Key Points
PRC1 and PRC2 have opposing activity in Eμ-myc lymphoma.
Inhibition of PRC2 leads to increased self-renewal in B-cell progenitors.
Abstract
Deregulation of polycomb group complexes polycomb repressive complex 1 (PRC1) and 2 (PRC2) is associated with human cancers. Although inactivating mutations in PRC2-encoding genes EZH2, EED, and SUZ12 are present in T-cell acute lymphoblastic leukemia and in myeloid malignancies, gain-of-function mutations in EZH2 are frequently observed in B-cell lymphoma, implying disease-dependent effects of individual mutations. We show that, in contrast to PRC1, PRC2 is a tumor suppressor in Eµ-myc lymphomagenesis, because disease onset was accelerated by heterozygosity for Suz12 or by short hairpin RNA–mediated knockdown of Suz12 or Ezh2. Accelerated lymphomagenesis was associated with increased accumulation of B-lymphoid cells in the absence of effects on apoptosis or cell cycling. However, Suz12-deficient B-lymphoid progenitors exhibit enhanced serial clonogenicity. Thus, PRC2 normally restricts the self-renewal of B-lymphoid progenitors, the disruption of which contributes to lymphomagenesis. This finding provides new insight regarding the functional contribution of mutations in PRC2 in a range of leukemias.
Introduction
Polycomb group (PcG) proteins are global transcriptional repressors first identified in Drosophila as silencers of Hox genes during development. Subsequent genomewide studies showed that PcG proteins regulate genes involved in diverse cellular functions.1,2 PcG proteins exist in 2 distinct protein complexes called polycomb repressive complex 1 (PRC1) and 2 (PRC2). Mammalian PRC1 components include Bmi1, Mel18, Cbx2, 4, 7, and 8, Scmh1 and 2, Phc1/Rae28, Phc2 and 3, Ring1A, and Ring1B; the complex is highly heterogeneous, and its precise makeup varies depending on the cellular and developmental context. Although Bmi1 is crucial for augmenting PRC1 activity, Ring1B is the enzyme that monoubiquitinates histone H2A at lysine 119 (H2AK119ub), a mark associated with transcriptional repression.3 PRC2 mediates tri-methylation of histone H3 at lysine 27 (H3K27me3), another repressive mark. The main components in PRC2 are Suz12, Eed, and the enzymatic components Ezh2 and/or Ezh1. Eed and Suz12 are essential for PRC2 complex stability, whereas accessory factors Jarid2, Rbbp4 and 7, Phf1, and Mtf2 are required to modulate PRC2 function.4
PRC1 and PRC2 interact to control transcriptional activity at target loci. In the hierarchical-recruitment model,5 PRC2-mediated H3K27me3 recruits PRC1 via the chromodomain of Cbx proteins, leading to H2AK119ub-induced transcriptional silencing. Studies have correlated PRC2 activity and H3K27me3 with PRC1 occupancy at target genes,1,6 providing support for a role of PRC2/H3K27me3 in recruiting PRC1. However, other observations suggest that the hierarchical model may not always hold. For example, PRC1 can bind nucleosomes lacking N-terminal histone tails in vitro and can be recruited to targets in the absence of PRC2.7,8 Moreover, mice with a heterozygous loss-of-function mutation in Suz12 display enhanced hematopoietic stem cell (HSC) activity,9 whereas mice lacking PRC1 components have functionally compromised HSCs.10,11
PcG genes are deregulated in many human cancers. EZH2 and BMI1 are overexpressed in some breast cancers and colon cancers, and increased expression of EZH2 is associated with more aggressive disease in prostate cancers.12 Discovery of a gain-of-function mutation in EZH2 in follicular and diffuse large B-cell lymphoma13-15 strengthened the argument that PcG genes are oncogenic. Conversely, loss-of-function mutations and deletions in EZH2, EED, and SUZ12 have been described in myelodysplastic syndromes and T-cell acute lymphoblastic leukemia (T-ALL).16-19 This suggests that PRC2 has a tumor suppressor role in specific hematological malignancies, a hypothesis supported in a mouse model where Ezh2 inactivation resulted in T-cell lymphoma.20 These studies suggest that aberrant PRC2 function contributes to tumorigenesis in a context-dependent manner and emphasize the need to define the underlying mechanisms via which altered PRC2 contributes to disease. Accordingly, we compared and contrasted the contribution of PRC1 and PRC2 to Myc-driven lymphomagenesis and show that, in contrast to PRC1, PRC2 behaves as a tumor suppressor by restricting self-renewal of B-cell progenitors.
Methods
Mice
Analysis of hematopoietic cells by flow cytometry
Single cell suspensions from bone marrow, spleen, and thymus were prepared in balanced salt solution (150 mM NaCl, 3.7 mM KCl, 2.5 mM CaCl2, 1.2 mM MgSO4, 7.4 mM HEPES.NaOH, 1.2 mM KH2PO4, and 0.8 mM K2HPO4) with 5% fetal calf serum. Whole blood was collected for automated cell count (Advia3120; Bayer). For fluorescence-activated cell sorter (FACS) analysis, erythrocytes were lysed in 150 mM NH4Cl, 0.1 mM EDTA, and 12 mM NaHCO3. Cells were stained with monoclonal antibodies to B220 (RA3-6B2), CD19 (1D3), CD25 (PC61), c-Kit (2B8), immunoglobulin M (IgM) (II/41), IgD (11-26), Mac-1 (M1-70), Gr-1 (RB6-8C5), Ter119 (Ter119), CD4 (GK1.5), CD8a (53-6.7), CD45.2 (104), CD45.1 (A20), Sca-1 (D7), CD34 (RAM34), CD150 (TC15-12F12.2), Flt3 (A2F10.1), IL7Ra (A7R34), Ly6D (49-H4), and Annexin V (#556420), sourced from BioLegend, BD Pharmingen, or eBiosciences. Propidium iodide or fluorogold was used for dead cell exclusion. The LSR I or II or Fortessa (BD Biosciences) instruments were used for analysis, whereas MoFlo (Beckman Coulter) and Aria (BD Biosciences) were used for sorting. FlowJo software (Treestar, Inc.) was used for data analysis.
Apoptosis and cell cycle analysis
5-Bromo-2′-deoxyuridine (BrdU) was administered intraperitoneally (0.1 mg/g body weight). BrdU incorporation in bone marrow and spleen was determined 1 hour after injection (BD Pharmingen). In vitro apoptosis assays of FACS-purified pro-B, pre-B, and sIg+ B cells were performed as described previously.23
Retrovirus production
The protocol for retrovirus production has been previously described.9 Briefly, retroviral supernatants were prepared by transfection of 293T cells with plasmids encoding viral envelope proteins and specific short hairpin RNAs (shRNAs) in the LTR-miR30-SV40–green fluorescent protein (GFP) (LMS) vector that target Suz12 (CGCTCTTACTGCTGAGCGTATA), Ezh2 (CGCTCTTACTGCTGAGCGTATA), or a proprietary scrambled sequence (Nons) designed by Open Biosystems.
Adoptive transfer of Eμ-myc fetal livers
Ter119-depleted E13.5 Eμ-myc fetal livers were transduced with retroviral supernatants containing LMS-Suz12, LMS-Ezh2, or LMS-Nons and cultured overnight at 37°C/5% CO2, as described previously.9,24 Cells (3-4 × 105) were injected intravenously into lethally irradiated CD45Ly5.1 recipients (11 Gy; 60Co source), and mice were monitored for lymphoma development.
In vitro culture of B lymphocytes
Unfractionated bone marrow cells and FACS-purified pro-B cells were either cultured on OP9 stroma supplemented with interleukin (IL)-7 (2% supernatant from an IL-7–producing cell line25 or 5 ng/mL murine rIL7; PeproTech) or in MethoCult M3630 (StemCell Technologies).
Immunoblotting
Cells and primary lymphomas were homogenized in radio-immunoprecipitation assay buffer (1% NP-40, 0.1% sodium dodecyl sulfate, 0.5% sodium decarboxylate, 150 mM NaCl, and 50 mM Tris-HCl) containing Complete Protease Inhibitors (Roche). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and blotted with antibodies against Suz12 (P-15; Santa Cruz Biotechnology, or D39F6; Cell Signaling Technology), Ezh2 (07-689; Millipore, or AC-22; Cell Signaling Technology), GFP (A-11122; Invitrogen), H3K27me3 (07-449; Millipore, or C36B11; Cell Signaling Technology), H2AK119ub (D27C4; Cell Signaling Technology), histone H3 (AS3; Millipore), and actin (I-19; Santa Cruz Biotechnology).
RNA extraction, cDNA synthesis, and quantitative reverse transcriptase-polymerase chain reaction
RNA was extracted using RNeasy Mini or Micro Columns with DNase I treatment (Qiagen), and reverse-transcribed into cDNA with oligo-dT priming (Promega, Madison, WI) using Superscript III reverse transcriptase (Invitrogen). Quantitative reverse transcriptase-polymerase chain reaction was performed using Taqman probes to c-myc (Mm00487804_m1), Pax5 (Mm00435501_m1), Ebf1 (Mm00395519_m1), Tcfe2a (Mm01175595_m1), and Hprt (Mm00446968_m1) in an ABI 7900HT PCR machine (Applied Biosystems). Relative gene expression was calculated using the 2−ΔΔCt method.
Statistical analysis
Data were analyzed using Prism GraphPad v6.0. A 2-tailed Student t test was performed in 2-group comparisons. When comparing multiple groups, 1-way analysis of variance followed by Tukey’s post hoc test was performed. A log-ranked (Mantel-Cox) test was used in survival studies. Progenitor frequencies from limiting dilution assays were determined using the extreme limiting dilution assay software tool.26
Expression profiling of Eμ-myc and Eμ-myc/Suz12Plt8/+ lymphomas
Total RNA from 7 Eμ-myc/Suz12+/+ and 5 Eμ-myc/Suz12Plt8/+ FACS-purified lymphomas were sequenced at the Australia Genome Research Facility. An average of 12.2 million single-end 100-bp sequence reads were obtained per lymphoma sample on an Illumina GA-II Sequencer. An average of 95% of the reads were successfully mapped to the mm9 mouse genome build using the Bioconductor package Rsubread.27 The RNA-Seq profiles were summarized using the feature Counts function to count the number of reads overlapping the exome of each gene. Differential expression analysis was performed using the Bioconductor package edgeR.28 Reads mapping to immunoglobulin, ribosomal RNA, and mitochondrial protein genes were filtered out of the analysis. Genes on the X and Y chromosomes were also removed to avoid any confounding factors due to lymphomas arising from mice of different sexes. Read counts were trimmed mean of M-values (TMM) normalized29 to adjust for compositional differences between samples. Genewise estimates of biological variation were obtained using empirical Bayes moderated dispersions.30 Statistical significance was assessed using an exact test for negative binomial distributed data.31 To prepare for gene set testing, the read counts were transformed to approximately standard normal deviates using the zscoreNBinom function of the edgeR package. A battery of gene sets from the Broad Institute’s curated C2 molecular signatures database32 was tested using the CAMERA function of the limma package.33 This is a competitive gene set test that tests whether the genes in the gene set are significantly more up- or downregulated compared with the other genes in the experiment. Human gene symbols were mapped to mouse orthologs using the Mouse Genome Database.34
Results
Suz12Plt8/+ mice have elevated numbers of B lymphocytes
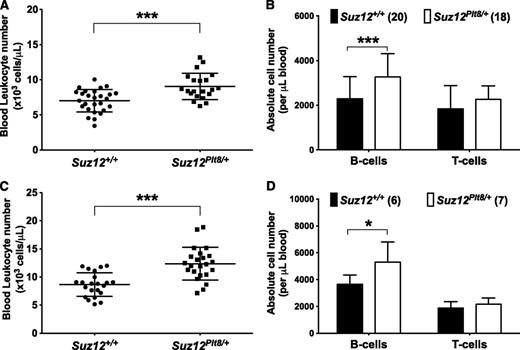
Previous studies have demonstrated that mice carrying a heterozygous loss-of-function allele of Suz12, Ezh2, or Eed have enhanced HSC activity and platelet production.9,24,35 However, the impact of these mutations has not been detailed in other hematopoietic cells types. Using Suz12Plt8/+ mice to model hypomorphic PRC2 function, we found a consistent elevation in blood leukocytes relative to Suz12+/+ mice at both 4 and 8 weeks of age (Figure 1), which was exclusively due to higher lymphocyte counts (supplemental Tables 1 and 2, available on the Blood Web site).
Suz12Plt8/+ mice have elevated numbers of B lymphocytes in the peripheral blood. Automated leukocyte counts from peripheral blood performed on (A) 4- and (C) 8-week-old Suz12+/+ and Suz12Plt8/+ mice. Enumeration of blood B and T lymphocytes in Suz12+/+ and Suz12Plt8/+ at (B) 4 and (D) 8 weeks of age. Data represent mean ± standard deviation. A 2-tailed Student t test was used for comparison between the genotypes (*P < .05; ***P < .001).
Suz12Plt8/+ mice have elevated numbers of B lymphocytes in the peripheral blood. Automated leukocyte counts from peripheral blood performed on (A) 4- and (C) 8-week-old Suz12+/+ and Suz12Plt8/+ mice. Enumeration of blood B and T lymphocytes in Suz12+/+ and Suz12Plt8/+ at (B) 4 and (D) 8 weeks of age. Data represent mean ± standard deviation. A 2-tailed Student t test was used for comparison between the genotypes (*P < .05; ***P < .001).
Progenitor cell analysis in Suz12Plt8/+ mice
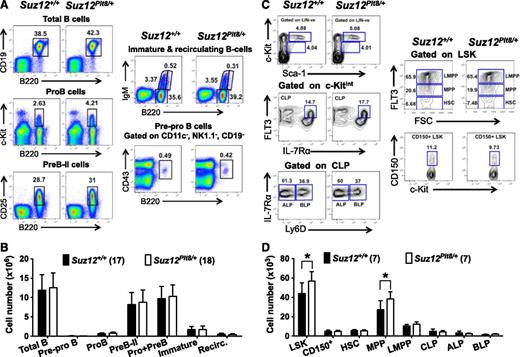
The elevated numbers of B cells in Suz12Plt8/+ mice prompted us to examine B-cell development in detail at 4 and 8 weeks of age. The numbers of cells at various stages of B-lymphoid maturation appeared unaltered at both ages, although a modest increase in total B-lymphoid cell numbers was evident in 8-week-old Suz12Plt8/+ mice (Figure 2A-B; supplemental Tables 3 and 4). Enumeration of B- and T-cell subsets in the spleen and thymus of Suz12Plt8/+ mice revealed no significant abnormalities (supplemental Tables 3 and 4). There was a modest increase in the number of LSK cells and multipotent progenitors (MPPs) in Suz12Plt8/+ mice at weaning (Figure 2D), although this was less pronounced in adult mice (supplemental Table 3). Enumeration of lymphoid-primed MPPs (LMPPs), and common lymphoid progenitors (CLPs) did not reveal significant differences between Suz12Plt8/+ and Suz12+/+ mice. Similarly, subfractionation of the CLP population into ALPs (Ly6D−; all-lymphoid progenitors) and BLPs (Ly6D+; B-cell primed progenitors)36 did not reveal any changes in Suz12Plt8/+ mice (Figure 2C-D; supplemental Table 3). To determine whether impaired PRC2 function influenced the expression of the master B-cell regulators Pax5, Ebf1, and E2A in immature progenitors, as observed in Bmi1 knockout mice,37 we compared their expression in purified HSC, MPP, LMPP, and CLP populations. There were no significant differences in the relative expression of these genes in all populations examined between Suz12+/+ and Suz12Plt8/+ mice (supplemental Figure 1).
Analysis of bone marrow progenitors and B-lymphocyte subsets in 4-week-old Suz12Plt8/+ mice. (A) Gating strategy and (B) number of viable cells for various B-lymphoid populations in the bone marrow of 4-week-old Suz12+/+ and Suz12Plt8/+ mice. (C) Gating strategy and (D) number of bone marrow progenitors in 4-week-old Suz12+/+ and Suz12Plt8/+ mice. Data represent mean ± standard deviation. A 2-tailed Student t test was used for comparison between the genotypes (*P < .05). Cell surface markers used to define various subsets are as follows: total B, B220+CD19+; pre-pro B, CD11c−NK1.1−CD19−B220+CD43+; pre-B-II, B220+CD19+cKit−CD25+IgM−; pro + pre-B, B220+CD19+cKit−IgM−; immature B, B220lowIgM+; recirculating B, B220highIgM+; LSK, lineage−Sca1+c-Kit+; CD150+, LSK CD150+; HSC, LSK Flt3−; MPP, LSK Flt3int; LMPP, LSK Flt3high; CLP, lineage− Sca-1+c-KitintIL-7Rα+Flt3+; ALP, CLP Ly6D−; BLP, CLP Ly6D+.
Analysis of bone marrow progenitors and B-lymphocyte subsets in 4-week-old Suz12Plt8/+ mice. (A) Gating strategy and (B) number of viable cells for various B-lymphoid populations in the bone marrow of 4-week-old Suz12+/+ and Suz12Plt8/+ mice. (C) Gating strategy and (D) number of bone marrow progenitors in 4-week-old Suz12+/+ and Suz12Plt8/+ mice. Data represent mean ± standard deviation. A 2-tailed Student t test was used for comparison between the genotypes (*P < .05). Cell surface markers used to define various subsets are as follows: total B, B220+CD19+; pre-pro B, CD11c−NK1.1−CD19−B220+CD43+; pre-B-II, B220+CD19+cKit−CD25+IgM−; pro + pre-B, B220+CD19+cKit−IgM−; immature B, B220lowIgM+; recirculating B, B220highIgM+; LSK, lineage−Sca1+c-Kit+; CD150+, LSK CD150+; HSC, LSK Flt3−; MPP, LSK Flt3int; LMPP, LSK Flt3high; CLP, lineage− Sca-1+c-KitintIL-7Rα+Flt3+; ALP, CLP Ly6D−; BLP, CLP Ly6D+.
Suz12 deficiency results in increased clonogenicity of B-lymphoid progenitors
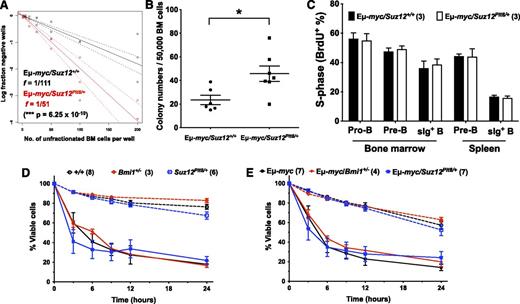
The frequency (f) of B-lymphoid progenitors with proliferative potential was assessed by limiting dilution from unfractionated bone marrow cells. Suz12Plt8/+ mice contained a higher number of clonogenic progenitors than Suz12+/+ controls (Figure 3A). This observation was independently verified when equal numbers of bone marrow cells from 3-week-old Suz12+/+ and Suz12Plt8/+mice were cultured in methylcellulose for development of B-lymphoid colonies (Figure 3B). Limiting dilution assay and methylcellulose cultures revealed no significant difference in clonogenicity of purified Suz12Plt8/+ pro-B cells (B220+CD19+c-Kit+IgM−) relative to control (Figure 3C-D). However, when cells from primary methylcellulose cultures were replated, Suz12Plt8/+ cells generated more secondary colonies than Suz12+/+ controls, and the difference in recloning potential was further enhanced on a third round of replating, in which Suz12+/+ cells generated very few colonies, whereas Suz12Plt8/+ cells replated robustly (Figure 3D). Increased replating potential of Suz12Plt8/+ pro-B cells was confirmed in a separate experiment in which 20 individual colonies from primary cultures were replated individually into secondary cultures. The frequency and the absolute number of secondary colonies were significantly higher in Suz12Plt8/+ pro-B cells than control (Figure 3E).
Suz12Plt8/+ pro-B cells have enhanced self-renewal potential. (A) Limiting dilution analysis of Suz12+/+ (n = 8) and Suz12Plt8/+ (n = 4) bone marrow cells was performed to compare the frequency (f) of B-lymphoid progenitors. Cells were cultured on OP-9 stroma with IL-7. The clonogenic cell frequency model fitted to each dilution series is shown by a solid straight line relating the log10 fraction of negative wells to the number of cells per well. Steeper slopes indicate higher frequencies of colony-forming cells. Broken lines show 95% confidence intervals. (B) Unfractionated bone marrow cells from 3-week-old Suz12+/+ (n = 6) and Suz12Plt8/+ (n = 5) mice were cultured in methylcellulose with IL-7, and the numbers of colonies were scored 7 days later. Data represent means ± standard error of the mean (SEM). A 2-tailed Student t test was performed (*P < .05). (C) Limiting dilution analysis showed no difference in progenitor frequencies of purified bone marrow pro-B cells (B220+CD19+c-Kit+IgM−) from 3-week-old Suz12+/+ and Suz12Plt8/+ mice. (D) Self-renewal potential of Suz12+/+ (n = 8) and Suz12Plt8/+ (n = 8) pro-B cells was determined by serial replating of colonies in methylcellulose. Data represent means ± SEM. A 2-tailed Student t test was performed (*P < .05; ***P < .001). (E) Comparison of the clonogenic potential of individual primary colonies from Suz12+/+ and Suz12Plt8/+ pro-B cells, expressed both in frequency and total secondary colony numbers. Data represent means ± SEM. A 2-tailed Student t test was performed (**P < .01; ***P < .001).
Suz12Plt8/+ pro-B cells have enhanced self-renewal potential. (A) Limiting dilution analysis of Suz12+/+ (n = 8) and Suz12Plt8/+ (n = 4) bone marrow cells was performed to compare the frequency (f) of B-lymphoid progenitors. Cells were cultured on OP-9 stroma with IL-7. The clonogenic cell frequency model fitted to each dilution series is shown by a solid straight line relating the log10 fraction of negative wells to the number of cells per well. Steeper slopes indicate higher frequencies of colony-forming cells. Broken lines show 95% confidence intervals. (B) Unfractionated bone marrow cells from 3-week-old Suz12+/+ (n = 6) and Suz12Plt8/+ (n = 5) mice were cultured in methylcellulose with IL-7, and the numbers of colonies were scored 7 days later. Data represent means ± standard error of the mean (SEM). A 2-tailed Student t test was performed (*P < .05). (C) Limiting dilution analysis showed no difference in progenitor frequencies of purified bone marrow pro-B cells (B220+CD19+c-Kit+IgM−) from 3-week-old Suz12+/+ and Suz12Plt8/+ mice. (D) Self-renewal potential of Suz12+/+ (n = 8) and Suz12Plt8/+ (n = 8) pro-B cells was determined by serial replating of colonies in methylcellulose. Data represent means ± SEM. A 2-tailed Student t test was performed (*P < .05; ***P < .001). (E) Comparison of the clonogenic potential of individual primary colonies from Suz12+/+ and Suz12Plt8/+ pro-B cells, expressed both in frequency and total secondary colony numbers. Data represent means ± SEM. A 2-tailed Student t test was performed (**P < .01; ***P < .001).
There was a modest decrease in global H3K27me3 level in Suz12Plt8/+ pro-B cells compared with Suz12+/+ pro-B cells, which coincided with a slight decrease in Suz12 and Ezh2 levels (supplemental Figure 2A). This effect was more pronounced in pro-B cells expressing shRNA to Suz12, in which a profound reduction in Suz12, Ezh2, and global H3K27me3 levels was evident (supplemental Figure 2B). The level of H2AK119ub, the histone mark deposited by PRC1, was not altered in Suz12-deficient cells relative to total protein (supplemental Figure 2).
PRC2 restricts Eμ-myc lymphomagenesis
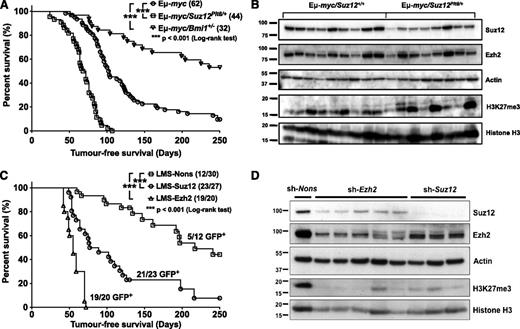
To address whether heterozygosity of PcG genes contributes to B-cell malignancy, Suz12Plt8/+ mice or mice with a heterozygous deletion in the gene encoding the PRC1 protein Bmi1 were crossed with Eµ-myc transgenic mice. Consistent with previous observations,38 Eμ-myc mice developed lymphoma with a median onset of 103 days, whereas loss of 1 allele of Bmi1 prolonged survival (Figure 4A). In contrast, Eμ-myc/Suz12Plt8/+ mice showed accelerated onset of disease with a median survival of 72 days (Figure 4A). This suggests that, unlike Bmi1, which promotes lymphomagenesis in Eμ-myc mice, Suz12 functions as a tumor suppressor.
PRC2 suppresses the development of B-cell lymphoma in Eμ-myc transgenic mice in a cell autonomous manner. (A) Lymphoma-free survival of Eμ-myc (circle), Eμ-myc/Suz12Plt8/+ (square), and Eμ-myc/Bmi1+/− (triangle) mice. (B) Immunoblot analysis of primary lymphomas from Eμ-myc and Eμ-myc/Suz12Plt8/+ mice. (C) Lymphoma-free survival of lethally irradiated Ly5.1 recipient mice reconstituted with Eμ-myc fetal liver cells expressing shRNA to Suz12 (circle), Ezh2 (triangle), and a Nons control (square). (D) Immunoblot analysis of Suz12, Ezh2, and H3K27me3 from FACS-sorted donor-derived lymphoma cells expressing the indicated shRNAs.
PRC2 suppresses the development of B-cell lymphoma in Eμ-myc transgenic mice in a cell autonomous manner. (A) Lymphoma-free survival of Eμ-myc (circle), Eμ-myc/Suz12Plt8/+ (square), and Eμ-myc/Bmi1+/− (triangle) mice. (B) Immunoblot analysis of primary lymphomas from Eμ-myc and Eμ-myc/Suz12Plt8/+ mice. (C) Lymphoma-free survival of lethally irradiated Ly5.1 recipient mice reconstituted with Eμ-myc fetal liver cells expressing shRNA to Suz12 (circle), Ezh2 (triangle), and a Nons control (square). (D) Immunoblot analysis of Suz12, Ezh2, and H3K27me3 from FACS-sorted donor-derived lymphoma cells expressing the indicated shRNAs.
Analysis of moribund Eμ-myc/Suz12Plt8/+ mice revealed lymphomas typical of those observed in Eμ-myc/Suz12+/+ mice. All moribund mice presented with lymphadenopathy, splenomegaly (supplemental Figure 3A) and increased leukocyte numbers, accompanied by normal red blood cell counts and mild thrombocytopenia (supplemental Figure 3D-F). All lymphomas were of B-lymphoid origin, and the proportion of pre-B, sIg+ B, or mixed pre-B/mature B lymphomas was consistent (supplemental Figure 3B). To verify malignancy, 2 × 106 splenocytes from lymphoma-bearing mice were transplanted into nonirradiated recipients. As documented for Eμ-myc disease,39 lymphomas developed quickly in all recipients, but the disease latency did not differ between recipients of Eμ-myc/Suz12+/+ and Eμ-myc/Suz12Plt8/+ lymphomas (supplemental Figure 3C). Eμ-myc/Suz12Plt8/+ lymphomas displayed a variable but on average modest reduction in Suz12 levels compared with Eμ-myc lymphomas, whereas there was no consistent difference in Ezh2 levels (Figure 4B; supplemental Figure 2C). Global H3K27me3 levels varied considerably between individual Eμ-myc/Suz12+/+ lymphomas, and a similar pattern was observed in Eμ-myc/Suz12Plt8/+ lymphomas, with no consistent differences between the 2 groups.
To independently confirm that reduced PRC2 activity accelerates Eμ-myc lymphoma, E13.5 CD45Ly5.2 Eμ-myc fetal liver cells were infected with GFP-tagged shRNAs targeting Suz12 (LMS-Suz12), Ezh2 (LMS-Ezh2), or a nonsilencing control (LMS-Nons) and transplanted into lethally irradiated CD45Ly5.1 recipients (supplemental Figure 4A). The activity of the shRNAs was confirmed using G1ME cells and primary pro-B-cell cultures (supplemental Figures 2B and 4B). Mice reconstituted with PRC2-deficient cells had higher blood leukocyte numbers at 4 and/or 8 weeks after transplantation (supplemental Figure 5A). Effective donor-derived contribution to mature blood cells was confirmed at 8 weeks after transplantation (supplemental Figure 5B). There was a preferential expansion in cells of the B-lymphoid lineage over T-lymphoid and myeloid lineages in recipients of LMS-Suz12– and LMS-Ezh2–expressing cells, which was not evident in the LMS-Nons control (supplemental Figure 5C-F).
Mice reconstituted with PRC2-deficient cells developed lymphomas faster than the control group: 19 of 20 mice from the LMS-Ezh2 group and 23 of 27 mice from the LMS-Suz12 group succumbed to lymphoma with a median onset of 100 days, whereas only 12 of 30 mice from the LMS-Nons group developed disease, with a median latency of 150 days (Figure 4C). All moribund mice developed either pre-B– or B-cell lymphomas at similar frequencies (supplemental Figure 5G) and displayed leukocytosis and thrombocytopenia similar to unmanipulated Eμ-myc mice (supplemental Figure 5H-J). Although only 5 of 12 LMS-Nons lymphomas expressed GFP, 21 of 23 LMS-Suz12 and 19 of 20 LMS-Ezh2 lymphomas were GFP+, indicating that PRC2-deficient cells were more lymphomagenic than the LMS-Nons control, which were no more likely to cause lymphoma than nontransduced (GFP−) cells. Immunoblotting analysis from FACS-purified lymphoma cells showed a significant reduction in Suz12, Ezh2, and global H3K27me3 levels in Suz12 and Ezh2 knockdown lymphomas relative to control (Figures 4D). These data confirm that PRC2 functions as a cell autonomous tumor suppressor of Eμ-myc lymphoma.
Expanded B lymphopoiesis in preneoplastic Eμ-myc/Suz12Plt8/+ mice
To understand the mechanisms behind accelerated lymphoma onset, we examined changes in cellular pathways in preneoplastic mice, a distinct phase in 3- to 4-week-old mice defined by the lack of transplantable tumor cells.40 Lymphomas failed to develop in mice transplanted with 106 splenocytes from 3- to 4-week-old Eμ-myc/Suz12Plt8/+ mice, confirming that a true preneoplastic phase also exists in these mice. Quantitative reverse transcriptase-polymerase chain reaction analysis from purified bone marrow pre-B cells showed that the Suz12Plt8 mutation did not influence c-myc RNA levels (supplemental Figure 6), excluding the possibility that accelerated lymphomagenesis in Eμ-myc/Suz12Plt8/+ mice was simply due to increased transgene expression.
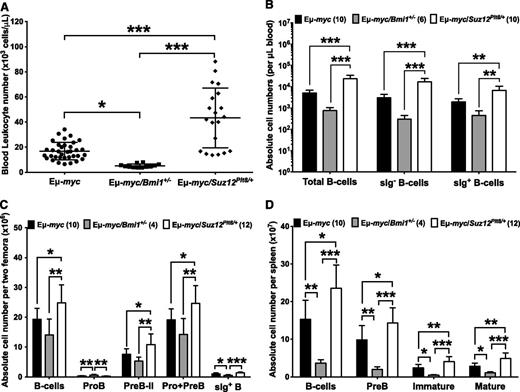
Preneoplastic Eμ-myc/Suz12Plt8/+ mice exhibited a threefold increase in blood leukocytes relative to control mice (Figure 5A), primarily due to increased lymphocytes (supplemental Table 2). In contrast, Eμ-myc/Bmi1+/− mice showed a significant reduction in total leukocytes (Figure 5A) and lymphocytes (supplemental Table 2). Analysis of nucleated blood cells revealed a marked increase in total B-lymphoid cell numbers in Eμ-myc/Suz12Plt8/+ mice relative to Eμ-myc littermates, whereas a significant reduction in B-lymphoid cells was observed in Eμ-myc/Bmi1+/− mice (Figure 5B). Although bone marrow cellularity was constant in the 3 groups (supplemental Table 4), Eμ-myc/Suz12Plt8/+ mice had an increased number of B-lineage cells compared with Eμ-myc mice, due to elevated numbers of sIg− precursors (Figure 5C; supplemental Table 4). In contrast, Eμ-myc/Bmi1+/− mice had fewer B-lymphoid cells than in Eμ-myc controls (Figure 5C; supplemental Table 4). Eμ-myc mice exhibited significant splenomegaly, both in weight and cellularity, which was further exacerbated in Eμ-myc/Suz12Plt8/+ mice, whereas the opposite was observed in Eμ-myc/Bmi1+/− mice (supplemental Table 4). This was attributable to an increased number of total splenic B-lymphoid cells and specific subsets including pre-B, immature, and mature B cells (Figure 5D). The numbers of T lymphocytes and myeloid cells were generally within the normal range in mice of all genotypes (supplemental Table 4).
Pre-neoplastic Eμ-myc/Suz12Plt8/+ mice have an expanded B-lymphoid compartment. (A) Automated enumeration of peripheral blood leukocytes from preneoplastic Eμ-myc (n = 17), Eμ-myc/Bmi1+/− (n = 8), and Eμ-myc/Suz12Plt8/+ (n = 12) mice. Immunophenotypic characterization of B-lymphocyte subsets from the (B) peripheral blood, (C) bone marrow, and (D) spleen of preneoplastic Eμ-myc, Eμ-myc/Bmi1+/−, and Eμ-myc/Suz12Plt8/+ mice. Data represent mean ± standard deviation. One-way analysis of variance followed by Tukey’s post hoc test was used for pairwise comparisons (*P < .05; **P < .01; ***P < .001). Cell surface markers used to define B-cell subsets are as follows: BM pro-B, B220+CD19+c-Kit+IgM−IgD−; BM pre-B-II, B220+CD19+c-Kit−CD25+IgM−IgD−; BM pro + pre-B, blood sIg− B, or spleen pre-B, B220+CD19+cKit−IgM−IgD−; spleen immature B, B220+CD19+c-Kit−IgM+IgD−, BM sIg+; spleen mature B, B220+CD19+c-Kit−IgM+IgD+.
Pre-neoplastic Eμ-myc/Suz12Plt8/+ mice have an expanded B-lymphoid compartment. (A) Automated enumeration of peripheral blood leukocytes from preneoplastic Eμ-myc (n = 17), Eμ-myc/Bmi1+/− (n = 8), and Eμ-myc/Suz12Plt8/+ (n = 12) mice. Immunophenotypic characterization of B-lymphocyte subsets from the (B) peripheral blood, (C) bone marrow, and (D) spleen of preneoplastic Eμ-myc, Eμ-myc/Bmi1+/−, and Eμ-myc/Suz12Plt8/+ mice. Data represent mean ± standard deviation. One-way analysis of variance followed by Tukey’s post hoc test was used for pairwise comparisons (*P < .05; **P < .01; ***P < .001). Cell surface markers used to define B-cell subsets are as follows: BM pro-B, B220+CD19+c-Kit+IgM−IgD−; BM pre-B-II, B220+CD19+c-Kit−CD25+IgM−IgD−; BM pro + pre-B, blood sIg− B, or spleen pre-B, B220+CD19+cKit−IgM−IgD−; spleen immature B, B220+CD19+c-Kit−IgM+IgD−, BM sIg+; spleen mature B, B220+CD19+c-Kit−IgM+IgD+.
Suz12 deficiency has no influence on apoptosis or cell cycle of Eμ-myc B-lymphoid cells
Preneoplastic Eμ-myc/Suz12Plt8/+ mice had an increased frequency of B-cell progenitors in their bone marrow as assessed by limiting dilution assay (Figure 6A), as well as an increased number of cells capable of forming B-cell colonies in methylcellulose (Figure 6B). The Suz12Plt8 mutation did not influence BrdU uptake in vivo in pro-B, pre-B, or sIg+ B cells in bone marrow or spleen of Eμ-myc mice (Figure 6C). Although bone marrow pre-B and sIg+ B cells from Eμ-myc mice died rapidly in the absence of cytokines, the rate of apoptosis was equivalent in Eμ-myc/Suz12Plt8/+ and Eμ-myc/Bmi1+/− mice (Figure 6D-E). It is therefore unlikely that changes in the cell cycle or apoptotic mechanisms contribute significantly to the increased number of B-cell progenitors evident in preleukemic Eμ-myc/Suz12Plt8/+ mice.
Proliferation or spontaneous apoptosis are unchanged in preneoplastic B-lymphoid cells between Eμ-myc/Suz12+/+ and Eμ-myc/Suz12Plt8/+ mice. (A) Limiting dilution analysis of unfractionated bone marrow cells from preneoplastic Eμ-myc/Suz12+/+ (n = 4) and Eμ-myc/Suz12Plt8/+ (n = 3) mice. (B) Unfractionated bone marrow cells from preneoplastic Eμ-myc (n = 6) and Eμ-myc/Suz12Plt8/+ (n = 7) mice were cultured in methylcellulose and scored 7 days later. Data represent means ± SEM. A 2-tailed Student t test was performed (*P < .05). (C) BrdU incorporation in Eμ-myc (n = 3) and Eμ-myc/Suz12Plt8/+ (n = 3) cells 1 hour after BrdU injection (0.1 mg/mg body weight). The percentage of BrdU+ cells in bone marrow pro-B, pre-B, and sIg+ B cells and in splenic pre-B and sIg+ B cells was determined by FACS. Data represent means ± standard deviation. A 2-tailed Student t test was used to determine statistical significance. In vitro survival assay was performed on cells from preneoplastic Eμ-myc, Eμ-myc/Bmi1+/−, and Eμ-myc/Suz12Plt8/+ mice. FACS-purified bone marrow (D) pre-B and (E) sIg+ B cells were cultured under conditions of cytokine deprivation. Cell viability was measured by Annexin-V and propidium iodide staining using flow cytometry. Three-week-old nontransgenic wildtype (+/+), Bmi1+/−, and Suz12Plt8/+mice were included as controls. Data represent means ± SEM at each time point. One-way analysis of variance followed by Tukey’s post hoc test was used to compare mice of the following genotypes: wildtype (+/+), Bmi1+/−, and Suz12Plt8/+, either carrying the Eμ-myc transgene or the corresponding nontransgenic controls.
Proliferation or spontaneous apoptosis are unchanged in preneoplastic B-lymphoid cells between Eμ-myc/Suz12+/+ and Eμ-myc/Suz12Plt8/+ mice. (A) Limiting dilution analysis of unfractionated bone marrow cells from preneoplastic Eμ-myc/Suz12+/+ (n = 4) and Eμ-myc/Suz12Plt8/+ (n = 3) mice. (B) Unfractionated bone marrow cells from preneoplastic Eμ-myc (n = 6) and Eμ-myc/Suz12Plt8/+ (n = 7) mice were cultured in methylcellulose and scored 7 days later. Data represent means ± SEM. A 2-tailed Student t test was performed (*P < .05). (C) BrdU incorporation in Eμ-myc (n = 3) and Eμ-myc/Suz12Plt8/+ (n = 3) cells 1 hour after BrdU injection (0.1 mg/mg body weight). The percentage of BrdU+ cells in bone marrow pro-B, pre-B, and sIg+ B cells and in splenic pre-B and sIg+ B cells was determined by FACS. Data represent means ± standard deviation. A 2-tailed Student t test was used to determine statistical significance. In vitro survival assay was performed on cells from preneoplastic Eμ-myc, Eμ-myc/Bmi1+/−, and Eμ-myc/Suz12Plt8/+ mice. FACS-purified bone marrow (D) pre-B and (E) sIg+ B cells were cultured under conditions of cytokine deprivation. Cell viability was measured by Annexin-V and propidium iodide staining using flow cytometry. Three-week-old nontransgenic wildtype (+/+), Bmi1+/−, and Suz12Plt8/+mice were included as controls. Data represent means ± SEM at each time point. One-way analysis of variance followed by Tukey’s post hoc test was used to compare mice of the following genotypes: wildtype (+/+), Bmi1+/−, and Suz12Plt8/+, either carrying the Eμ-myc transgene or the corresponding nontransgenic controls.
Gene expression analysis of PRC2-deficient lymphomas
A gene level differential expression analysis revealed that 35 genes upregulated (supplemental Table 5) and 32 genes downregulated (supplemental Table 6) in Eμ-myc/Suz12Plt8/+ lymphomas. Competitive gene set analysis33 was used to interpret the differential expression patterns in terms of molecular pathways, using the curated gene set collection of the molecular signatures database.32 Of the 3235 gene sets tested, 111 were significantly altered in Eμ-myc/Suz12Plt8/+ lymphomas (supplemental Table 7). Many of the gene sets were derived from hematological and epithelial tumor studies. For example, genes involved in the progression from benign adenoma to malignant hepatocellular carcinomas were upregulated (P = .010),41 whereas genes that are normally upregulated during the transition from pro-B to pre-B cells were downregulated in Eμ-myc/Suz12Plt8/+ lymphomas (P = .036).42
Discussion
Although a role for deregulation of PRC2 in multiple cancer contexts is compelling, the diversity of molecular lesions in PRC2 components implies that PRC2 can contribute to tumorigenesis via multiple mechanisms. To better understand the role of PRC1 and PRC2 in hematological malignancies, we compared the effect of compromising each complex in the Eμ-myc transgenic mouse model of B lymphoma. In striking contrast to the previously established effects of heterozygosity of the PRC1 gene Bmi1, which delays disease onset,38 a loss-of-function allele of Suz12 accelerated B lymphoma in the Eμ-myc model. Similar observations were found in chimeric mice reconstituted with Suz12 or Ezh2 knockdown Eμ-myc fetal liver cells. It is known that Eμ-myc B-lymphoid cells have increased rates of cell cycling and deregulated apoptosis23,40 ; however, these parameters were not measurably different in PRC2-compromised Eμ-myc mice. The enhanced self-renewal capacity of B-lymphoid progenitors evident in Suz12Plt8/+ mice is likely responsible for the expansion of the B-cell lineage in these animals and the accelerated disease onset in Eμ-myc mice that have impaired PRC2 activity.
Although studies in Drosophila led to the proposal of the initiator-mediator model, which posits that PRC1 is sequentially recruited to PRC2 targets to execute gene silencing,5 PRC1 and PRC2 clearly have opposing functions in Eµ-myc–driven lymphomagenesis. The concept that the actions of PRC1 and PRC2 can diverge from the initiator-mediator model in specific circumstances is supported by several in vitro studies. For example, PRC1 can be recruited to targets in PRC2-deficient ES cells during X inactivation,7 and genomewide chromatin immunoprecipitation studies showed PRC1 and PRC2 can occupy distinct loci.43 The recent identification of a novel PRC1 complex that monoubiquitinates histone H2A independent of H3K27me3/PRC2 in mouse ES cells44 provides a potential mechanism for PRC2-independent gene regulation by PRC1. These observations imply that polycomb complexes regulate gene expression via multiple mechanisms in context-dependent manners.
Previously, we demonstrated that PRC1 and PRC2 regulate distinct targets in HSCs.24 Although these genes represent a small proportion of total PRC-responsive genes, they can have a profound influence on HSC functions. The contrasting roles of PRC1 and PRC2 in Eμ-myc–driven lymphomagenesis suggest that these complexes may also regulate distinct targets in lymphoid progenitors. Another alternative is that some target genes are particularly sensitive to the dosage of either PRC1 or PRC2 or to individual complex components. For example, the tumor suppressor gene Cdkn2a is very sensitive to changes in the level of PRC1,11 but it is less responsive to inhibition of PRC2.24 Our results suggest that inhibition of PRC2 results in enhanced self-renewal in lineage-committed progenitors that speeds up the course of Eμ-myc disease. Further analysis of the genomic occupancy and activity of PRC1 and PRC2 will be required to determine why inhibition of these complexes results in such distinct outcomes.
Mutations that disrupt PRC2 have been identified in human lymphoid malignancy, but the precise role of these mutations remains unclear. Although activating mutations in EZH2 are common in diffuse large B-cell lymphoma,13-15 loss-of-function mutations and deletions of EZH2 and SUZ12 have been described in human T-ALL,18,19 and recently, Ezh2 was shown to be critical for T-ALL suppression in mouse models.20 These observations mirror those obtained with our results in Eµ-myc lymphoma, which emphasizes that PRC2 function is indeed context dependent. EZH2 expression is high in B-lymphoid progenitors, declines during B-cell maturation, and then is upregulated again during affinity maturation of activated germinal center B cells.45 Thus, although PRC2 activity may restrict the proliferative potential of B-lymphoid progenitors via effects on self-renewal, it may be that gain-of-function mutations work to stimulate proliferation specifically in more mature B cells.
Loss of EZH2 has also been identified in myelodysplastic syndrome.16 Intriguingly, ectopic expression of Ezh2 in mice also results in myeloid malignancies,46 and PRC2 is required for the maintenance of self-renewal in myeloid leukemias driven by mixed-lineage leukemia fusion genes.47 These studies collectively show that maintenance of PRC2 activity within a defined normal range is essential, as either reduced or excess activity predisposes to malignancy. Modulation of epigenetic regulators, including EZH2, is an exciting and rapidly developing area in cancer therapy.48,49 It is important to consider that PRC2 is clearly acting as a tumor suppressor in some contexts, which has important implications for how to approach this complex therapeutically.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jason Corbin, Dina Stockwell, Jackie Gilbert, Lauren Wilkins, Mathew Salzone, and Sally Richards for technical assistance. The authors also thank Yifang Hu and Wei Shi for bioinformatics assistance.
This work was supported by program grants (1016647 and 575500), a project grant (1011663), National Health and Medical Research Council (NHMRC) fellowships (to W.S.A., I.J.M., M.E.B., D.J.H., and G.K.S.), fellowships from the Australia Research Council (to M.E.B. and S.L.N.), a NHMRC–Institut National de la Santé et de la Recherche Médicale Postdoctoral Training fellowship (to R.S.A.), Australian postgraduate awards (to S.C.W.L., B.P., H.S.L., and A.L.), Independent Research Institutes Infrastructure Support Scheme grant 361646 from the NHRMC, the Australian Cancer Research Fund, and a Victorian State Government Operational Infrastructure Support grant.
Authorship
Contribution: S.C.W.L., W.S.A., and I.J.M. designed experiments and wrote the manuscript; S.C.W.L., C.D.H., H.S.L., and R.S.A. performed experiments and analyzed data; B.P., A.L., and G.K.S. performed bioinformatics analyses; S.L.N. and M.E.B. provided reagents; D.J.H., S.L.N., M.E.B., and G.K.S. provided critical feedback to the manuscript; and W.S.A. and I.J.M. edited the manuscript and supervised research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ian J. Majewski, 1G Royal Parade, Parkville, VIC 3052, Australia; e-mail: majewski@wehi.edu.au.
References
Author notes
W.S.A. and I.J.M. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal