure 5A). Although both vCyclin and vIL-6 were identified in the PU-H71 pull-downs (Table 2), some Hsp90-binding proteins are not destabilized upon inhibition of Hsp90,18 so vCyclin and vIL-6 appear to fall into this category. Similarly, binding to Hsp90 by LANA, with a decrease of the viral protein following treatment with Hsp90 inhibitors, has been reported,10 but we did not observe this decrease with PU-H71.
PU-H71 inhibits NF-κB with minimal lytic reactivation. (A) Immunoblot analysis of BC3 cells treated with 2 µM PU-H71 at the indicated time points shows that vFLIP is eliminated by 48 hours, whereas there is no consistent decrease in vCyclin, LANA, or vIL-6, as determined by quantification of band intensity and normalized to β-actin and untreated control cells a time 0 (shown below each immunoblot lane). (B) Immunoblot analysis for the panel of viral genes indicated was done using additional PEL cell lines, treated with 2 μM PU-H71 for 24 hours. (C) JSC-1 cells were treated with TPA or PU-H71, and the percentages of green fluorescent protein (GFP)+ and RFP+ cells determined by counting under a fluorescent microscope. (D) Expression levels of latent (LANA, vFLIP, vIL-6) and lytic (ORF50, ORF59, and K8.1) transcripts was studied by quantitative reverse-transcription polymerase chain reaction analysis of extracts from BC3 cells treated with PU-H71 for the indicated time points. The experiment was performed 3 times in triplicate, and representative results are shown. A variable but global decrease of these transcripts was observed (for analysis of other cell lines, see supplemental Figure 7A). (E) Expression of lytic genes induced by TPA was inhibited by PU-H71, as shown after treatment with the compounds for 24 hours.
PU-H71 inhibits NF-κB with minimal lytic reactivation. (A) Immunoblot analysis of BC3 cells treated with 2 µM PU-H71 at the indicated time points shows that vFLIP is eliminated by 48 hours, whereas there is no consistent decrease in vCyclin, LANA, or vIL-6, as determined by quantification of band intensity and normalized to β-actin and untreated control cells a time 0 (shown below each immunoblot lane). (B) Immunoblot analysis for the panel of viral genes indicated was done using additional PEL cell lines, treated with 2 μM PU-H71 for 24 hours. (C) JSC-1 cells were treated with TPA or PU-H71, and the percentages of green fluorescent protein (GFP)+ and RFP+ cells determined by counting under a fluorescent microscope. (D) Expression levels of latent (LANA, vFLIP, vIL-6) and lytic (ORF50, ORF59, and K8.1) transcripts was studied by quantitative reverse-transcription polymerase chain reaction analysis of extracts from BC3 cells treated with PU-H71 for the indicated time points. The experiment was performed 3 times in triplicate, and representative results are shown. A variable but global decrease of these transcripts was observed (for analysis of other cell lines, see supplemental Figure 7A). (E) Expression of lytic genes induced by TPA was inhibited by PU-H71, as shown after treatment with the compounds for 24 hours.
Induction of lytic reactivation has been reported as a consequence of NF-κB inhibition.27 To assess effects on the lytic viral cycle, we used the modified recombinant KSHV.219–infected PEL cell line JSC-1, which expresses green fluorescent protein under the control of the cellular EF-1 α promoter and red fluorescent protein (RFP) under the control of the early lytic PAN promoter of KSHV.28 Treatment of these cells with PU-H71 turned on RFP expression in only a very small fraction of cells (3%), in contrast to treatment with the lytic phase inducer 12-O-tetradecanoylphorbol-13-acetate (TPA) (42%) (Figure 5C).
We analyzed PU-H71 treated cells for levels of latent and lytic viral transcripts. Real-time quantitative reverse-transcription polymerase chain reaction analysis of whole cell extracts from PU-H71–treated cells showed a variable reduction of the latent transcripts evaluated in BC3 cells (Figure 5D) and other PEL cell lines (supplemental Figure 7A). Note that LANA, vFLIP, and vCYC are controlled by the same promoter, and vFLIP and vCYC are encoded in the same transcript, but the decrease in RNA level did not translate into consistently decreased LANA or vCYC (Figure 5A-B). Similarly, vIL-6 protein levels did not change, although transcriptional levels were variable (Figure 5A-B). There were also variable changes in the 2 lytic transcripts examined in other PEL cell lines. The global decrease in the major latent viral transcripts can be explained by the inhibition of several transcription factors by PU-H71 as well as by effects on chromatin remodeling proteins and ribonucleoproteins, also identified by PU-H71 affinity capture (Table 1). For example, the shared vFLIP/vCYC/LANA promoter is highly dependent on SP1,29 a transcription factor that is an established Hsp90 client protein.30 Indeed, immunoblotting revealed a significant decrease in SP1 levels after PU-H71 treatment in PEL cells (supplemental Figure 7B). The reduction of vFLIP messenger RNA is partial and transient, so it may be only partially responsible for the dramatic decrease in vFLIP protein levels, and there is probably also posttranslational downregulation of vFLIP.
We assessed the role of Hsp90 in lytic reactivation. There was a reduction of the late lytic transcripts K8.1 and ORF59 and a minor induction of the immediate–early transcript ORF50 (Figure 5C), with similar but variable trends observed in other PELs (supplemental Figure 7A), confirming that this compound induces only minimal lytic reactivation. Furthermore, we found that PU-H71 inhibits TPA-induced lytic gene expression (Figure 5D).
PU-H71 confers a significant survival advantage to PEL-xenografted mice
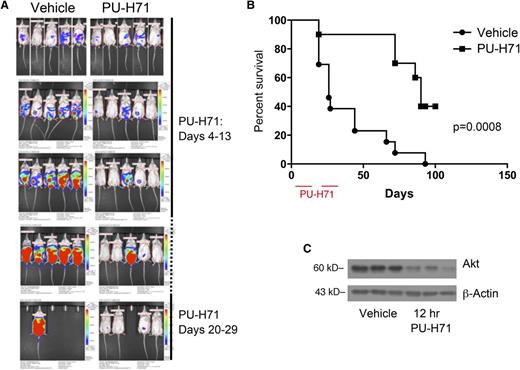
To evaluate PU-H71 in a preclinical model, we used the BC3-NF-κB-luc xenograft model in NOD-severe combined immunodeficiency mice in 3 independent trials. Mice were randomized after xenograft engraftment into vehicle (n = 10) and treatment cohorts (n = 13). The PU-H71 cohort was treated on a daily basis by IP injection at 75 mg/kg per day for 2 10-day intervals separated by a week.
Representative in vivo imaging results are shown in Figure 6A. A pattern of protection conferred by PU-H71 treatment was obvious from quantification of tumor burden in these mice. Kaplan-Meier analysis indicated that PU-H71 treatment conferred a significant survival advantage (median survival time = 90 days for treated, 26 days for vehicle) to xenografted mice (Figure 6B; P = .0008).
PU-H71 induces tumor responses in mice with PEL. (A) In vivo imaging of mice that received an IP injection of BC3/NFκB-luc cells (day 0) and subsequently were untreated (left) or mice that received PU-H71 IP at a dose of 75 mg/kg per day on days 4-13 and 20-29. (B) Kaplan-Meier survival curves for treated and control mice showed prolonged survival in the treatment arm; mice were euthanized when the developed ascites leading to a >10% increase of body weight in a 1-week period; therefore, survival is directly related to tumor growth and independent of possible reporter effects. (C) Established tumors from 3 mice in the vehicle arm were collected 12 hours after administration of 1 75 mg/kg dose of PU-H71 (right lanes); protein extracts were compared with those from tumors in untreated controls (left lanes). Immunoblot analysis shows a decrease in vivo of a classic Hsp90 client.
PU-H71 induces tumor responses in mice with PEL. (A) In vivo imaging of mice that received an IP injection of BC3/NFκB-luc cells (day 0) and subsequently were untreated (left) or mice that received PU-H71 IP at a dose of 75 mg/kg per day on days 4-13 and 20-29. (B) Kaplan-Meier survival curves for treated and control mice showed prolonged survival in the treatment arm; mice were euthanized when the developed ascites leading to a >10% increase of body weight in a 1-week period; therefore, survival is directly related to tumor growth and independent of possible reporter effects. (C) Established tumors from 3 mice in the vehicle arm were collected 12 hours after administration of 1 75 mg/kg dose of PU-H71 (right lanes); protein extracts were compared with those from tumors in untreated controls (left lanes). Immunoblot analysis shows a decrease in vivo of a classic Hsp90 client.
We performed ex vivo analysis to verify PU-H71 activity in the PEL xenografts. Whole cell extracts of ascites from vehicle cohort mice treated with PU-H71 12 hours before euthanasia were compared with ascites from untreated mice. We confirmed target inhibition ex vivo by immunoblotting for AKT (Figure 6C) because of the lack of a robust commercial antibody for vFLIP in xenograft analysis.
PU-H71 synergizes with a pan-BCL2 family inhibitor in PEL cells
Based on the large number of proteins involved in apoptosis by PU-H71 affinity-capture in PEL cells, we hypothesized that inhibition of apoptosis is critically important and that targeting antiapoptotic proteins could be a viable approach for the treatment of KSHV-associated malignancies. We used obatoclax (GX15-070), a small-molecule BH-3 mimetic that antagonizes BCL2, BCL-XL, BCL-W, and MCL1,31 to treat PEL cells. This compound was shown to successfully inhibit viability of a panel of 5 PEL cell lines with submicromolar GI50 (Figure 7A). To assess synergy, cell lines were treated with a range of concentrations of both PU-H71 and obatoclax. We chose doses for each drug that had partial toxicity. We used an ATP-content assay to assess viability rather than the MTT assay shown in Table 1, which gave somewhat higher GI50s. As shown in Figure 7B, some cell lines (BC3 and BC2) had much greater synergy. We further assessed synergy for BC3, by treating with 6 concentrations of PU-H71, obatoclax, the combination, or vehicle, for 48 and 72 hours. We determined the combinatorial index32 and found that it was synergistic at both time points (Figure 7C-D). The fraction affected–combinatorial index plot showed that the combination is highly synergistic for all the effect points at 72 (Figure 7D) and 48 hours (not shown).
PU-H71 synergizes with a pan-BCL2 family inhibitor. (A) The indicated PEL cell lines were treated with pan-BCL2-family inhibitor obatoclax for 72 hours, and the dose response curves are shown. The fraction of cells killed relative to the untreated control is represented on the y-axis; the obatoclax (Obx) dose is shown in the x-axis. Plots were calculated using Compusyn Software and represent best-fit models of the data point. The R value for the dose-effect curves ranged between 0.94 and 0.97 in all the cell lines. (B) These PEL cells were treated with PUH71, Obx, and a combination of both for 72 hours. Doses were chosen based to the sensitivity of each cell line, and were as follows: BC3, PUH71 0.078125 μM; Obx, 0.15625 μM; BC2, PUH71 and Obx 0.3125 μM; and BC5 and BCBL1, PUH71 and Obx 0.625 μM. (C) BC3 cells treated with PU-H71 or Obx for 48 hours were analyzed for synergy. The isobologram indicates GI50, GI75, and GI90 values (for PUH71, GI50, GI75, and GI90 are 0.29256, 0.83922, and 2.38530 μM, respectively; for Obx, 0.14949, 0.24479, and 0.40084 μM), indicating strong synergy with combination index values of ∼0.2. (D) The fraction affected–combinatorial index plot for the combination of Obx and PU-H71 at 72 hours. The fraction of cells killed is represented on the x-axis (Fa) and the combinatorial index is represented on the y-axis (CI). The dotted line represents an additive effect and divides the antagonistic zone (above) and synergistic zone (below). The points represent actual combination data points and the blue line represents the computer simulation for the relationship.
PU-H71 synergizes with a pan-BCL2 family inhibitor. (A) The indicated PEL cell lines were treated with pan-BCL2-family inhibitor obatoclax for 72 hours, and the dose response curves are shown. The fraction of cells killed relative to the untreated control is represented on the y-axis; the obatoclax (Obx) dose is shown in the x-axis. Plots were calculated using Compusyn Software and represent best-fit models of the data point. The R value for the dose-effect curves ranged between 0.94 and 0.97 in all the cell lines. (B) These PEL cells were treated with PUH71, Obx, and a combination of both for 72 hours. Doses were chosen based to the sensitivity of each cell line, and were as follows: BC3, PUH71 0.078125 μM; Obx, 0.15625 μM; BC2, PUH71 and Obx 0.3125 μM; and BC5 and BCBL1, PUH71 and Obx 0.625 μM. (C) BC3 cells treated with PU-H71 or Obx for 48 hours were analyzed for synergy. The isobologram indicates GI50, GI75, and GI90 values (for PUH71, GI50, GI75, and GI90 are 0.29256, 0.83922, and 2.38530 μM, respectively; for Obx, 0.14949, 0.24479, and 0.40084 μM), indicating strong synergy with combination index values of ∼0.2. (D) The fraction affected–combinatorial index plot for the combination of Obx and PU-H71 at 72 hours. The fraction of cells killed is represented on the x-axis (Fa) and the combinatorial index is represented on the y-axis (CI). The dotted line represents an additive effect and divides the antagonistic zone (above) and synergistic zone (below). The points represent actual combination data points and the blue line represents the computer simulation for the relationship.
Discussion
This study is the first to identify the entire teHsp90 interactome in PEL cells and demonstrates specific targeting of a viral oncoprotein by an oncocomplex-specific Hsp90 inhibitor. We show that a teHsp90 inhibitor can successfully be used to block pathogenic pathways in viral malignancies and reveal synergistic interactions through teHsp90 interactome analysis. Our results are consistent with previous studies showing that Hsp90 exists in a specific conformation in tumor cells, described here as teHsp90, which is selectively bound and inhibited by PU-H71, and that is enriched in complexes containing dysregulated oncoproteins.18 Specifically, using PU-H71 affinity capture and proteomic analysis, we were able to identify proteins in pathways known to be essential for KSHV-mediated oncogenesis, including NF-κB, apoptosis, and autophagy. This analysis also revealed other proteins and additional pathways that may play a role in KSHV oncogenesis.
vFLIP is a critical viral oncoprotein and Hsp90 has been shown to be present in vFLIP and IKKγ-containing complexes. Although we did not find vFLIP in the unbiased proteomic approach, it was previously shown18 that Hsp90 interacts with nodal proteins in signaling pathways, and affinity purification may identify only key components of an Hsp90-addicted protein pathway. Nevertheless, we confirmed the interaction of teHsp90 and vFLIP and demonstrated that teHsp90 inhibition effectively abolishes the oncogenic vFLIP-IKKγ signaling complex. Complex elimination decreased downstream signaling and inducted autophagy and apoptosis. Notably, although the closest cellular homolog of vFLIP is cFLIPS, we were only able to document teHsp90 binding to vFLIP and cFLIPL. Because vFLIP and cFLIPL can induce NF-κB activity in PEL cells, these results confirm the selectivity of binding of teHsp90 to active proteins in particular cellular contexts.
The number of proteins involved in apoptosis, including proapoptotic, antiapoptotic, and effector molecules (Figure 1), found to be associated with teHsp90 was remarkable, and not seen in other tumors so far.18 Because the presence of a virus is likely to be an important apoptotic trigger, it follows that teHsp90 in virus-associated malignancies maintains proteins involved in apoptosis in complexes or configurations that prevent cell death. These observations explain the exquisite sensitivity of KSHV- and EBV-infected lymphoma cell lines to treatment with PU-H71. We hypothesized that based on the large number of apoptosis pathway proteins bound to teHsp90, we could also kill PEL cells with an inhibitor of antiapoptotic proteins. In fact, we found that all PEL cell lines tested were highly sensitive to treatment with a pan-BCL2 inhibitor obatoclax, and that, at least in BC3 cells, there is dramatic synergy when treating cells with PU-H71 and obatoclax. This result indicates that an understanding of the PU-H71/teHsp90 interactome in cancer cells can inform future therapeutic choices and inhibitor combinations.
The inhibition of autophagy is a major function for vFLIP.22 vFLIP directly binds ATG3 and prevents it from processing LC3.22 We identified ATG3 by PU-H71 affinity capture, as well as additional proteins involved in autophagy. The PI3K pathway has also been shown to inhibit starvation-induced autophagy in macrophages,33 suggesting that the AKT/PI3K pathway may contribute to induction of autophagy in PEL cells by PU-H71. We observed a decrease of Akt protein levels in cells treated with PU-H71, both in vitro and in vivo. Other proteins in this pathway were also identified in the teHsp90 interactome.
KS is a tumor of endothelial cell origin in which angiogenesis plays a critical role. Remarkably, among the pathways that were identified in the teHsp90 interactome are those associated with vascular endothelial growth factor and Ras-related C3 botulinum toxin substrate. These 2 pathways have been implicated as central in angiogenesis and KS pathogenesis.34,35 PU-H71 was also found to bind several lytic proteins. This is most likely a result of the extremely high levels of lytic proteins in the few lytic cells in cell culture. It is also likely that binding by teHsp90 to complexes containing lytic proteins is very robust, and possibly direct, which could be tested using recombinant proteins for direct binding to Hsp90 and dissociation in vitro. PU-H71 also inhibits expression of lytic viral transcripts. Consistent with this, another study found inhibition of KSHV viral replication by Hsp90 inhibitors.36 This antiviral activity could provide further benefit for the treatment of individuals with KS, in which lytic genes may contribute to disease pathogenesis.37
Analysis in mice demonstrated that PU-H71 can be used successfully to prolong disease-free survival. Although posttreatment relapses were observed, they may be overcome by extending treatment.38 Thus, treatment with PU-H71 in combination with other specific inhibitors may be required for an effective clinical response. PU-H71 has demonstrated efficacy in several in vivo models of malignancy,39 with phase 1 clinical trials ongoing.
In summary, we used PU-H71 affinity capture and proteomics to elucidate the Hsp90-associated oncoproteome of a KSHV-associated PEL cell line. This unbiased analysis revealed many pathways associated with KSHV pathogenesis, lending support to the power of this methodology. This approach demonstrated a novel mechanistic function for teHsp90 inhibitors wherein they can be successfully used to target viral oncoproteins. We demonstrated clear potential for PU-H71 in the treatment of KSHV-associated diseases, and our analysis revealed a number of teHsp90-interacting proteins and pathways that will provide the basis for exploring additional new therapeutic approaches and combinations.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by grants from the National Institutes of Health/National Cancer Institute (R01-CA68939) (E.C.), (P30CA008748) (E.C. and G.C.), (P30CA008748) (Microchemistry and Proteomics Core Laboratory of Memorial Sloan-Kettering Cancer Center), and T32 AI007621 Immunology and Microbial Pathogenesis Program Training Grant (U.N).
Authorship
Contribution: U.N. designed and performed in vitro and mouse studies, analyzed data, and wrote and edited paper; P.L. designed and performed GI50 and apoptosis experiments; R.L.G. performed validation pull-downs and synergy experiments; J.V. analyzed mouse data; G.B. developed vFLIP transgenic mice and isolated mouse B cells; A.R. performed targeted pull-down; T.T. prepared compound; H.E.-B. analyzed proteomics data; M.C. assisted with quantitative reverse-transcription polymerase chain reaction for lytic genes; R.B. provided resources for mouse studies; A.M. supervised synergy experiments; L.C. designed and analyzed synergy experiments; G.C. supervised and designed targeted pull-down; Y.L.W. supervised and designed GI50 and apoptosis experiments; and E.C. supervised and designed in vitro and mouse experiments, analyzed pull-down data, and wrote and edited paper.
Conflict-of-interest disclosure: Memorial Sloan-Kettering Cancer Center holds the intellectual rights to the purine-scaffold Hsp90 inhibitors. Samus Therapeutics, of which G.C. has partial ownership, has licensed PU-H71. The remaining authors declare no competing financial interests.
The current affiliation for P.L. and Y.L.W. is Department of Pathology, University of Chicago, Chicago, IL. The current affiliation for J.V. is Host Response to Cancer Laboratory, School of Medical Science, Griffith University, Gold Coast, Australia.
Correspondence: Ethel Cesarman, Department of Pathology and Laboratory Medicine, Weill Cornell Medical College, 1300 York Ave, Room C410, New York, NY 10065; e-mail: ecesarm@med.cornell.edu; and Y. Lynn Wang, Department of Pathology, University of Chicago, 5841 S Maryland Ave, Room N-316A , Mail Code 1089, Chicago, IL 60637; e-mail: lyw2013@uchicago.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal