Key Points
The PAFc subunit, Cdc73, is required for the proliferation and proper epigenetic regulation of proleukemic oncogenes in AML cells.
Disrupting the MLL-PAFc interaction selectively inhibits the growth of MLL-associated leukemic cells without altering normal hematopoietic stem cell function.
Abstract
MLL rearrangements are common in leukemia and considered an adverse risk factor. Through interactions with the polymerase-associated factor complex (PAFc), mixed lineage leukemia (MLL) fusion proteins activate genes critical for blocking differentiation, such as HOXA9. Here we investigate whether the MLL-PAFc interaction can be exploited therapeutically using both genetic and biochemical approaches. We tested the genetic requirement of the PAFc in acute myeloid leukemia (AML) using a conditional allele of the PAFc subunit, Cdc73. We show that the PAFc is indiscriminately necessary for the proliferation of AML cells through the epigenetic regulation of proleukemogenic target genes, such as MEIS1 and Bcl2. To investigate the therapeutic potential of targeting the MLL-PAFc interaction, we engineered a dominant negative fragment of MLL capable of binding to the PAFc. Disruption of the MLL-PAFc interaction selectively inhibits the proliferation of MLL leukemic cells without affecting cells transformed by an unrelated E2A-HLF fusion protein. Using in vivo hematopoietic reconstitution assays, we demonstrate that disruption of the MLL-PAFc does not alter normal hematopoietic stem cell function. Together, our data show a selective growth inhibition of MLL-associated leukemic cells and tolerance of normal hematopoiesis to disruption of the MLL-PAFc interaction establishing the MLL-PAFc interaction as an attractive therapeutic target.
Introduction
Human leukemias harboring mixed lineage leukemia (MLL) translocations constitute greater than 70% of infant acute lymphoid leukemia (ALL), between 35% and 50% of infant acute myeloid leukemias (AML) and about 10% of adult AML. MLL-associated leukemias are particularly aggressive and usually associated with an adverse prognosis.1 As such, the event-free survival rates of ALL patients with MLL translocations is markedly reduced compared with ALL patients with germline MLL, demonstrating the necessity for better treatment options for patients with MLL rearrangements.2-4
Recent work has focused on identifying the molecular mechanisms driving MLL fusion protein–mediated transcriptional activation that has revealed new potential therapeutic targets. Investigation of the role of the fusion partner in MLL-rearranged leukemia has revealed that some of the most common MLL translocation partner proteins (AF4, AF9, ENL, and ELL) recruit a transcriptional activation complex that includes p-TEFb (cyclin T1 and CDK9) and promotes transcriptional elongation through polymerase II (Pol II) C-terminal repeat domain phosphorylation at serine 2.5-8 These and other MLL fusion proteins also interact with the histone H3K79 methyltransferase DOT1L,9,10 which together with positive transcription elongation factor b confer deregulated transcriptional activation of MLL target genes.
Notably, H3K4 and H3K79 methyltransferase activity associated with MLL and DOT1L, respectively, are regulated by the polymerase-associated factor complex (PAFc). Both H3K4 and H3K79 methylation are associated with actively transcribed chromatin and require the presence of monoubiquitinated histone H2B (H2Bub) as a prerequisite for deposition.11-13 H2Bub is dependent on the PAFc, which directly recruits the RAD6/BRE1/2 E3 ubiquitin ligase complex necessary for H2Bub.14,15 Consistent with this epigenetic hierarchy, loss of PAFc in yeast and human cells results in not only defective H2Bub, but also in H3K4 and H3K79 methylation.12,14,16,17 This is explained, in part, by recent findings that MLL is recruited through direct interactions with PAFc to target loci.18,19
The PAFc transcriptional activation complex physically interacts with the C-terminal domain of RNA Pol II and is integral to the transcriptional elongation of a subset of genes including HOX genes.17,19-21 This transcriptional machinery is directly engaged by MLL, which delivers H3K4 methyltransferase activity to target genes (or H3K79 methyltransferase activity in the case of some MLL fusion proteins). The MLL-PAFc interaction is mediated by sequences flanking the CxxC domain of MLL that are invariably retained in MLL fusion proteins; as such, MLL fusion proteins also interact with PAFc.19 MLL fusion protein–mediated transcriptional activity synergizes with the PAFc-Pol II initiation complex leading to augmented transcriptional activation and ultimately leukemia. Deletion of MLL sequence necessary for interaction with the PAFc destroys MLL fusion-mediated leukemogenesis, implying this interaction may be a worthy drug target. Here we explore the therapeutic value of disrupting the MLL-PAFc interaction using both genetic and biochemical techniques in AML cells using differing mechanisms of transformation. Although AML cells are dependent on the PAFc for full proliferation, targeted disruption of the MLL-PAFc interaction selectively inhibits the growth of MLL fusion protein–transformed cells. Importantly, normal hematopoiesis is not affected by disruption of the MLL-PAFc interaction, suggesting that this interface is a promising therapeutic target.
Materials and methods
Mice
Hrpt2 floxed mice have been described previously.22 Six- to 8-week-old female C57Bl/6 mice were purchased from Tacomic Farms (Hudson, NY). All animal studies were approved by the University of Michigan Committee on Use and Care of Animals and Unit for Laboratory Medicine. Additional Methods can be found in the supplemental data on the Blood Web site.
Cell lines
Hrpt2 (Cdc73) floxed cell lines were generated by isolating bone marrow cells from female Hrpt2 floxed mouse femurs and tibias 5 days after intraperitoneal injection of 5-fluorouracil (Sigma) at 150 mg/kg. Lin−c-kit+ cells were isolated using the EasySep Mouse hematopoietic progenitor cell enrichment kit (Stem Cell Technologies) following manufacturer’s instructions and grown overnight in prestimulation media (Iscove modified Dulbecco medium [Gibco], 15% fetal bovine serum [StemCell Technologies], Pen/Strep [100 U/mL; Gibco], interleukin-3 [IL-3; 10 ng/mL], IL-6 [10 ng/mL], and stem cell factor (SCF) [100 ng/mL] (R&D Systems)]. Cells were transduced on consecutive days with MSCV-neo-F-MLL-AF9 (described previously19 ) and murine stem cell virus (MSCV)-neo-F-E2A-HLF (described previously6 ) packaged retrovirus in the presence of polybrene (4 μ/mL) by spinoculation for 90 minutes at 3200 rpm. Retroviral packaging was achieved by transient transfection of Plat-E cells with appropriate retroviral vectors using Fugene 6 (Promega). Established cell lines were weaned from SCF and secondarily transduced with MSCV-puro-CreER retrovirus on consecutive days. Cells recovered in prestimulation media (without SCF and IL-6) for 2 days before selection in puromycin (2 μg/mL; Sigma) for 2 weeks. Resultant cell lines (MA-Cdc73fl-CreER, EH-Cdc73-CreER, MA-CreER, and EH-CreER) were maintained in growth media (Iscove modified Dulbecco medium containing Pen/Strep, 15% fetal bovine serum, and IL-3 [10 ng/mL]).
Plasmids
MSCV-neo-F-MLL-AF9 has been described.19 MSCV-neo-E2A-HLF was kindly provided by Dr. Michael Cleary (Stanford University). The estrogen-binding domain was fused to CRE to generate MSCV-puro-CreER. A small fragment of MLL containing the PAF1 interaction surface fused to a Myc tag and 2 nuclear localization signals (MLLDN) construct was generated by inserting a stop codon in the MSCV-puro-Myc-NLS-NLS-CxxC-RD2 construct after amino acid 1173 of MLL using the QuikChange XL site-directed mutagenesis kit (Stratagene). MSCV-puro-Myc-NLS-NLS-CxxC-RD2 was generated by polymerase chain reaction (PCR)-based amplification of XhoI-Myc-NLS-NLS-CxxC-RD2-HpaI from the template vector CMV-Myc-NLS-NLS-CxxC-RD2 (described previously19 ). This product was ligated into MSCV-puro and MigR1 using the XhoI and HpaI restriction sites.
Genotyping
Cdc73 floxed mice and cell lines were genotyped using the following PCR conditions: denaturing, 94°C for 30 seconds; annealing, 55°C for 30 second; and elongation, 68°C for 1 minute, 30 seconds for 31 cycles. Taq polymerase (Invitrogen) was used with 2 mM MgCl2, 1 mM dNTP, and 500 nM primers following manufacturer’s instructions. Primers include Hrpt2 allele F: TCCTTTCCATTGTGCAGCTGGTTG, Hrpt2 allele R: TGCCAGTGCAAGAACCTCATCCTA, and Hrpt2 flox: ATTCCAACTGGCTTCCAAGCAG.
IP and western blotting
A total of 293 cells were seeded at 1 × 106 cells on 10-cm tissue culture plates 1 day before transfection. Cells were transiently transfected with expression plasmids using Fugene 6 (Promega). Cells were lysed in BC-300 buffer (20 mM tris[hydroxymethyl]aminomethane-HCl [pH 7.4], 10% glycerol, 300 mM KCl, 0.1% NP-40) and immunoprecipitations were performed overnight with anti-Myc agarose resin (Clontech) or anti-HA affinity matrix (Roche). Immunoprecipitin (Ips) were washed 4 times with BC-300 buffer and proteins were eluted by boiling in sodium dodecyl sulfate–loading buffer. Proteins were visualized by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blotting with anti-Myc (Abcam ab9132) anti-HA (Abcam ab9110), anti-PAF1 (Bethyl A300-172A), and anti-CDC73 (Bethyl A300-170A).
Results
Proliferation of AML cells is dependent on Cdc73
We previously established a direct interaction between the PAFc and MLL fusion proteins. To investigate the therapeutic value of disrupting the PAFc in AML, we established leukemic cell lines from bone marrow–derived from conditional Hrpt2 knockout mice.22 These mice harbor floxed Hrpt2 alleles coding for the Parafibromin protein, the mammalian homolog of the Cdc73 protein found in yeast. We generated leukemic cell lines through retroviral transduction of lin−ckit+Hrpt2 floxed bone marrow cells with the MLL-AF9 and E2A-HLF fusion oncogenes and 4-hydroxy tamoxifen (4-OHT)-inducible CreER, referred to hereafter as MA-Cdc73fl-CreER and EH-Cdc73fl-CreER, respectively. These oncogenes were chosen because of their differing mechanisms of transformation; MLL fusions transform through mechanisms dependent on HOX transcription factors, whereas E2A-HLF transforms through inhibition of apoptosis.23 Treatment with 4-OHT for 48 hours leads to near complete excision of the Cdc73fl allele and significant loss of protein expression (Figure 1A; supplemental Figure 1). To examine the importance of Cdc73 in leukemic cell proliferation, the MA-Cdc73fl-CreER, EH-Cdc73fl-CreER, and wild-type cell lines expressing CreER and transformed with MLL-AF9 and E2A-HLF were grown in IL-3–conditioned media in the presence or absence of 4-OHT. Loss of Cdc73 results in decreased proliferation of both MLL-AF9 and E2A-HLF cells (Figure 1B-C). Notably, a more pronounced proliferative defect was observed in MLL-AF9 cells compared with E2A-HLF cells that may reflect increased dependency on the PAFc (Figure 1B-C). Control leukemic cells expressing CreER showed no response to 4-OHT, indicating this phenotype is a direct result of loss of Cdc73 irrespective of oncogene (Figure 1B-C). Additional studies were performed with leukemic cell lines containing a floxed Cdc73 locus but lacking CreER expression whose proliferation was not altered by 4-OHT treatment, confirming the proliferation defect is due to loss of Cdc73 (supplemental Figure 2). CDC73 makes direct contact with the CTR9, RTF1, and LEO1 subunits of the PAFc,24 and mutation or loss of CDC73 results in decreased expression and association with other subunits of PAFc and reduced binding of PAFc to RNAPII.19,20,25 Thus, Cdc73, and likely the entire PAFc, is required for the proliferation of AML cells independent of transformation mechanism but may be particularly important in MLL-associated leukemic cells.
Loss of Cdc73 leads to decreased proliferation of leukemia cells. (A) Western blotting was performed on whole cell lysate for the MA-Cdc73fl-CreER and EH-Cdc73fl-CreER cell lines treated with EtOH or 5 nM 4-OHT for 48 hours. Cdc73 protein is reduced after 48 hours of 4-OHT treatment. β-actin serves as a loading control. (B-C) MA-Cdc73fl-CreER, EH-Cdc73fl-CreER, and control cell lines (MA-CreER, EH-CreER) were grown in liquid culture in the presence of either EtOH or 5 nM 4-OHT and counted daily for 5 days. The total cell number for each day is plotted for each condition (mean ± standard deviation [SD]; n = 2 independent experiments; NS = not significant (P > .05); 2-way analysis of variance [ANOVA]). (D-E) qRT-PCR performed on either the MA-Cdc73fl-CreER and MA-CreER or EH-Cdc73fl-CreER and EH-CreER cell lines 48 hours after EtOH or 4-OHT treatment. Expression levels for Hoxa9, Meis1, and Bcl2 are shown relative to each cell line treated with EtOH (mean ± SD; n = 2 independent experiments. ****P < .001; **P < .005; *P < .05; NS, not significant (P > .05); unpaired 2-tailed Student t test).
Loss of Cdc73 leads to decreased proliferation of leukemia cells. (A) Western blotting was performed on whole cell lysate for the MA-Cdc73fl-CreER and EH-Cdc73fl-CreER cell lines treated with EtOH or 5 nM 4-OHT for 48 hours. Cdc73 protein is reduced after 48 hours of 4-OHT treatment. β-actin serves as a loading control. (B-C) MA-Cdc73fl-CreER, EH-Cdc73fl-CreER, and control cell lines (MA-CreER, EH-CreER) were grown in liquid culture in the presence of either EtOH or 5 nM 4-OHT and counted daily for 5 days. The total cell number for each day is plotted for each condition (mean ± standard deviation [SD]; n = 2 independent experiments; NS = not significant (P > .05); 2-way analysis of variance [ANOVA]). (D-E) qRT-PCR performed on either the MA-Cdc73fl-CreER and MA-CreER or EH-Cdc73fl-CreER and EH-CreER cell lines 48 hours after EtOH or 4-OHT treatment. Expression levels for Hoxa9, Meis1, and Bcl2 are shown relative to each cell line treated with EtOH (mean ± SD; n = 2 independent experiments. ****P < .001; **P < .005; *P < .05; NS, not significant (P > .05); unpaired 2-tailed Student t test).
PAFc synergizes with MLL fusion proteins to activate transcription.18,19 Consistent with this, we observed a greater than twofold reduction in the expression of 2 critical downstream target genes of MLL fusion proteins, Hoxa9 and Meis1, and the E2A-HLF target, Bcl2,26 following Cdc73 excision (Figure 1D-E). The expression of these gene targets was only slightly changed in both wild-type CreER and 4-OHT control cell lines, confirming the importance of the PAFc in MLL fusion transcriptional activation (Figure 1D-E; supplemental Figure 3).
MLL fusion proteins require PAFc for proper gene targeting
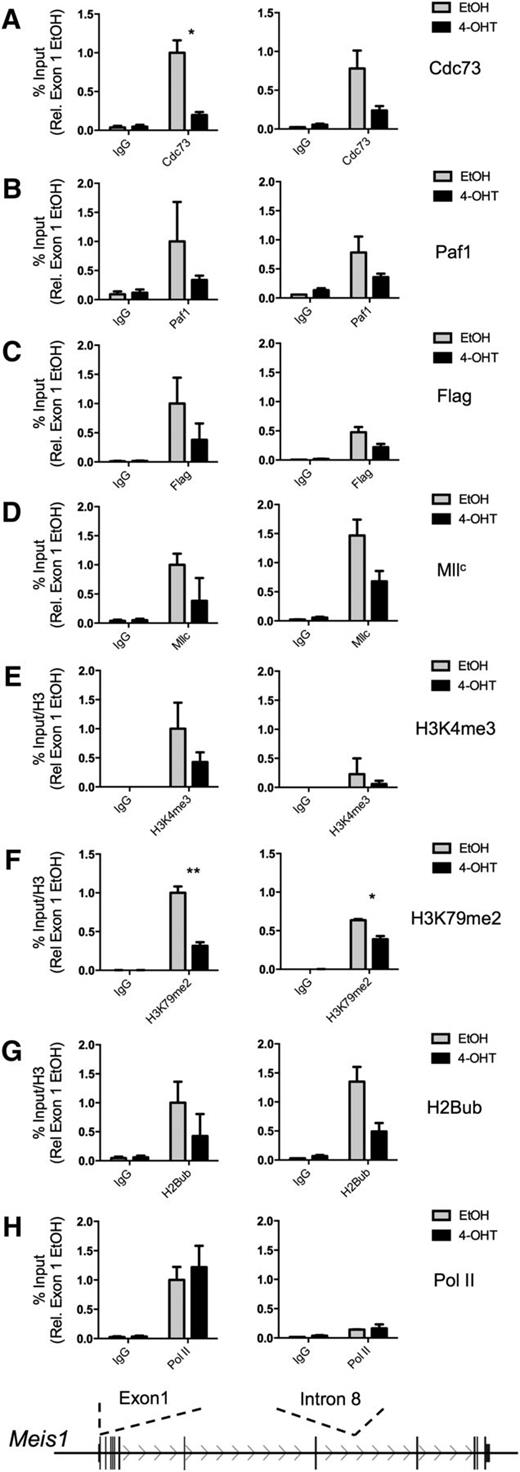
The loss of Cdc73 protein following 4-OHT treatment allowed the use of the MA-Cdc73fl-CreER cell line to study the PAFc and MLL fusion protein association with transcriptional targets with and without Cdc73. Using increased 4-OHT concentrations to achieve complete loss of the Cdc73 protein (supplemental Figure 4), MA-Cdc73fl-CreER cells were treated with EtOH or 4-OHT for 48 hours before fixation for chromatin IP (ChIP) assays on the Meis1 locus, an essential downstream target of MLL fusion proteins.27 Meis1 is improperly transcribed in MLL-AF9–transformed cells following loss of Cdc73 (Figure 1D). Excision of Cdc73 results in a marked reduction of Cdc73 protein associated with the Meis1 locus (Figure 2A; supplemental Figure 5). Consistent with Cdc73 playing an essential role in the structural integrity of the entire PAFc,27 4-OHT treatment also led to decreased Paf1 binding, suggesting loss of Cdc73 results in the improper localization of the entire PAFc to target loci (Figure 2B). Next, we examined the localization of Mll and the MLL-AF9 fusion proteins. We found reduced wild-type Mll binding to the Meis1 locus following loss of Cdc73, consistent with our earlier findings (Figure 2D).19 Similarly, MLL-AF9, detected with flag antibody, showed reduced chromatin occupancy in the absence of PAFc (Figure 2C). We observed decreased H2Bub across the coding region of the Meis1 locus as well as reduced H3K4me3 and H3K79me2 at the transcriptional start site following loss of Cdc73 (Figure 2E-G), consistent with PAFc playing an essential role in the establishment of these epigenetic marks through recruitment of Mll H3K4 and Dot1 H3K79 methyltransferase activity to the Meis1 locus. Only minor changes were detected for H3K4me1 and H3K4me2 following loss of Cdc73, suggesting these do not contribute to changes in gene expression of Meis1 (supplemental Figure 5). We observed little change in RNA Pol II at the transcriptional start site, consistent with the recruitment of PAFc following that of Pol II (Figure 2H). Together, these data demonstrate the importance of PAFc in the localization of not only wild-type Mll but also MLL fusion proteins to target loci and suggest transcriptional defects associated with inhibition of PAFc are due to improper recruitment to target loci.
Cdc73 is necessary for proper recruitment of PAFc and MLL fusion proteins to target loci. ChIP assays were performed on the MA-Cdc73fl-CreER cell line treated with either EtOH (solid black lines) or 100 nM 4-OHT (dotted black lines) for 48 hours. The Meis1 locus (transcribed from left to right) is shown schematically with the sites of primer probe sets recognizing exon 1 and intron 8 of Meis1. IPs performed in EtOH-treated cells are shown in gray, and IPs performed in 4-OHT cells are shown in black. ChIP experiments were performed for (A) Cdc73, (B) Paf1, (C) Flag (MLL-AF9 fusion protein), (D) MllC, (E) H3K4me3, (F) H3K79me2, (G) H2Bub, and (H) RNA Pol II. All binding was calculated as a percentage of 1% input chromatin—except in E, F, and G—that are then divided by total histone H3 and presented in relation to binding at exon 1 in EtOH-treated cells (mean ± SD; n = 2 independent experiments performed in duplicate; *P < .05, **P < .01). Additional ChIP data are presented in supplemental Figure 5. Ig, immunoglobulin.
Cdc73 is necessary for proper recruitment of PAFc and MLL fusion proteins to target loci. ChIP assays were performed on the MA-Cdc73fl-CreER cell line treated with either EtOH (solid black lines) or 100 nM 4-OHT (dotted black lines) for 48 hours. The Meis1 locus (transcribed from left to right) is shown schematically with the sites of primer probe sets recognizing exon 1 and intron 8 of Meis1. IPs performed in EtOH-treated cells are shown in gray, and IPs performed in 4-OHT cells are shown in black. ChIP experiments were performed for (A) Cdc73, (B) Paf1, (C) Flag (MLL-AF9 fusion protein), (D) MllC, (E) H3K4me3, (F) H3K79me2, (G) H2Bub, and (H) RNA Pol II. All binding was calculated as a percentage of 1% input chromatin—except in E, F, and G—that are then divided by total histone H3 and presented in relation to binding at exon 1 in EtOH-treated cells (mean ± SD; n = 2 independent experiments performed in duplicate; *P < .05, **P < .01). Additional ChIP data are presented in supplemental Figure 5. Ig, immunoglobulin.
Targeted disruption of MLL-PAFc selectively inhibits the growth of MLL leukemia cells
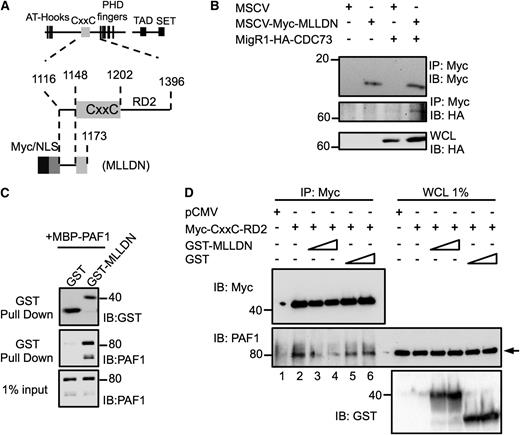
Our previous work identified a direct interaction between the PAF1 subunit of the PAFc with the pre-CxxC region of MLL.19 We asked whether expression of a small fragment of MLL containing the pre-CxxC region of MLL could function as a dominant negative to inhibit MLL association with the PAFc. To this end, we engineered an expression plasmid that includes MLLDN (Figure 3A). As expected, transient cotransfection of hemagglutinin (HA)-tagged CDC73 revealed MLLDN can efficiently coimmunoprecipitate the PAFc, confirming an interaction between the MLLDN and the PAFc in cells (Figure 3B). To confirm that MLLDN functions as a dominant negative, we bacterially expressed and purified GST-MLLDN that binds directly to PAF1 (Figure 3C). We transiently transfected 293 cells with F/HA-CxxC-RD2, which is known to bind efficiently to the PAFc (Figure 3D).19 Although F/HA-CxxC-RD2 successfully coimmunoprecipitated endogenous PAF1 (lane 2), addition of purified MLLDN, but not the glutathione S-transferase (GST) tag alone, led to reduced binding of PAF1 to the F/HA-CxxC-RD2 in a dose-dependent manner, confirming a dominant negative function (Figure 3D).
A fragment of MLL functions as a dominant negative to disrupt the MLL-PAFc interaction. (A) Schematic diagram of the MLL protein and functional domains with magnified view of the CxxC domain and the RD2 region with illustration of the engineered MLLDN fragment. Amino acid numbers for regions of MLL are indicated. (B) Expression of MLLDN was confirmed to coimmunoprecipitate CDC73 by transient transfection of 293 cells. (C) Purified GST-tagged MLLDN bound directly and specifically to MBP-tagged full-length PAF1 in GST pull-down experiments. (D) Coimmunoprecipitation assays performed in 293 cells demonstrate the MLL fragment Myc-CxxC-RD2 associates with endogenous PAF1 (lane 2). Increasing doses of MLLDN (1.5 and 3.0 μM, lanes 3 and 4) disrupt the interaction between the CxxC-RD2 and PAF1, whereas increasing doses of the GST tag (1.5 and 3.0 μM, lanes 5 and 6) do not. Arrow indicates PAF1 protein. One of 2 representative experiments is shown.
A fragment of MLL functions as a dominant negative to disrupt the MLL-PAFc interaction. (A) Schematic diagram of the MLL protein and functional domains with magnified view of the CxxC domain and the RD2 region with illustration of the engineered MLLDN fragment. Amino acid numbers for regions of MLL are indicated. (B) Expression of MLLDN was confirmed to coimmunoprecipitate CDC73 by transient transfection of 293 cells. (C) Purified GST-tagged MLLDN bound directly and specifically to MBP-tagged full-length PAF1 in GST pull-down experiments. (D) Coimmunoprecipitation assays performed in 293 cells demonstrate the MLL fragment Myc-CxxC-RD2 associates with endogenous PAF1 (lane 2). Increasing doses of MLLDN (1.5 and 3.0 μM, lanes 3 and 4) disrupt the interaction between the CxxC-RD2 and PAF1, whereas increasing doses of the GST tag (1.5 and 3.0 μM, lanes 5 and 6) do not. Arrow indicates PAF1 protein. One of 2 representative experiments is shown.
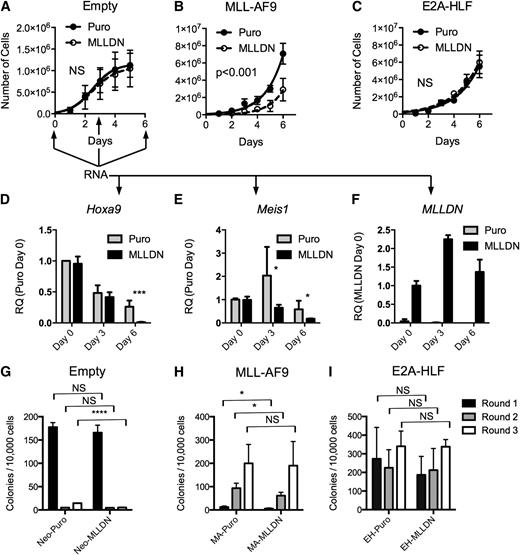
To assess the function of MLLDN, we performed bone marrow retroviral transduction and proliferation assays with the MLL-AF9 and E2A-HLF fusion proteins. Lin−c-kit+ bone marrow cells were transduced with combinations of MLL-AF9 or E2A-HLF fusion proteins and MSCV-puro or MSCV-MLLDN. Cells were selected for 1 day and grown in liquid prestimulation media to determine the rate of proliferation. Expression of MLLDN had little effect on the proliferation of isolated lin− hematopoietic progenitor cells (Figure 4A). Strikingly, bone marrow cells transduced with the MLL-AF9 fusion protein displayed a strong sensitivity to disruption of the MLL-PAFc complex following cotransduction with MLLDN (Figure 4B). In contrast, expression of MLLDN showed no effect on E2A-HLF–transduced bone marrow cells; these grew at a similar rate as control cells cotransduced with E2A-HLF and empty vector (Figure 4C). RNA was collected from MSCVNeo/Puro (control) and MSCVNeo/MLLDN cotransduced cells at days 0, 3, and 6 to assess gene expression following disruption of the MLL-PAFc interaction in normal hematopoietic progenitors. A gradual downregulation of Hoxa9 expression was observed in control cells during ex vivo culture that was accelerated in the presence of MLLDN (Figure 4D). Expression of MLLDN also resulted in decreased Meis1 expression, which was observed as early as day 3 (Figure 4E). MLLDN expression was confirmed to be stable during the duration of the experiment (Figure 4F), suggesting MLLDN mediated inhibition of Hox and Meis1 expression is tolerated in normal untransformed bone marrow cells. Bone marrow transduction and replating assays displayed similar results as the liquid growth curves with a reduction in colony formation observed specifically in bone marrow cells expressing MLL-AF9 and MLLDN, whereas E2A-HLF–mediated colony formation was unaffected by expression of MLLDN (Figure 4H-I). The negative effect of MLLDN on MLL-AF9 transformation was evident during the first and second round of the colony assay, but less pronounced by the third round of replating, suggestive of a strong selective pressure against the expression of MLLDN during MLL-AF9–mediated transformation. Downregulation of MLLDN expression specifically in MLL-AF9–transformed cells was confirmed by quantitative reverse transcription PCR (qRT-PCR) (supplemental Figure 6). Notably, the number of first-round colonies was not affected by cells cotransduced with empty MSCVNeo with and without MLLDN, suggesting that normal hematopoiesis is not affected by disruption of the MLL-PAFc interaction (Figure 4G).
MLLDN expression specifically inhibits the initiation of MLL leukemic cells. (A-C) Proliferation assays were performed with lin−c-kit+ mouse bone marrow cells grown in double selective media following transduction with combinations of the MLL-AF9 or E2A-HLF fusion proteins and empty MSCV puro or MLLDN after 1 day in selective media (mean ± SD; n = 2 independent experiments performed in duplicate (P > .05); 2-way ANOVA). (D-E) qRT-PCR was performed on RNA collected from either Neo-Puro– or Neo-MLLDN–transduced bone marrow cells at days 0, 3, and 6 following plating described in (A-C). Arrows indicate time points for RNA collection. Expression levels for Hoxa9 and Meis1 are shown relative to Neo-Puro–transduced cells at day 0. MLLDN expression is shown relative to Neo-MLLDN–transduced cells at day 0 (mean ± SD; n = 2 independent experiments performed in triplicate; ***P < .001, *P < .05; unpaired 2-tailed Student t test). (G-I) Colony-forming assays were performed using the methods described in (A-C). Double-transduced cells were grown in double-selective semisolid media for 7 days for each round. Colony numbers from each successive round of replating are shown. Results suggest a selective sensitivity of MLL-AF9–transformed cells to disruption of the MLL-PAFc interaction. (G) Cells transduced with empty MSCV-neo and either MSCV puro or MSCV-MLLDN. (H) Cells transduced with MSCV-MLL-AF9 and either MSCV puro or MSCV-MLLDN. (I) Cells transduced with MSCV-E2A-HLF and either MSCV puro or MSCV-MLLDN (mean ± SD; n = 2 independent experiments performed in duplicate; *P < .05, ****P < .0001 (P > .05); unpaired 2-tailed Student t test).
MLLDN expression specifically inhibits the initiation of MLL leukemic cells. (A-C) Proliferation assays were performed with lin−c-kit+ mouse bone marrow cells grown in double selective media following transduction with combinations of the MLL-AF9 or E2A-HLF fusion proteins and empty MSCV puro or MLLDN after 1 day in selective media (mean ± SD; n = 2 independent experiments performed in duplicate (P > .05); 2-way ANOVA). (D-E) qRT-PCR was performed on RNA collected from either Neo-Puro– or Neo-MLLDN–transduced bone marrow cells at days 0, 3, and 6 following plating described in (A-C). Arrows indicate time points for RNA collection. Expression levels for Hoxa9 and Meis1 are shown relative to Neo-Puro–transduced cells at day 0. MLLDN expression is shown relative to Neo-MLLDN–transduced cells at day 0 (mean ± SD; n = 2 independent experiments performed in triplicate; ***P < .001, *P < .05; unpaired 2-tailed Student t test). (G-I) Colony-forming assays were performed using the methods described in (A-C). Double-transduced cells were grown in double-selective semisolid media for 7 days for each round. Colony numbers from each successive round of replating are shown. Results suggest a selective sensitivity of MLL-AF9–transformed cells to disruption of the MLL-PAFc interaction. (G) Cells transduced with empty MSCV-neo and either MSCV puro or MSCV-MLLDN. (H) Cells transduced with MSCV-MLL-AF9 and either MSCV puro or MSCV-MLLDN. (I) Cells transduced with MSCV-E2A-HLF and either MSCV puro or MSCV-MLLDN (mean ± SD; n = 2 independent experiments performed in duplicate; *P < .05, ****P < .0001 (P > .05); unpaired 2-tailed Student t test).
To test the effect of MLLDN on leukemic maintenance, we introduced MLLDN into established leukemic cell lines. Previously established MLL-AF9, MLL-GAS7, E2A-HLF, and AML-ETO9a cell lines were transduced with MigR1-MLLDN or empty control vector. Green fluorescent protein (GFP)+ cells were sorted and plated in methylcellulose to assess colony formation. Again, expression of MLLDN selectively inhibited the growth of MLL-transformed cell lines without affecting growth of E2A-HLF cells (Figure 5A-B). AML-ETO9a–transformed cells showed intermediate sensitivity to MLLDN compared with MLL-transformed cells, consistent with recent data suggesting collaboration between AML-ETO and Meis1 (Figure 5A).28 Analysis of MLL fusion target genes from the resulting MLL-AF9 colony assays demonstrated reduced expression of key target genes such as Hoxa9 and Meis1, consistent with targeted disruption of the MLL-PAFc interaction (Figure 5C). We used GFP as a surrogate for MLLDN expression as this is expressed off a single transcript with MLLDN because of an internal ribosomal entry site in the MigR1 vector. A selective outgrowth of GFP-negative cells was observed, specifically in MLL-AF9 cells following transduction with MigR1-MLLDN compared with cells transduced with empty MigR1 as determined by microscopy and mean fluorescence values derived from flow cytometry (Figure 5D,F). In contrast, E2A-HLF cells maintained GFP expression after transduction with either empty MigR1 or MigR1-MLLDN, suggestive of a strong selective pressure against expression of the MLLDN specifically in MLL leukemic cells (Figure 5E,G). Expression of MLLDN did not alter disease latency in an MLL-AF9 leukemia mouse model, likely because of strong selective pressure and the extended duration of the experiment. In support, retrieved MLL-AF9 leukemic cells displayed low expression of MLLDN, as determined by GFP expression, compared with MigR1-transduced cells (supplemental Figure 7). Next, we wanted to confirm the proliferative defect observed in MLL fusion–transformed cells was due specifically to expression of MLLDN. To this end, we engineered MigR1 to express an unrelated sequence from MLL, the first PHD finger of MLL (PHD1). Expression of MigR1-PHD1 in both MA or EH cells resulted in no change in colony-forming units and displayed no reduction in GFP expression, confirming the proliferative defect observed in MA cells is specific to expression of MLLDN (supplemental Figure 8). Collectively, these data strongly suggest that targeted disruption of the MLL-PAFc interaction through expression of MLLDN selectively blocks both MLL fusion–mediated leukemic initiation and maintenance.
The maintenance of MLL leukemic cells is inhibited by disruption of MLL-PAFc. (A) Colony assays were performed with GFP+-sorted MLL-AF9, MLL-GAS7, E2A-HLF, and AML-ETO9a cell lines transduced with MigR1 or MigR1-MLLDN. The number of colonies is shown relative to each cell line transduced with empty MigR1 (mean ± SD; MLL-AF9 [n = 4], MLL-GAS7 [n = 2], E2A-HLF [n = 4], and AML-ETO9a [n = 2] independent experiments; **P < .0001, *P < .05 (P > .05); unpaired 2-tailed Student t test). (B) Representative INT-stained plates from the colony assay described in (A) are shown. (C) qRT-PCR was performed for Hoxa9 and Meis1 using RNA isolated from the MLL-AF9 colonies transduced with either MigR1 or MigR1 MLLDN described in (A). Expression is shown relative to the MLL-AF9 cell line transduced with MigR1 (mean ± SD; n = 2 independent experiments; *P < .05, **P < .01; unpaired 2-tailed Student t test). (D-E) Representative colonies from colony assays performed with either the MLL-AF9 or E2A-HLF cell line transduced with MigR1 or MLLDN are shown in both phase contrast and GFP fluorescence. Scale bars = 1 mm. Images were acquired using a 4× lens and Olympus BX-51 microscope with Olympus DP controller software. (F-G) Quantitation of the mean fluorescence from both the MLL-AF9 and E2A-HLF colony assays is shown as determined by flow cytometry performed at the time of sort (precolony) and at the conclusion of the experiment (postcolony, 7 days).
The maintenance of MLL leukemic cells is inhibited by disruption of MLL-PAFc. (A) Colony assays were performed with GFP+-sorted MLL-AF9, MLL-GAS7, E2A-HLF, and AML-ETO9a cell lines transduced with MigR1 or MigR1-MLLDN. The number of colonies is shown relative to each cell line transduced with empty MigR1 (mean ± SD; MLL-AF9 [n = 4], MLL-GAS7 [n = 2], E2A-HLF [n = 4], and AML-ETO9a [n = 2] independent experiments; **P < .0001, *P < .05 (P > .05); unpaired 2-tailed Student t test). (B) Representative INT-stained plates from the colony assay described in (A) are shown. (C) qRT-PCR was performed for Hoxa9 and Meis1 using RNA isolated from the MLL-AF9 colonies transduced with either MigR1 or MigR1 MLLDN described in (A). Expression is shown relative to the MLL-AF9 cell line transduced with MigR1 (mean ± SD; n = 2 independent experiments; *P < .05, **P < .01; unpaired 2-tailed Student t test). (D-E) Representative colonies from colony assays performed with either the MLL-AF9 or E2A-HLF cell line transduced with MigR1 or MLLDN are shown in both phase contrast and GFP fluorescence. Scale bars = 1 mm. Images were acquired using a 4× lens and Olympus BX-51 microscope with Olympus DP controller software. (F-G) Quantitation of the mean fluorescence from both the MLL-AF9 and E2A-HLF colony assays is shown as determined by flow cytometry performed at the time of sort (precolony) and at the conclusion of the experiment (postcolony, 7 days).
Expression of MLLDN does not alter normal hematopoiesis
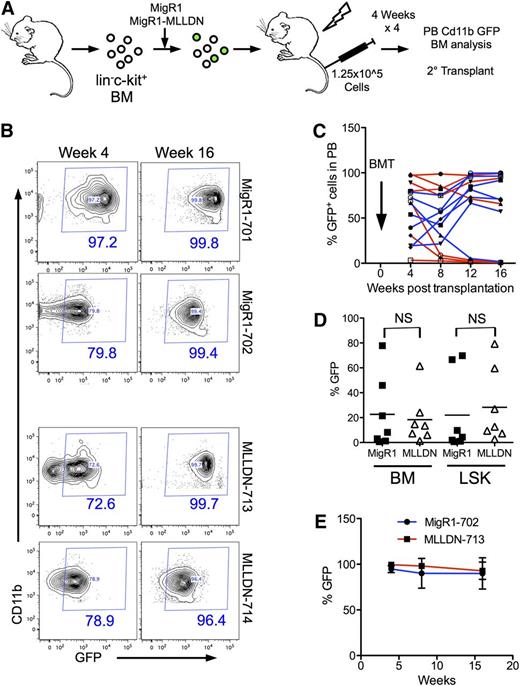
We next assessed the impact of disrupting the MLL-PAFc interaction on normal hematopoiesis. In vivo hematopoietic reconstitution assays were performed by rescuing lethally irradiated mice with bone marrow cells transduced with MigR1 or MigR1-MLLDN and tracking GFP expression in peripheral blood (PB) Cd11b+ cells (Figure 6A). The reconstitutions assays were performed in a competitive manner by injecting unsorted GFP+ and GFP− cells into syngeneic recipients. We observed persistent GFP expression in the PB of a subset of mice injected with MigR1- and MigR1-MLLDN–expressing cells through 16 weeks after reconstitution (Figure 6B). Tracking all mice in each cohort revealed a similar ratio of mice successfully reconstituted with GFP+ cells. We observed a similar pattern of GFP expression in Cd11b+ PB cells with 57% of MigR1- and 85% of MLLDN-reconstituted mice displaying greater than 50% GFP+ cells in the PB 16 weeks after irradiation (Figure 6C). The persistent GFP expression in the presence of competitive GFP− cells suggests the presence of MLLDN does not inhibit reconstitution of the hematopoietic compartment. After 16 weeks, cells were isolated from both cohorts for analysis of GFP in the whole bone marrow and lin−Sca1+c-kit+ (LSK) cell population (supplemental Figure 9). Isolation of bone marrow cells at 16 weeks revealed similar levels of GFP expression in both the whole bone marrow and LSK cell population from MigR1- and MigR1-MLLDN–injected mice consistent with MLLDN not altering the repopulating function of primitive hematopoietic cells (Figure 6D). Analysis of lineage cells including Cd11b+ Gr1+ myeloid cells, B220+ B cells, and TCRβ+ T cells displayed little change in GFP expression between the 2 cohorts (supplemental Figure 10). To rigorously test hematopoietic reconstitution potential in the presence of MLLDN, isolated bone marrow cells from GFP+ primary transplant recipients were used for secondary transplants into lethally irradiated syngeneic recipients. Again, both MigR1- and MLLDN-transduced cells successfully reconstituted the hematopoietic system of recipient mice at similar efficiencies as evidenced by persistent GFP expression through 16 weeks after transplantation (Figure 6E). These data would suggest that disruption of the MLL-PAFc interaction does not significantly alter normal hematopoietic stem cell function.
In vivo hematopoietic reconstitution is not altered by MLLDN. (A) Schematic representation of reconstitution experiment. The hematopoietic system of lethally irradiated mice was competitively reconstituted with unsorted lin−c-kit+ bone marrow cells transduced with either MigR1 or MigR1-MLLDN. Cd11b+ cells of the peripheral blood were analyzed for GFP positivity by flow cytometry every 4 weeks for 16 weeks. (B) GFP expression in the peripheral blood at 4 and 16 weeks postinjection of representative mice reconstituted with GFP+ bone marrow cells expressing either MigR1 or MigR1 MLLDN. The percentage of cells within the GFP+ gate is shown in blue. (C) The percentage of GFP+ cells from each individual mouse from the MigR1 (red) and MigR1 MLLDN (blue) cohort are plotted over time. BMT, bone marrow transplantation. (D) The percentage of GFP+ cells from whole bone marrow cells and LSK populations are plotted for each mouse from the MigR1 and MLLDN cohorts (mean shown by horizontal line; n = 7 (P > .05); unpaired 2-tailed Student t test). (E) Secondary transplantation assays performed by injecting primary transplanted bone marrow from the MigR1-702 and MLLDN-713 mice into 5 sublethally irradiated recipients for each group. GFP expression detected in the Cd11b+ peripheral blood cells is shown for each cohort through 16 weeks posttransplantation.
In vivo hematopoietic reconstitution is not altered by MLLDN. (A) Schematic representation of reconstitution experiment. The hematopoietic system of lethally irradiated mice was competitively reconstituted with unsorted lin−c-kit+ bone marrow cells transduced with either MigR1 or MigR1-MLLDN. Cd11b+ cells of the peripheral blood were analyzed for GFP positivity by flow cytometry every 4 weeks for 16 weeks. (B) GFP expression in the peripheral blood at 4 and 16 weeks postinjection of representative mice reconstituted with GFP+ bone marrow cells expressing either MigR1 or MigR1 MLLDN. The percentage of cells within the GFP+ gate is shown in blue. (C) The percentage of GFP+ cells from each individual mouse from the MigR1 (red) and MigR1 MLLDN (blue) cohort are plotted over time. BMT, bone marrow transplantation. (D) The percentage of GFP+ cells from whole bone marrow cells and LSK populations are plotted for each mouse from the MigR1 and MLLDN cohorts (mean shown by horizontal line; n = 7 (P > .05); unpaired 2-tailed Student t test). (E) Secondary transplantation assays performed by injecting primary transplanted bone marrow from the MigR1-702 and MLLDN-713 mice into 5 sublethally irradiated recipients for each group. GFP expression detected in the Cd11b+ peripheral blood cells is shown for each cohort through 16 weeks posttransplantation.
Discussion
In this study, we explored the therapeutic value of targeted disruption of the PAFc in AML using both a Cdc73 conditional knockout mouse model and biochemical methods. Consistent with an essential requirement for Cdc73 in complex eukaryotes,22 we found Cdc73 to be essential for the proliferation of AML cells independent of a transforming oncogene; however, MLL fusion protein–transformed cells display a greater sensitivity to loss of Cdc73 (Figure 1). Interestingly, although it appears that Cdc73 (and likely PAFc) is required in most cell types, the MLL-PAFc interaction appears to play a more crucial role in MLL fusion–transformed leukemia cells compared with other AML subtypes and for normal hematopoiesis. These data point to a potential area for therapeutic development at the MLL-PAFc interaction surface for the treatment of patients bearing MLL translocations.
MLL and MLL fusion protein chromatin occupancy
Studies of Cdc73 conditional knockout MLL-AF9 cell lines suggested that loss of Cdc73 led to an inefficient localization of the entire PAFc as evidenced by reduced Paf1 binding at the Meis1 locus (Figure 2). This is in line with data showing loss of CDC73 results in decreased expression levels of both PAF1 and LEO1.19,25 In addition, Roeder and colleagues recently generated an interaction map of the PAFc that indicated that CDC73 directly bound to CTR9, RTF1, and LEO1.24 Together these data support the importance of CDC73 in maintaining the integrity of the entire PAFc at target loci.
Strikingly, we observed a robust loss of Mll at the Meis1 locus following loss of Cdc73 (Figure 2). Several structural domains and protein-protein interactions aid in Mll localization, including the Menin-LEDGF-MLL trimeric interaction29 as well as AT-hook and CxxC domain binding to the minor groove and unmethylated CpG islands of DNA, respectively.30 Additionally, the third PHD finger of MLL possesses affinity for H3K4me2/me318 and aids in MLL recruitment. Our data are consistent with a highly coordinated multicontact recruitment model in which several protein-protein (including the MLL-PAFc interaction) and protein-DNA interactions are required to act in concert for proper target recognition by MLL and MLL fusion proteins. Because mutant MLL proteins unable to bind Menin, PAFc, or H3K4me3 display chromatin-binding defects, MLL may require several protein-protein and protein-DNA contact points for stable binding at gene loci.
Targeted disruption of MLL-PAFc minimally affects normal hematopoietic proliferation
A major hurdle in development of cancer therapies is the toxic nature of these chemicals on normal tissues. Such is the case with many of the classic DNA intercalating agents used to ablate rapidly proliferating cells. To analyze the effects of a specific disruption of the MLL-PAFc interaction, we introduced a small fragment of MLL into cells that acts as a dominant negative by binding to the PAF1 subunit of the PAFc, thus disrupting the MLL-PAFc interaction and allowing specific characterization of the MLL-PAFc interaction (Figure 3). Using this targeted approach, we observed decreased MLL-AF9–mediated transformation following disruption of the MLL-PAFc interaction (Figures 4 and 5). Strikingly, this effect was leukemia cell type–specific, because no change in E2A-HLF–mediated transformation was observed. Importantly, using rigorous bone marrow reconstitution assays, we were able to assess the importance of the MLL-PAFc interaction on the function of hematopoietic stem cells. We observed a robust reconstitution of the hematopoietic compartment by hematopoietic stem cells expressing MLLDN, suggesting this is tolerated by hematopoietic stem cells (Figure 6). This is surprising given the importance of Hox genes on hematopoietic development and the role of both MLL and PAFc in positively regulating Hox expression.17-19 One possible explanation for this is an increased “addiction” to Hox gene expression in MLL fusion leukemias. As such, MLL-associated leukemic cells may display increased sensitivity to even minor decreases in Hox expression that only moderately affect normal hematopoietic cells. Consistent with this idea, we do not observe a complete disruption of the MLL-PAFc interaction in the presence of the dominant negative fragment (Figure 3). This likely results in reduced but not ablated Hox expression in the targeted cells, which was observed in transduced hematopoietic progenitor cells expressing MLLDN (Figure 4). The resistance of the E2A-HLF cells to disruption of the MLL-PAFc interaction serves as additional evidence that the MLL-PAFc interaction plays a more central role in MLL-associated leukemic cells. These data also support the concept that although AML cells require the Cdc73 protein for growth, AML cells dependent on Hox and Meis1 gene upregulation for survival are increasingly dependent on the MLL-PAFc interaction and represent a potential molecular point of susceptibility for therapeutic development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Jiaying Tan, Jingya Wang, and Dr Ronald Craig for experimental advice and Dr Yali Dou (University of Michigan) for the MllC antibody.
This work was supported by grants from the National Institutes of Health (R01 CA151425) (J.L.H.), (R00 CA158136) (A.G.M.), and an American Society of Hematology Scholar Award (A.G.M.).
Authorship
Contribution: A.G.M. designed and performed experiments, analyzed data, and wrote the manuscript; W.C., M.J., and E.M.G. performed experiments and analyzed data; and I.M. and J.L.H. designed experiments, analyzed data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew G. Muntean, 7520B Medical Science Research Building I, 1301 Catherine Road, Ann Arbor, MI 48109-5602; e-mail: andrewmu@umich.edu.

![Figure 1. Loss of Cdc73 leads to decreased proliferation of leukemia cells. (A) Western blotting was performed on whole cell lysate for the MA-Cdc73fl-CreER and EH-Cdc73fl-CreER cell lines treated with EtOH or 5 nM 4-OHT for 48 hours. Cdc73 protein is reduced after 48 hours of 4-OHT treatment. β-actin serves as a loading control. (B-C) MA-Cdc73fl-CreER, EH-Cdc73fl-CreER, and control cell lines (MA-CreER, EH-CreER) were grown in liquid culture in the presence of either EtOH or 5 nM 4-OHT and counted daily for 5 days. The total cell number for each day is plotted for each condition (mean ± standard deviation [SD]; n = 2 independent experiments; NS = not significant (P > .05); 2-way analysis of variance [ANOVA]). (D-E) qRT-PCR performed on either the MA-Cdc73fl-CreER and MA-CreER or EH-Cdc73fl-CreER and EH-CreER cell lines 48 hours after EtOH or 4-OHT treatment. Expression levels for Hoxa9, Meis1, and Bcl2 are shown relative to each cell line treated with EtOH (mean ± SD; n = 2 independent experiments. ****P < .001; **P < .005; *P < .05; NS, not significant (P > .05); unpaired 2-tailed Student t test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/11/10.1182_blood-2013-02-486977/4/m_1914f1.jpeg?Expires=1767761304&Signature=y0bqbcRC-0hbfiq4fw3SptBVHOWKt~b2MRbcioHubhAv0CcHtNzFcC5E~0iSfHPi7hEE~pt5ZBrQE8zHd5yJ7SrWEbSzk29hoB7aH0KXuyaZujFPnAuWIxhU0G4FBA20hY1dLCGHg9vmcBcU1caARec961dvmyTcGKnGzAnGmww30txNt30JrQsQQPCVdTW~aJCGylmiygy0mZtwuoaQd9qc94llL~UG4Grbzk6rTO2XZ8CtByqs7Xj9GNzm~gJ5d7IcxkpIeYPRNQwdPvawB2SY-BV~p4bQY7XUPnO8raqawlNCRvkZbcga8BjxkIM~ta5NZ3JZoTUzSuOLQB6CEQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. The maintenance of MLL leukemic cells is inhibited by disruption of MLL-PAFc. (A) Colony assays were performed with GFP+-sorted MLL-AF9, MLL-GAS7, E2A-HLF, and AML-ETO9a cell lines transduced with MigR1 or MigR1-MLLDN. The number of colonies is shown relative to each cell line transduced with empty MigR1 (mean ± SD; MLL-AF9 [n = 4], MLL-GAS7 [n = 2], E2A-HLF [n = 4], and AML-ETO9a [n = 2] independent experiments; **P < .0001, *P < .05 (P > .05); unpaired 2-tailed Student t test). (B) Representative INT-stained plates from the colony assay described in (A) are shown. (C) qRT-PCR was performed for Hoxa9 and Meis1 using RNA isolated from the MLL-AF9 colonies transduced with either MigR1 or MigR1 MLLDN described in (A). Expression is shown relative to the MLL-AF9 cell line transduced with MigR1 (mean ± SD; n = 2 independent experiments; *P < .05, **P < .01; unpaired 2-tailed Student t test). (D-E) Representative colonies from colony assays performed with either the MLL-AF9 or E2A-HLF cell line transduced with MigR1 or MLLDN are shown in both phase contrast and GFP fluorescence. Scale bars = 1 mm. Images were acquired using a 4× lens and Olympus BX-51 microscope with Olympus DP controller software. (F-G) Quantitation of the mean fluorescence from both the MLL-AF9 and E2A-HLF colony assays is shown as determined by flow cytometry performed at the time of sort (precolony) and at the conclusion of the experiment (postcolony, 7 days).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/11/10.1182_blood-2013-02-486977/4/m_1914f5.jpeg?Expires=1767761304&Signature=vo99i6aNsLwhd5IwMzW~pJKxm0agnFiDG4PjR-vRcni00ScjEzKecJdIH3YaypbWosSEM1OhuVybYpq0S~Ve9XkrproUZ0uDKN1A5b0rB1p9Qiehb3p4lsM4lQfEYV7hSLEaNKfiwW6mINRrDHpOim7aFGNVoR4yOV3QrsAN~51XOA~yfivoQNze~pWqN2BZ3nUJG24XTnujcdsx-enttE4MTYUioR2pDAlUmrQSjlVI-GHiVFk0GdUfjw3TZoHR5MyNwcP9a9gxc9lG4x5kM0oc4A98kE~xMK~0KQXjd2eRcg7K7tIhifoceWbd7ULiMPXhVCGBjc3EC-Nuh2bq6g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)