Key Points
p210 BCR/ABL interacts with β-catenin in the bone marrow transplantation model for chronic myelogenous leukemia.
Loss of the interaction results in an altered disease phenotype, suggesting a role for β-catenin in chronic phase disease.
Abstract
We have identified a ubiquitin-binding domain within the NH2-terminal sequences of p210 BCR/ABL and determined that the binding site co-localizes with the binding site for β-catenin. The domain does not support the auto- or trans-kinase activity of p210 BCR/ABL or its ability to interact with GRB2 and activate ERK1/2 signaling. Expression of p210 BCR/ABL, but not a β-catenin–binding mutant, in hematopoietic cells is associated with the accumulation of p-β-catenin (Tyr654) and increased TCF/LEF-mediated transcription. In a bone marrow transplantation model, the interaction between β-catenin and p-β-catenin (Tyr654) is detectable in mice transplanted with p210 BCR/ABL, but not the mutant. Whereas mice transplanted with p210 BCR/ABL exhibit myeloid disease with expansion of monocytes and neutrophils, mice transplanted with the mutant predominantly exhibit expansion of neutrophils, polycythemia, and increased lifespan. The increased disease latency is associated with expansion of megakaryocyte-erythrocyte progenitors, a decrease in common myeloid progenitors, and reduced β-catenin signaling in the bone marrow of the diseased mice. These observations support a model in which p210 BCR/ABL may influence lineage-specific leukemic expansion by directly binding and phosphorylating β-catenin and altering its transcriptional activity. They further suggest that the interaction may play a role in chronic phase disease progression.
Introduction
Blast crisis is the terminal stage of chronic myelogenous leukemia (CML), and recent studies have identified a granulocyte-macrophage progenitor (GMP) population with an anomalous self-renewal capacity in blast crisis patients.1 Since deregulated self-renewal has been recognized as an important feature of leukemic progression, this population may function as the leukemic stem cells during blast crisis.2,3 A similar GMP population was identified in a bone marrow transplantation (BMT) model for CML and was shown to be sufficient for leukemic initiation.4
The GMP populations that are proposed to function as leukemic stem cells in mice and in blast crisis patients contain abnormally high levels of β-catenin activity.1,4 In mice, renewal of the hematopoietic stem cell population requires activation of the β-catenin pathway and increased expression of target genes such as c-Myc and cyclin D1.5,6 Although the mechanism by which p210 BCR/ABL activates β-catenin is unclear, the two proteins have been shown to directly interact.7 The interaction is dependent upon the tyrosine kinase activity of p210 BCR/ABL, and β-catenin can serve as a direct substrate for p210 BCR/ABL tyrosine kinase activity. When phosphorylated on tyrosine by p210 BCR/ABL, β-catenin is transcriptionally activated and is impaired in its ability to interact with the cytoplasmic Axin/GSK3β complex.7 Deletion of β-catenin in donor hematopoietic stem cells reduces the ability of these cells to induce p210 BCR/ABL–mediated leukemia in the BMT model, suggesting that the interaction between these two proteins may support disease progression.8
In a recent study, we have shown that BCR interacts with components of the ESCRT1 complex in the late endosome and regulates epidermal growth factor receptor turnover.9 Components of ESCRT1 recognize ubiquitinated cargo on the endosomal membrane and mediate their internalization during formation of the multivesicular body.10,11 Since the two components of the ESCRT1 complex to which BCR binds contain ubiquitin-binding domains (UBDs),12 we examined BCR and p210 BCR/ABL for the presence of a UBD. In this study, we show that both BCR and p210 BCR/ABL contain a UBD within their NH2-terminus and that the UBD co-localizes with the binding site for β-catenin. Deletion of this domain impairs β-catenin binding and alters disease progression in the BMT model for CML. These observations represent direct in vivo evidence that the interaction between β-catenin and p210 BCR/ABL supports disease progression.
Methods
Molecular constructs and yeast 2-hybrid analysis
The pAX142 mammalian expression vector has been described previously.13 pAX142-bcr-abl and pAX142-bcr-abl(ΔUBD) contain HA-epitope tagged, full-length p210 BCR/ABL and p210 BCR/ABL with an internal deletion of residues 180-191, respectively. The yeast 2-hybrid constructs used for mapping were constructed in the pGBT9 vector. The MSCV-IRES-gfp retroviral vector is as described by the manufacturer (Addgene, Cambridge, MA). MSCV-p210 Bcr-Abl-IRES-gfp and MSCV-p210 Bcr-Abl(ΔUBD)-IRES-gfp contain complementary DNAs encoding full-length p210 BCR/ABL and p210 BCR/ABL with an internal deletion of residues 180-191, respectively. All yeast 2-hybrid analyses were performed as previously described.14
Cell culture
293T and Phoenix-ecotropic cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (FBS) (Gemini, Woodland, CA). K562 cells were cultured in RPMI-1640 supplemented with 20% FBS (BD Biosciences, Franklin Lakes, NJ). Ba/F3 cells were maintained in RPMI-1640 supplemented with 10% FBS (Gemini, Woodland, CA) and 10% WEHI-conditioned media. High-titer retrovirus was generated by using Phoenix-Ecotropic packaging cells (American Type Culture Collection, Manassas, VA). Viral supernatant was collected at 48 and 72 hours posttransfection and stored at −80°C. Stocks were titered by infection of NIH3T3 cells followed by flow cytometry to detect GFP expression.
Protein expression and co-immunoprecipitation
Western blot analysis and co-immunoprecipitations were performed as previously described.9 Antibodies used include anti-HA (HA11; Sigma, St. Louis, MO); anti–c-ABL, anti-BCR, anti-GRB2, anti–phospho-c-ABL (Tyr245), anti-CRKL (32H4), anti–phospho-CRKL (Tyr207), and anti–cyclin D1 (Cell Signaling, Danvers, MA); anti–phospho-tyrosine (PY20) and anti-β-catenin (BD Biosciences, Franklin Lakes, NJ); anti–β-catenin (Tyr654) (Abcam, Cambridge, MA); and agarose-conjugated anti–c-abl and anti–c-myc (Santa Cruz Biotechnology, Santa Cruz, CA). The E1 inhibitor UBEI-41 (Biogenova, Rockville, MD) was used at a concentration of 50 µM.
Cell fractionation
Nuclear and cytosolic fractions were prepared as described with modifications.12 Briefly, Ba/F3 cells were lysed in hypotonic lysis buffer with protease inhibitors. Dounce homogenizer was used to disrupt the cells but keep the nuclei intact. The lysates were centrifuged at 375g for 5 minutes. The precipitations (nuclear fractions) were washed with hypotonic lysis buffer with 0.1% nonyl phenoxypolyethoxylethanol 5 times; the supernatants (cytosolic fractions) were further centrifuged at 150 000g for 40 minutes to remove any membrane contamination.
Reporter assays
Reporter assays were performed as described.12 Briefly, Ba/F3 cells were transfected with TOPflash or FOPflash plasmids (Millipore, Temecula, CA). pSV-β-gal plasmid was co-transfected into the cells as an internal control to normalize transfection efficiency. After 24 hours, cells were lysed with reporter lysis buffer (Promega, Madison, WI), and luciferase activity was measured by using a luminometer (Analytical Luminescence Laboratory, Sparks, MD). For reporter assays in the progenitors, common myeloid progenitors (CMPs) and megakaryocyte-erythrocyte progenitors (MEPs) were sorted by flow cytometry and cultured as previously described.15,16 The progenitors were then co-transduced with lentiviral particles containing a TCF/LEF reporter and a Renilla control to normalize transfection efficiency (Qiagen, Valencia, CA). Firefly and Renilla luciferase activities were measured with a Dual-Luciferase Reporter Assay system following the manufacturer’s instructions (Promega, Madison, WI).
Ubiquitin affinity purification assay
Whole-cell lysates from transfected 293T cells or untransfected K562 cells were collected in ubiquitin-binding lysis buffer (50 mM tris(hydroxymethyl)aminomethane; pH, 7.5), 5 mM adenosine triphosphate, 10 mM MgCl2, 0.2 mM dithiothreitol, and 0.5% nonyl phenoxypolyethoxylethanol) supplemented with a protease inhibitor and phosphatase inhibitor cocktail (Calbiochem, San Diego, CA). Lysates were then incubated with either agarose-conjugated immunoglobulin G (Santa Cruz Biotechnology, Santa Cruz, CA) or agarose-conjugated ubiquitin (BIOMOL, Plymouth Meeting, PA) overnight with rotation at 4°C. Lysates were precipitated by centrifugation at 1000 rpm for 5 minutes, and the precipitates were washed 3 times with ice-cold phosphate-buffered saline supplemented with 5 mM adenosine triphosphate, protease inhibitor, and phosphatase inhibitor cocktail (Calbiochem, San Diego, CA). Proteins that co-precipitated with the ubiquitin were identified by western blot.
Bone marrow transduction and transplantation
Primary and secondary bone marrow transplantation were done as previously described.17 All experiments were performed on 6- to 12-week-old female BALB/c mice (Jackson Laboratories, Bar Harbor, ME). All animal care, housing, and experimentation were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of University of Medicine and Dentistry of New Jersey-New Jersey Medical School. Histopathology was performed as previously described.17
Evaluation of disease progression by flow cytometry
Mice were euthanized through CO2 inhalation, and hematopoietic organs were harvested. White blood cell (WBC) surface marker staining was carried out as previously described.17 For progenitor analysis, bone marrow cells were harvested at death and stained with lineage-specific biotinylated antibodies as previously described.17 For analysis of β-catenin expression, cells were stained overnight with an eFluor660-conjugated antibody against β-catenin (eBioscience, San Diego, CA).
Results
Identification of a UBD in BCR and p210 BCR/ABL
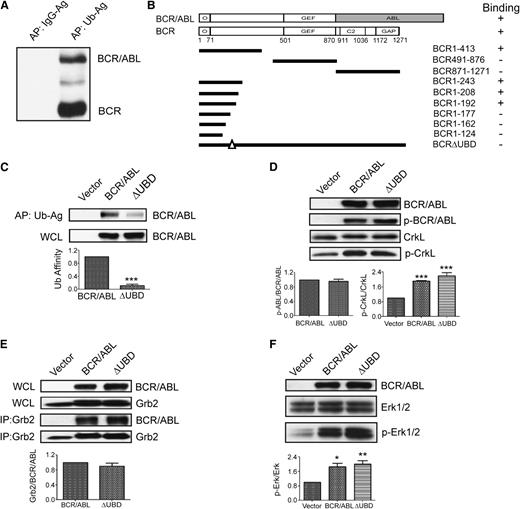
Since we have previously shown that BCR interacts with two components of the ESCRT1,9 both of which contain UBDs, we determined whether BCR and/or p210 BCR/ABL also contain a UBD. Whole-cell lysates from K562 cells were subjected to affinity precipitation using agarose-conjugated ubiquitin or agarose-conjugated immunoglobulin G (as a negative control). Precipitates were then examined by western blot by using an antibody that recognizes the NH2-terminus of BCR and p210 BCR/ABL. Two predominant bands were observed at 160 and 210 kDa, which correspond with the predicted molecular mass of BCR and p210 BCR/ABL, respectively (Figure 1A). To confirm and map the interaction, a ubiquitin complementary DNA was cloned into a yeast 2-hybrid vector18 and was examined for binding with BCR and p210 BCR/ABL. This confirmed the presence of a UBD in both BCR and p210 BCR/ABL, and further mapping with a panel of BCR truncation mutants in yeast localized the UBD to a region of BCR (residues 180-191) that is retained in all three BCR/ABL variants (Figure 1B). Deletion of the UBD did not impair the interaction with the isolated oligomerization domain of BCR, which was used as a positive control for binding in yeast (not shown). We then transiently expressed HA-tagged p210 BCR/ABL or a mutant that lacks the UBD (p210 BCR/ABL(ΔUBD)) in 293T cells and performed an affinity precipitation by using agarose-conjugated ubiquitin. Whereas we were readily able to affinity precipitate p210 BCR/ABL in this manner, the interaction between ubiquitin and the mutant was significantly weaker (Figure 1C).
Identification of the β-catenin binding site in the NH2-terminus of BCR and p210 BCR/ABL. (A) Whole-cell lysates (WCLs) were collected from K562 cells and used for affinity precipitations (APs) using agarose-conjugated immunoglobulin G (IgG-Ag) or agarose-conjugated ubiquitin (Ub-Ag). Precipitated proteins were resolved and identified by western blot by using an antibody that recognizes the NH2-terminus of BCR and p210 BCR/ABL (BCR/ABL). (B) Yeast 2-hybrid analysis was used to map the UBD to residues 180-191 of BCR and p210 BCR/ABL. The upper schematics show the domain structure of the full-length BCR and p210 BCR/ABL constructs that were used for the study (shaded area indicates the ABL sequences). The solid lines below indicate the predicted translational products of the truncation mutants that were used to map the UBD. Binding is indicated as plus (+) or minus (–). (C) 293T cells were transfected with p210 BCR/ABL (BCR/ABL), the ΔUBD mutant, or cognate vector (vector). Lysates were collected and examined by western blot for expression (WCL, lower panel) or used for affinity precipitations using agarose-conjugated ubiquitin (upper panel). Affinity-precipitated BCR/ABL was normalized to input and then expressed as a percentage of wild-type BCR/ABL (Ub affinity). (D) WCLs were collected and examined by western blot for expression of the constructs (BCR/ABL), autophosphorylated p210 BCR/ABL (p-BCR/ABL), CrkL, or phosphorylated CrkL (p-CrkL) as indicated. (E) WCLs were examined by western blot for expression of the transfected constructs or endogenous GRB2. Lysates were used for immunoprecipitations (IPs) using a GRB2 antibody, and immunoprecipitates were examined by western blot using antibodies for p210 BCR/ABL (BCR/ABL) or GRB2 as indicated. (F) WCLs were examined by western blot for expression of the transfected constructs, total ERK1/2, and activated ERK1/2 (p-ERK1/2) as indicated. All data shown represent the average of at least 3 independent experiments. P values were calculated by using a paired Student t test or a one-way analysis of variance (ANOVA) followed by Tukey tests. O, oligomerization domain; GEF, guanine nucleotide exchange factor domain; C2, C2 domain; GAP, GTPase-activating domain. *P < .05; **P < .01; ***P < .001.
Identification of the β-catenin binding site in the NH2-terminus of BCR and p210 BCR/ABL. (A) Whole-cell lysates (WCLs) were collected from K562 cells and used for affinity precipitations (APs) using agarose-conjugated immunoglobulin G (IgG-Ag) or agarose-conjugated ubiquitin (Ub-Ag). Precipitated proteins were resolved and identified by western blot by using an antibody that recognizes the NH2-terminus of BCR and p210 BCR/ABL (BCR/ABL). (B) Yeast 2-hybrid analysis was used to map the UBD to residues 180-191 of BCR and p210 BCR/ABL. The upper schematics show the domain structure of the full-length BCR and p210 BCR/ABL constructs that were used for the study (shaded area indicates the ABL sequences). The solid lines below indicate the predicted translational products of the truncation mutants that were used to map the UBD. Binding is indicated as plus (+) or minus (–). (C) 293T cells were transfected with p210 BCR/ABL (BCR/ABL), the ΔUBD mutant, or cognate vector (vector). Lysates were collected and examined by western blot for expression (WCL, lower panel) or used for affinity precipitations using agarose-conjugated ubiquitin (upper panel). Affinity-precipitated BCR/ABL was normalized to input and then expressed as a percentage of wild-type BCR/ABL (Ub affinity). (D) WCLs were collected and examined by western blot for expression of the constructs (BCR/ABL), autophosphorylated p210 BCR/ABL (p-BCR/ABL), CrkL, or phosphorylated CrkL (p-CrkL) as indicated. (E) WCLs were examined by western blot for expression of the transfected constructs or endogenous GRB2. Lysates were used for immunoprecipitations (IPs) using a GRB2 antibody, and immunoprecipitates were examined by western blot using antibodies for p210 BCR/ABL (BCR/ABL) or GRB2 as indicated. (F) WCLs were examined by western blot for expression of the transfected constructs, total ERK1/2, and activated ERK1/2 (p-ERK1/2) as indicated. All data shown represent the average of at least 3 independent experiments. P values were calculated by using a paired Student t test or a one-way analysis of variance (ANOVA) followed by Tukey tests. O, oligomerization domain; GEF, guanine nucleotide exchange factor domain; C2, C2 domain; GAP, GTPase-activating domain. *P < .05; **P < .01; ***P < .001.
The UBD is not required to support p210 BCR/ABL tyrosine kinase activity
To determine whether the UBD supports the tyrosine kinase activity of p210 BCR/ABL, we expressed p210 BCR/ABL, p210 BCR/ABL(ΔUBD), or cognate vector in 293T cells and then performed western blots with antibodies that recognize total and autophosphorylated p210 BCR/ABL (Figure 1D, upper 2 panels). We detected no significant difference in the level of autophosphorylated p210 BCR/ABL. We also examined the same lysates for levels of total and phosphorylated CrkL, a known substrate for p210 BCR/ABL transkinase activity (Figure 1D, lower 2 panels). The level of phosphorylated CrkL was significantly elevated in cells that express p210 BCR/ABL, and an equivalent level of phosphorylation was observed in cells that express p210 BCR/ABL(ΔUBD). Autophosphorylation of p210 BCR/ABL creates a docking site for GRB2, which in turn activates ERK1/2 signaling. Thus, we also compared p210 BCR/ABL and p210 BCR/ABL(ΔUBD) for their ability to interact with GRB2, and activate ERK1/2. Both p210 BCR/ABL and p210 BCR/ABL(ΔUBD) interact with endogenous GRB2 (Figure 1E) and activate ERK1/2 (Figure 1F) to a similar extent.
The UBD co-localizes with the binding site for β-catenin
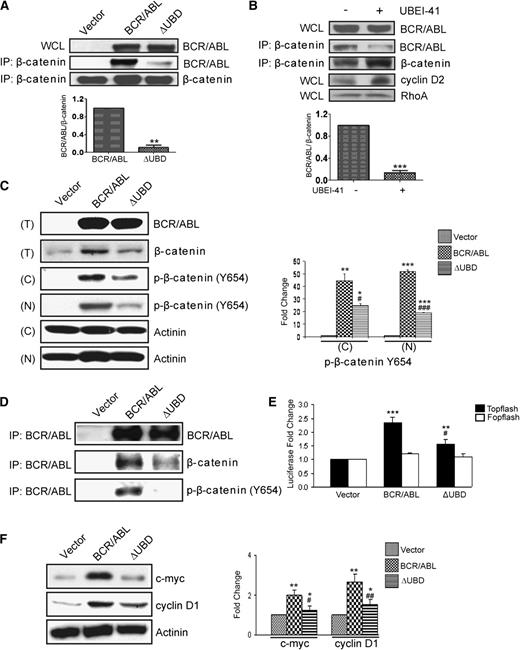
It has been previously shown that β-catenin interacts with an amino-terminal fragment of BCR (residues 1-202),19 and a subsequent study showed that this binding is retained in p210 BCR/ABL.7 Since the UBD is contained within this region, we determined whether β-catenin and ubiquitin share a docking site within p210 BCR/ABL. We overexpressed p210 BCR/ABL or p210 BCR/ABL(ΔUBD) in 293T cells and performed immunoprecipitations by using an antibody that recognizes β-catenin (Figure 2A). We were readily able to co-immunoprecipitate β-catenin and p210 BCR/ABL, consistent with previous reports. However, the interaction between β-catenin and the ubiquitin-binding mutant was substantially impaired. Since β-catenin is a ubiquitinated protein and may bind to p210 BCR/ABL in a ubiquitin-dependent manner, we repeated the co-immunoprecipitation in the presence of a ubiquitin E1 inhibitor (Figure 2B). As expected, the level of total β-catenin was increased when cells were treated with the inhibitor, as were the levels of cyclin D2, another known target of ubiquitin-mediated degradation. In contrast, the levels of a protein that is not ubiquitinated (RhoA), remained unchanged. Despite the increased cellular levels of β-catenin, the interaction between p210 BCR/ABL and β-catenin was reduced in the presence of the E1 inhibitor, suggesting that the interaction may be ubiquitin dependent.
Deletion of the β-catenin binding site in p210 BCR/ABL diminishes the accumulation of p-β-catenin (Tyr654) and TCF/LEF-mediated transcription. (A) 293T cells were transiently transfected with p210 BCR/ABL (BCR/ABL), the ΔUBD mutant, or cognate vector. WCLs were collected and examined by western blot for expression of the constructs. Lysates were also used for IPs using a β-catenin antibody, and immunoprecipitates were examined by western blot using antibodies for β-catenin and p210 BCR/ABL. (B) 293T cells were transfected with p210 BCR/ABL in the presence or absence of the E1 inhibitor. WCLs were collected and examined by western blot for expression of p210 BCR/ABL (BCR/ABL), cyclin D2, or RhoA. Lysates were also used for IPs using a β-catenin antibody, and immunoprecipitates were examined by western blot using antibodies for β-catenin and BCR/ABL. (C) Ba/F3 cells were infected with retroviral particles that encode MSCV-bcr-abl/p210-IRES-gfp (BCR/ABL), MSCV-bcr-abl/p210(ΔUBD)-IRES-gfp (ΔUBD), or cognate vector. GFP+ cells were then sorted and cultured. Total cell lysates (T) were collected and examined by western blot for expression of the constructs and β-catenin. Nuclear (N) and cytosolic (C) lysates were prepared and examined by western blot as indicated. Quantified data for p-β-catenin (Tyr654) expression are shown on the right. (D) WCLs were also used for IPs using a BCR/ABL antibody, and immunoprecipitates were examined by western blot using antibodies for β-catenin and BCR/ABL as indicated. (E) TOP- and FOPflash plasmids were transfected into the stably infected Ba/F3 cells. Twenty-four hours after transfection, cells were analyzed for luciferase activity. (F) WCLs from stably infected Ba/F3 cells were examined by western blot for expression of cyclin D1 and c-Myc. All quantitative data shown are averages ± standard deviations of at least 3 independent experiments. P values were calculated by using a paired Student t test or a one-way ANOVA followed by Tukey tests. *,#P < .05; **,##P < .01; ***,###P < .001. *Indicates significance relative to vector; #indicates significance relative to p210 BCR/ABL.
Deletion of the β-catenin binding site in p210 BCR/ABL diminishes the accumulation of p-β-catenin (Tyr654) and TCF/LEF-mediated transcription. (A) 293T cells were transiently transfected with p210 BCR/ABL (BCR/ABL), the ΔUBD mutant, or cognate vector. WCLs were collected and examined by western blot for expression of the constructs. Lysates were also used for IPs using a β-catenin antibody, and immunoprecipitates were examined by western blot using antibodies for β-catenin and p210 BCR/ABL. (B) 293T cells were transfected with p210 BCR/ABL in the presence or absence of the E1 inhibitor. WCLs were collected and examined by western blot for expression of p210 BCR/ABL (BCR/ABL), cyclin D2, or RhoA. Lysates were also used for IPs using a β-catenin antibody, and immunoprecipitates were examined by western blot using antibodies for β-catenin and BCR/ABL. (C) Ba/F3 cells were infected with retroviral particles that encode MSCV-bcr-abl/p210-IRES-gfp (BCR/ABL), MSCV-bcr-abl/p210(ΔUBD)-IRES-gfp (ΔUBD), or cognate vector. GFP+ cells were then sorted and cultured. Total cell lysates (T) were collected and examined by western blot for expression of the constructs and β-catenin. Nuclear (N) and cytosolic (C) lysates were prepared and examined by western blot as indicated. Quantified data for p-β-catenin (Tyr654) expression are shown on the right. (D) WCLs were also used for IPs using a BCR/ABL antibody, and immunoprecipitates were examined by western blot using antibodies for β-catenin and BCR/ABL as indicated. (E) TOP- and FOPflash plasmids were transfected into the stably infected Ba/F3 cells. Twenty-four hours after transfection, cells were analyzed for luciferase activity. (F) WCLs from stably infected Ba/F3 cells were examined by western blot for expression of cyclin D1 and c-Myc. All quantitative data shown are averages ± standard deviations of at least 3 independent experiments. P values were calculated by using a paired Student t test or a one-way ANOVA followed by Tukey tests. *,#P < .05; **,##P < .01; ***,###P < .001. *Indicates significance relative to vector; #indicates significance relative to p210 BCR/ABL.
The UBD supports phosphorylation of β-catenin on Tyr654
It has been shown previously that β-catenin is phosphorylated on tyrosine in cells that express p210 BCR/ABL.7 Thus, we determined whether phosphorylation is dependent upon an interaction between β-catenin and p210 BCR/ABL. For this analysis, we infected Ba/F3 cells with bicistronic retroviral vectors that encode p210 BCR/ABL or p210 BCR/ABL(ΔUBD) along with GFP. GFP+ cells were sorted and western blots were performed to confirm equal expression of the proteins (Figure 2C, upper panel). Elevated levels of total β-catenin were observed in lysates that expressed p210 BCR/ABL, but not the mutant. Lysates were separated into nuclear and cytoplasmic fractions and examined by western blot for expression of p-β-catenin (Tyr654). We observed elevated levels of p-β-catenin (Tyr654) in both the nuclear and cytoplasmic fractions in cells expressing p210 BCR/ABL, consistent with previous results. Levels are significantly lower in the nuclear and cytoplasmic fractions from cells that express the mutant. To confirm that the interaction between p210 BCR/ABL and β-catenin is detectable in these cells, immunoprecipitations were performed (Figure 2D). Although we were able to detect an interaction between both β-catenin and p-β-catenin (Tyr654) and p210 BCR/ABL, these interactions were significantly reduced with the mutant.
We then determined whether loss of the interaction between p210 BCR/ABL and β-catenin affects TCF/LEF signaling (Figure 2E). In the Ba/F3 cells, the luciferase activity of the TCF/LEF-responsive TOPflash plasmid was significantly increased in the presence of p210 BCR/ABL but to a much lesser extent in the presence of the mutant. It has been shown previously that two known transcriptional targets of β-catenin, cyclin D1 and c-myc, are upregulated by p210 BCR/ABL activity.7,14,20 To determine whether loss of the UBD has any effect on these known β-catenin transcriptional targets, we examined lysates for levels of expression of cyclin D1 and c-Myc. Whereas levels of both were elevated in p210 BCR/ABL–expressing cells, this was significantly reduced in cells that express the mutant (Figure 2F).
β-catenin binding contributes to disease progression in a BMT model for CML
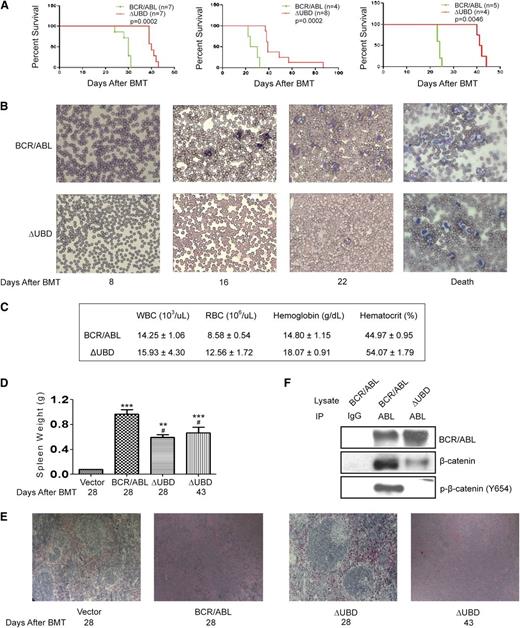
To determine whether the interaction with β-catenin supports disease progression, we compared p210 BCR/ABL and the binding mutant in the murine BMT model. Consistent with previous studies,21-23 all p210 BCR/ABL–transplanted mice (3 independent experiments; n = 16 total) became moribund within 23 to 31 days of transplantation displaying cachexia, a decrease in activity, and increased respiration (Figure 3A). An examination of peripheral blood smears revealed extensive leukocytosis beginning on day 16 post-BMT, with a predominant expansion of the myeloid population (Figure 3B). Complete blood counts performed at death confirmed increased numbers of WBCs, while red blood cell (RBC) and hematocrit levels were normal (Figure 3C). All mice had splenomegaly at death (Figure 3D), and histologic examination showed disruption of both the white and red pulp by infiltrating leukocytes and foci of extramedullary hematopoeisis (Figure 3E).
β-catenin binding contributes to disease progression in a BMT model for CML. (A) Survival of mice transplanted with p210 BCR/ABL or p210 BCR/ABL(ΔUBD). Kaplan-Meier curves were generated from 3 independent experiments as indicated. Mantel-Cox tests of the 3 survival curves yielded values of P = .0002 (χ2 = 13.5), P = .0002 (χ2 = 13.9), and P = .0046 (χ2 = 8.0), respectively. (B) Blood smears were performed weekly to monitor disease progression. (C) Results of complete blood counts performed at death. Data shown represent an average of 3 mice and are presented with standard deviations. (D) Spleens that were harvested at time of death were weighed. P values were calculated by using a one-way ANOVA followed by Tukey tests. (E) Spleens were harvested at the indicated times, and processed slides were stained with hematoxylin and eosin. Images were visualized by using a Nikon Eclipse 50i microscope (Nikon, Tokyo, Japan) equipped with a 10×/0.25 numerical aperture and were acquired by using a Go-5 camera (QImaging, Surrey, Canada) and QCapture Pro 6.0 software. (F) WCLs were prepared from spleens collected at time of death. Lysates were immunoprecipitated with a BCR/ABL antibody and examined by western blot as indicated. IgG indicates normal rabbit IgG-conjugated agarose; Abl indicates agarose-conjugated anti-abl antibody. **P < .01; ***P < .001 compared with vector; #P < .05 compared with p210 BCR/ABL.
β-catenin binding contributes to disease progression in a BMT model for CML. (A) Survival of mice transplanted with p210 BCR/ABL or p210 BCR/ABL(ΔUBD). Kaplan-Meier curves were generated from 3 independent experiments as indicated. Mantel-Cox tests of the 3 survival curves yielded values of P = .0002 (χ2 = 13.5), P = .0002 (χ2 = 13.9), and P = .0046 (χ2 = 8.0), respectively. (B) Blood smears were performed weekly to monitor disease progression. (C) Results of complete blood counts performed at death. Data shown represent an average of 3 mice and are presented with standard deviations. (D) Spleens that were harvested at time of death were weighed. P values were calculated by using a one-way ANOVA followed by Tukey tests. (E) Spleens were harvested at the indicated times, and processed slides were stained with hematoxylin and eosin. Images were visualized by using a Nikon Eclipse 50i microscope (Nikon, Tokyo, Japan) equipped with a 10×/0.25 numerical aperture and were acquired by using a Go-5 camera (QImaging, Surrey, Canada) and QCapture Pro 6.0 software. (F) WCLs were prepared from spleens collected at time of death. Lysates were immunoprecipitated with a BCR/ABL antibody and examined by western blot as indicated. IgG indicates normal rabbit IgG-conjugated agarose; Abl indicates agarose-conjugated anti-abl antibody. **P < .01; ***P < .001 compared with vector; #P < .05 compared with p210 BCR/ABL.
Mice transplanted with cells that express p210 BCR/ABL(ΔUBD) (3 independent experiments; n = 19 total) began to exhibit signs of overt illness much later than those transplanted with p210 BCR/ABL, and the range of ages at which the mice became moribund (40 to 88 days) was significantly longer (Figure 3A). The peripheral blood smears also revealed extensive leukocytosis at death, with a prominent expansion of the myeloid population (Figure 3B), but evidence of leukocytosis was not seen until after day 22. Although the WBC count at death was equivalent to that in the p210 BCR/ABL–transplanted mice, both RBC and hematocrit levels were significantly higher and fell outside the normal range (Figure 3C). On day 28 when p210 BCR/ABL mice were becoming moribund, mice transplanted with the mutant also exhibited splenomegaly (Figure 3D), but splenic architecture (Figure 3E) remained relatively intact. On day 43 when mutant transplanted mice were becoming moribund, the splenic architecture was completely disrupted, but the extent of the splenomegaly was still lower than that in the p210 BCR/ABL–transplanted mice. Analysis of genomic DNA by Southern blot revealed that the disease is polyclonal in both the mutant and p210 BCR/ABL–transplanted mice (supplemental Figure 1). One predominant clone and 2 to 4 minor clones were observed for each diseased animal. No difference in the number of clones was noted between the p210 BCR/ABL– and mutant-transfected mice, suggesting that the difference in disease progression cannot be attributed to a difference in engraftment efficiency.
Recipients that received marrow infected with the MIG vector (3 independent experiments; n = 20 total) had normal blood smears and remained disease free through 6 months post-BMT (not shown).
To confirm that the interaction between p210 BCR/ABL and p-β-catenin (Tyr654) is detectable in the diseased mice, lysates collected from spleens at death were examined by co-immunoprecipitation (Figure 3F). Whereas we were readily able to detect the interaction with both β-catenin and p-β-catenin (Tyr654) in p210 BCR/ABL–transplanted mice, the interactions were significantly disrupted in the mutant-transplanted mice.
Comparisons at equivalent points after transplantation reveal differences in disease progression
In order to directly compare disease progression, tail vein blood was collected on days 14, 21, 25, and 28 post-BMT and was subjected to immunophenotyping (Table 1). On day 14, an equivalent amount of GFP+ cells was evident in both the p210 BCR/ABL– and mutant-transplanted mice, suggesting that the difference in disease progression could not be attributed to a difference in engraftment. Cells were also examined for the expression of myeloid (CD11b), T-cell (CD3), and B-cell (B220) markers. At this early time point, both the p210 BCR/ABL– and mutant-transplanted mice were already starting to exhibit an equivalent amount of myeloproliferative disease, but there was no evidence of B- or T-cell expansion. Consistent with the blood smears, the percentage of CD11b+ cells rose steadily between day 14 and day 28 (point of death) in the peripheral blood of mice transplanted with p210 BCR/ABL. In mice transplanted with the mutant, however, the percentage of CD11b+ cells in the peripheral blood did not increase between day 14 and day 25, and further expansion was not observed until day 28.
Immunophenotyping of disease progression
| . | % of Total cells . | |||
|---|---|---|---|---|
| GFP+ . | GFP+/CD11b+ . | GFP+/CD3+ . | GFP+/B220+ . | |
| Day 14 | ||||
| Vector (n = 5) | 11.49 ± 1.36 | 5.99 ± 1.35 | 0.46 ± 0.04 | 1.24 ± 0.37 |
| BCR/ABL (n = 5) | 30.31 ± 9.17 | 28.15 ± 8.41 | 0.20 ± 0.07 | 1.08 ± 0.61 |
| ΔUBD (n = 5) | 26.07 ± 3.34 | 23.84 ± 1.20 | 0.26 ± 0.02 | 1.29 ± 1.03 |
| Day 21 | ||||
| Vector (n = 5) | 7.46 ± 0.92 | 3.17 ± 0.72 | 0.21 ± 0.17 | 1.06 ± 1.17 |
| BCR/ABL (n = 5) | 63.09 ± 12.00 | 38.62 ± 12.00 | 0.92 ± 1.45 | 5.17 ± 1.62 |
| ΔUBD (n = 5) | 44.43 ± 15.28 | 19.35 ± 2.09 | 0.79 ± 0.51 | 4.62 ± 3.65 |
| Day 25 | ||||
| Vector (n = 5) | 9.59 ± 5.93 | 4.14 ± 1.45 | 0.14 ± 0.08 | 1.68 ± 1.29 |
| BCR/ABL (n = 5) | 64.50 ± 9.25 | 55.34 ± 8.34 | 0.25 ± 0.09 | 4.42 ± 3.12 |
| ΔUBD (n = 5) | 39.46 ± 19.52 | 18.64 ± 8.46 | 0.59 ± 0.21 | 2.25 ± 1.43 |
| Day 28 | ||||
| Vector (n = 5) | 11.22 ± 3.95 | 5.37 ± 3.09 | 0.66 ± 0.35 | 2.76 ± 0.75 |
| BCR/ABL (n = 5) | 87.10 ± 2.65 | 76.04 ± 5.10 | 0.37 ± 0.17 | 4.26 ± 2.05 |
| ΔUBD (n = 5) | 80.16 ± 5.90 | 42.25 ± 17.11 | 0.64 ± 0.29 | 5.28 ± 1.99 |
| . | % of Total cells . | |||
|---|---|---|---|---|
| GFP+ . | GFP+/CD11b+ . | GFP+/CD3+ . | GFP+/B220+ . | |
| Day 14 | ||||
| Vector (n = 5) | 11.49 ± 1.36 | 5.99 ± 1.35 | 0.46 ± 0.04 | 1.24 ± 0.37 |
| BCR/ABL (n = 5) | 30.31 ± 9.17 | 28.15 ± 8.41 | 0.20 ± 0.07 | 1.08 ± 0.61 |
| ΔUBD (n = 5) | 26.07 ± 3.34 | 23.84 ± 1.20 | 0.26 ± 0.02 | 1.29 ± 1.03 |
| Day 21 | ||||
| Vector (n = 5) | 7.46 ± 0.92 | 3.17 ± 0.72 | 0.21 ± 0.17 | 1.06 ± 1.17 |
| BCR/ABL (n = 5) | 63.09 ± 12.00 | 38.62 ± 12.00 | 0.92 ± 1.45 | 5.17 ± 1.62 |
| ΔUBD (n = 5) | 44.43 ± 15.28 | 19.35 ± 2.09 | 0.79 ± 0.51 | 4.62 ± 3.65 |
| Day 25 | ||||
| Vector (n = 5) | 9.59 ± 5.93 | 4.14 ± 1.45 | 0.14 ± 0.08 | 1.68 ± 1.29 |
| BCR/ABL (n = 5) | 64.50 ± 9.25 | 55.34 ± 8.34 | 0.25 ± 0.09 | 4.42 ± 3.12 |
| ΔUBD (n = 5) | 39.46 ± 19.52 | 18.64 ± 8.46 | 0.59 ± 0.21 | 2.25 ± 1.43 |
| Day 28 | ||||
| Vector (n = 5) | 11.22 ± 3.95 | 5.37 ± 3.09 | 0.66 ± 0.35 | 2.76 ± 0.75 |
| BCR/ABL (n = 5) | 87.10 ± 2.65 | 76.04 ± 5.10 | 0.37 ± 0.17 | 4.26 ± 2.05 |
| ΔUBD (n = 5) | 80.16 ± 5.90 | 42.25 ± 17.11 | 0.64 ± 0.29 | 5.28 ± 1.99 |
Immunophenotyping was performed as described in Methods.
Comparison of disease phenotype at death
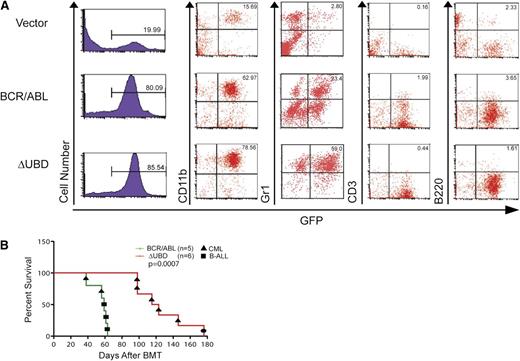
To further compare the disease phenotype, mice were randomly selected for immunophenotyping when they became moribund (Table 2 and Figure 4A). In order to determine whether there is a qualitative difference in myeloid expansion cells, they were also examined for the expression of a granulocyte marker (Gr1+). At death, all of the mice had predominantly myeloproliferative disease, although several had slightly elevated B-cell counts. Although the percentage of GFP+CD11b+ cells was equivalent, the percentage of cells that were GFP+Gr1+ was significantly higher in mice transplanted with the mutant than in mice transplanted with p210 BCR/ABL.
Immunophenotyping at death
| . | % of Total cells . | ||||
|---|---|---|---|---|---|
| GFP+ . | GFP+/CD11b+ . | GFP+/Gr1+ . | GFP+/CD3+ . | GFP+/B220+ . | |
| Peripheral blood | |||||
| Vector | 8.69 ± 6.51 | 4.78 ± 1.44 | 1.81 ± 0.07 | 0.046 ± 0.04 | 0.002 ± 0.005 |
| BCR/ABL | 82.12 ± 8.53 | 69.20 ± 16.69 | 16.78 ± 2.63 | 0.51 ± 0.22 | 6.72 ± 6.35 |
| ΔUBD | 78.78 ± 18.49 | 69.89 ± 16.48 | 39.58 ± 7.15 | 0.33 ± 0.20 | 3.75 ± 1.49 |
| Bone marrow | |||||
| Vector | 9.80 ± 9.09 | 7.41 ± 3.97 | 4.11 ± 1.96 | 0.64 ± 0.30 | 0.003 ± 0.002 |
| BCR/ABL | 69.19 ± 17.23 | 55.38 ± 16.17 | 16.3 ± 3.58 | 0.82 ± 0.60 | 3.61 ± 3.58 |
| ΔUBD | 69.91 ± 16.76 | 61.24 ± 17.15 | 28.88 ± 6.87 | 0.68 ± 0.36 | 2.75 ± 1.61 |
| Spleen | |||||
| Vector | 5.67 ± 3.74 | 4.74 ± 1.73 | 2.37 ± 0.88 | 1.85 ± 0.59 | 0.005 ± 0.003 |
| BCR/ABL | 65.12 ± 9.07 | 39.97 ± 17.11 | 15.95 ± 2.36 | 2.94 ± 1.44 | 4.61 ± 2.29 |
| ΔUBD | 72.09 ± 14.96 | 60.53 ± 17.14 | 34.5 ± 7.23 | 1.46 ± 0.74 | 3.16 ± 1.71 |
| . | % of Total cells . | ||||
|---|---|---|---|---|---|
| GFP+ . | GFP+/CD11b+ . | GFP+/Gr1+ . | GFP+/CD3+ . | GFP+/B220+ . | |
| Peripheral blood | |||||
| Vector | 8.69 ± 6.51 | 4.78 ± 1.44 | 1.81 ± 0.07 | 0.046 ± 0.04 | 0.002 ± 0.005 |
| BCR/ABL | 82.12 ± 8.53 | 69.20 ± 16.69 | 16.78 ± 2.63 | 0.51 ± 0.22 | 6.72 ± 6.35 |
| ΔUBD | 78.78 ± 18.49 | 69.89 ± 16.48 | 39.58 ± 7.15 | 0.33 ± 0.20 | 3.75 ± 1.49 |
| Bone marrow | |||||
| Vector | 9.80 ± 9.09 | 7.41 ± 3.97 | 4.11 ± 1.96 | 0.64 ± 0.30 | 0.003 ± 0.002 |
| BCR/ABL | 69.19 ± 17.23 | 55.38 ± 16.17 | 16.3 ± 3.58 | 0.82 ± 0.60 | 3.61 ± 3.58 |
| ΔUBD | 69.91 ± 16.76 | 61.24 ± 17.15 | 28.88 ± 6.87 | 0.68 ± 0.36 | 2.75 ± 1.61 |
| Spleen | |||||
| Vector | 5.67 ± 3.74 | 4.74 ± 1.73 | 2.37 ± 0.88 | 1.85 ± 0.59 | 0.005 ± 0.003 |
| BCR/ABL | 65.12 ± 9.07 | 39.97 ± 17.11 | 15.95 ± 2.36 | 2.94 ± 1.44 | 4.61 ± 2.29 |
| ΔUBD | 72.09 ± 14.96 | 60.53 ± 17.14 | 34.5 ± 7.23 | 1.46 ± 0.74 | 3.16 ± 1.71 |
Immunophenotyping was performed as described in Methods.
Comparison of immunophenotypes at death in mice transplanted with p210 BCR/ABL or p210 BCR/ABL(ΔUBD). (A) WBCs were collected from p210 BCR/ABL–, p210 BCR/ABL(ΔUBD)–, and vector-transplanted mice when they became moribund. Cells were examined by flow cytometry for GFP expression and then stained for CD11b, Gr1, B220, and CD3 as indicated. Data shown are for peripheral blood. (B) Survival curves for secondary transplantations. A Mantel-Cox test yielded a value of P = .0007 (χ2 = 11.5). The black circle indicates a mouse that died with splenomegaly and hepatomegaly. However, GFP+ cells were negative for major lineage markers, including CD11b, Gr1, B220, CD3, and Ter119. B-ALL, B-cell acute lymphoblastic leukemia.
Comparison of immunophenotypes at death in mice transplanted with p210 BCR/ABL or p210 BCR/ABL(ΔUBD). (A) WBCs were collected from p210 BCR/ABL–, p210 BCR/ABL(ΔUBD)–, and vector-transplanted mice when they became moribund. Cells were examined by flow cytometry for GFP expression and then stained for CD11b, Gr1, B220, and CD3 as indicated. Data shown are for peripheral blood. (B) Survival curves for secondary transplantations. A Mantel-Cox test yielded a value of P = .0007 (χ2 = 11.5). The black circle indicates a mouse that died with splenomegaly and hepatomegaly. However, GFP+ cells were negative for major lineage markers, including CD11b, Gr1, B220, CD3, and Ter119. B-ALL, B-cell acute lymphoblastic leukemia.
Because the myeloproliferative disease in the mutant-transplanted mice is phenotypically different than that in the p210 BCR/ABL–transplanted mice, secondary transplantations were performed to confirm that the mice had succumbed to a malignancy (Figure 3B). Of the 5 primary tumors examined from the p210 BCR/ABL–transplanted mice, 2 transmitted a myeloproliferative disease while the remaining 3 developed a B-cell acute lymphoblastic leukemia phenotype with slightly longer disease latency. In contrast, 5 of the 6 primary tumors examined from the mutant-transplanted mice transmitted a myeloproliferative disease that was indistinguishable from that in the primary recipients. The sixth mouse succumbed after 180 days with no evidence of leukemic expansion.
Loss of β-catenin binding alters progenitor expansion and TCF/LEF signaling
In order to determine whether the impairment in disease progression could be attributed to differences in progenitor expansion, GFP+ cells were examined from the bone marrow of p210 BCR/ABL– and p210 BCR/ABL(ΔUBD)–transplanted mice at death (Figure 5A-B). Functional assays on the isolated populations confirmed that the colony-forming activity of the progenitor populations was consistent with what had been previously validated for other mouse strains (supplemental Figure 2).15 Although the total number of progenitors was equivalent between the two populations, the number of CMPs was significantly lower in the mutant-transplanted mice, while the number of MEPs was significantly higher. The increase in MEPs was consistent with the elevated RBCs and hematocrit that we observed in the mutant-transplanted mice. Cell cycle analysis was performed on the progenitor populations (Figure 5C). Relative to the p210 BCR/ABL–transplanted mice, both the CMPs and GMPs from the mutant-transplanted mice showed significantly elevated levels of apoptosis (sub-G1 phase) and decreased levels of proliferation (S phase). A significant increase in S-phase MEPs was also observed in mutant-transplanted mice. Although loss of β-catenin binding did not alter the level of total β-catenin in the MEPs and CMPs (Figure 5D), TCF/LEF signaling was significantly lower in progenitor populations isolated from the mutant-transplanted mice (Figure 5E).
Loss of β-catenin binding limits TCF/LEF signaling and self-renewal of myeloid progenitors and sensitizes the cells to apoptosis. Bone marrow cells isolated from diseased mice at death were used for immunophenotypic and cell cycle analysis of progenitor populations. (A) Representative fluorescence-activated cell sorter staining profiles of progenitor populations. (B) Percentages of each progenitor populations (GMP, CMP, and MEP) relative to total GFP+ cells. Values were derived from at least 4 mice per group and are represented as averages. Data show significant decreases of CMP in BCR/ABL(ΔUBD) mice relative to BCR/ABL mice (P < .01), whereas a significant increase in MEPs was found in BCR/ABL (ΔUBD) mice (P < .05). (C) Cell cycle analysis of CMPs, GMPs, and MEPs in p210 BCR/ABL mice and p210 BCR/ABL(ΔUBD) mice. Values were derived from at least 4 mice per group and are represented as averages. A significant increase of sub G1 cells (P < .001) and decrease of S phase cells (P < .001) were found in the CMPs of BCR/ABL (ΔUBD) mice relative to BCR/ABL mice. A significant increase of sub G1 cells (P < .001) and decrease of S phase cells (P < .01) were found in the GMPs of BCR/ABL (ΔUBD) mice relative to BCR/ABL mice. A significant increase in S phase cells (P < .05) was found in the MEPs of BCR/ABL (ΔUBD) mice relative to BCR/ABL mice. (D) Analysis of β-catenin expression in CMPs and MEPs isolated from p210 BCR/ABL and p210 BCR/ABL(ΔUBD) mice. Data are representative of 3 mice evaluated at death. (E) Comparison of TCF/LEF reporter activity in CMPs and MEPs isolated from diseased mice. Progenitors were isolated at death, cultured, and infected with lentiviral particles containing a TCF/LEF reporter along with Renilla control. Seventy-two hours after transfection, cells were analyzed for luciferase and Renilla activity. Luciferase levels were normalized to the Renilla control, and then p210 BCR/ABL was expressed as fold activation relative to p210 BCR/ABL(ΔUBD). Data shown represent the average of 3 mice at death. **P < .01; ***P < .001. FSC: foward scatter; SSC: side scatter.
Loss of β-catenin binding limits TCF/LEF signaling and self-renewal of myeloid progenitors and sensitizes the cells to apoptosis. Bone marrow cells isolated from diseased mice at death were used for immunophenotypic and cell cycle analysis of progenitor populations. (A) Representative fluorescence-activated cell sorter staining profiles of progenitor populations. (B) Percentages of each progenitor populations (GMP, CMP, and MEP) relative to total GFP+ cells. Values were derived from at least 4 mice per group and are represented as averages. Data show significant decreases of CMP in BCR/ABL(ΔUBD) mice relative to BCR/ABL mice (P < .01), whereas a significant increase in MEPs was found in BCR/ABL (ΔUBD) mice (P < .05). (C) Cell cycle analysis of CMPs, GMPs, and MEPs in p210 BCR/ABL mice and p210 BCR/ABL(ΔUBD) mice. Values were derived from at least 4 mice per group and are represented as averages. A significant increase of sub G1 cells (P < .001) and decrease of S phase cells (P < .001) were found in the CMPs of BCR/ABL (ΔUBD) mice relative to BCR/ABL mice. A significant increase of sub G1 cells (P < .001) and decrease of S phase cells (P < .01) were found in the GMPs of BCR/ABL (ΔUBD) mice relative to BCR/ABL mice. A significant increase in S phase cells (P < .05) was found in the MEPs of BCR/ABL (ΔUBD) mice relative to BCR/ABL mice. (D) Analysis of β-catenin expression in CMPs and MEPs isolated from p210 BCR/ABL and p210 BCR/ABL(ΔUBD) mice. Data are representative of 3 mice evaluated at death. (E) Comparison of TCF/LEF reporter activity in CMPs and MEPs isolated from diseased mice. Progenitors were isolated at death, cultured, and infected with lentiviral particles containing a TCF/LEF reporter along with Renilla control. Seventy-two hours after transfection, cells were analyzed for luciferase and Renilla activity. Luciferase levels were normalized to the Renilla control, and then p210 BCR/ABL was expressed as fold activation relative to p210 BCR/ABL(ΔUBD). Data shown represent the average of 3 mice at death. **P < .01; ***P < .001. FSC: foward scatter; SSC: side scatter.
Discussion
Several independent studies have established that sequences that reside within the first exon of BCR, and which are retained in p190, p210, and p230 BCR/ABL, are required to support transforming activity.24-26 Loss of these sequences is generally associated with diminished tyrosine kinase activity, suggesting that one function of the BCR-encoded sequences is to directly regulate the catalytic activity of Abl. We have confirmed that the binding site for β-catenin lies within the first exon of BCR in a region that is conserved in all BCR/ABL variants. Deletion of the binding site has no effect on the auto- or trans-kinase activity of p210 BCR/ABL nor does it interfere with oligomerization or Grb2-mediated activation of ERK1/2, both of which are known to support p210 BCR/ABL transformation.27 This suggests that BCR-encoded activities can facilitate disease progression through mechanisms other than supporting tyrosine kinase activity and identify the β-catenin interaction as an important example of such an activity.
In this study, we have shown that a p210 BCR/ABL mutant that is impaired in its ability to interact with β-catenin shows an altered disease phenotype and increased disease latency in the BMT model. Whereas high numbers of CMPs are detected in the bone marrow of mice transplanted with p210 BCR/ABL, this expansion is more limited in mice transplanted with the mutant. Cell cycle analysis confirmed that the CMPs in the mice transplanted with the mutant had reduced proliferation and enhanced sensitivity to apoptosis. This suggests that in the BMT model, expansion of CMPs by p210 BCR/ABL is dependent upon a direct interaction with β-catenin. Analysis of lineage-specific markers reveals that myeloid expansion, which is driven by the CMPs, is qualitatively different in the mutant-transplanted mice. Thus, the leukemic expansion consists primarily of neutrophils, which likely accounts for the increased lifespan. Not only are the CMPs reduced in number, their lineage commitment is also altered. The progenitor analysis also indicated that the interaction with β-catenin may limit the expansion of MEPs. Elevated numbers of MEPs were found in the marrow of mutant-transplanted mice, which likely accounts for the polycythemia we observed in these mice. Although no significant difference was observed in total GMP, the cell cycle analysis indicated reduced proliferative potential and increased sensitivity to apoptosis in the mutant-transplanted mice. This differential effect on multiple progenitor populations suggests that the interaction between p210 BCR/ABL and β-catenin may also be influencing the lineage commitment of the leukemic stem cell during the chronic phase of disease progression.
Previous studies have suggested that p210 BCR/ABL can directly phosphorylate β-catenin on tyrosine and that this modification sequesters β-catenin from Axin and prevents its ubiquitin-mediated degradation.7 Although we have observed expression of p-β-catenin (Tyr654) in diseased mice expressing p210 BCR/ABL, but not the mutant, we did not observe any measurable difference in total β-catenin levels. This suggests that the phosphorylation of β-catenin on tyrosine alone may be sufficient to alter transcriptional activity and the disease outcome. Consistent with this, we have observed that loss of the interaction resulted in reduced TCF/LEF signaling in MEPs and CMPs isolated from diseased mice. In patient samples, elevated β-catenin expression has been observed in blast crisis but not in the chronic phase.1 Since the BMT model is thought to more closely resemble the chronic phase, our observations suggest that the interaction between p210 BCR/ABL and β-catenin may support disease progression at an earlier stage than previously appreciated and may represent a viable target for clinical intervention.
Although it has been previously reported that β-catenin interacts with p210 BCR/ABL,7 our study suggests that this interaction may be partially dependent upon marking of the protein with ubiquitin. One possibility is that p210 BCR/ABL may also interact with monoubiquitinated β-catenin after it is recruited to the Axin/GSK3β complex and stabilize it against degradation. However, since pools of ubiquitinated β-catenin that are both stable and transcriptionally active have been detected in the nucleus of tumor cells, the interaction that we have detected may be unrelated to the Axin/GSK3β complex.28 Since ubiquitin is highly conserved and since UBDs do not have a high level of intrinsic binding specificity, it seems likely that p210 BCR/ABL may be binding to both the ubiquitin tag and directly to β-catenin itself. This is thought to be the case for many UBD-containing proteins, which need to recognize their targets over the background noise of a large cellular roster of ubiquitinated proteins.29
Acquired resistance to tyrosine kinase inhibitors and minimal residual disease have emerged as growing challenges in the treatment of patients with CML. This has fueled the search for additional p210 BCR/ABL–encoded activities that support disease progression, particularly in the leukemic stem cell compartment. The recent identification of a GMP population with aberrant, β-catenin-mediated, self-renewal capacity in patients with breast cancer suggests that the interaction between p210 BCR/ABL and β-catenin may be a clinically significant target.1 Our observation that the disruption of the interaction between p210 BCR/ABL and β-catenin in the BMT model results in a greater disease latency with a qualitatively different expansion of the myeloid lineage supports this possibility.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Public Health Service grant CA097066 (I.P.W.) from the National Cancer Institute. N.L.P. is the recipient of a fellowship from the New Jersey Commission for Cancer Research.
Authorship
Contribution: R.C., T.H., and G.M.M. designed and performed all experiments; N.L.P. and I.T. assisted in the bone marrow transplantation assays, bone marrow isolations, and the flow cytometry; and I.P.W. and H.L.O designed experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ian P. Whitehead, Cancer Center H Level, New Jersey Medical School, 205 South Orange Ave, Newark, NJ 07101; e-mail: whiteip@umdnj.edu.
References
Author notes
R.C. and T.H. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal