Key Points
Fibrin polymerization was observed for the first time at the single-molecule level by total internal reflection fluorescence microscopy.
Live observation of fibrin polymerization with a single-molecule fluorescence intensity calibration revealed the real-time growth kinetics.
Abstract
Individual fluorescently labeled fibrin(ogen) molecules and their assembly to make a clot were observed by total internal reflection fluorescence microscopy (TIRFM). We used the bleaching of the fluorescent labels to determine the number of active fluorophores attached nonspecifically to each molecule. From the total intensity of bleaching steps, as single-molecule signature events, and the distribution of active labeling, we developed a new single-molecule intensity calibration, which accounts for all molecules, including those “not seen.” Live observation of fibrin polymerization in TIRFM by diffusive mixing of thrombin and plasma revealed the real-time growth kinetics of individual fibrin fibers quantitatively at the molecular level. Some fibers thickened in time to thousands of molecules across, equivalent to hundreds of nanometers in diameter, whereas others reached an early stationary state at smaller diameters. This new approach to determine the molecular dynamics of fiber growth provides information important for understanding clotting mechanisms and the associated clinical implications.
Introduction
Much is known about the properties and functions of fibrinogen, including fibrin polymerization. Most studies of fibrin clotting visualized large clot structures either at low resolution by confocal microscopy or at higher resolution by electron microscopy (at certain time points), or used indirect biophysical methods.1-6 As a result, little is known about the microscopic and molecular structural origins of the clot, especially about the mechanisms in the early stages of polymerization and the molecular kinetics.1,7 More recently, some aspects of assembly before the gel point were visualized by deconvolution and spinning disk confocal microscopy,8,9 but the smallest structures observed were protofibrils. Information about clotting, especially in the early stages that determine clot properties and the gel point, has clinical relevance, because the gel point is used diagnostically and disorders of clotting or fibrinolysis accompany or cause many pathologic conditions, including myocardial infarction and stroke.
Here we visualize and characterize individual fibrinogen molecules by total internal reflection fluorescence microscopy (TIRFM) and investigate the dynamics of fibrin assembly and fibrin fiber properties at the single-molecule level. In TIRFM, because of low background, sensitivity is high for events near the coverslip, and single molecules can be detected.10-12 By analyzing the bleaching of single fibrinogen molecules, we characterize the distribution of labeling and develop a new molecular calibration for the fluorescence intensity of fibers, revealing aspects of the real-time growth of individual fibers at the molecular level.
Methods
Approval was obtained from the University of Pennsylvania Institutional Review Board for these studies.
Purified human fibrinogen from Hyphen BioMed was labeled nonspecifically with tetramethyl-rhodamine and Alexa Fluor 488 (supplemental Methods, available on the Blood Web site; see supplemental Materials link at the top of the online article) in parallel samples to assure that none of the observations were the result of artifacts because of dye attachment.
For single-molecule TIRFM observation, 1 mg/mL fibrinogen stocks were diluted 1000× in 20mM HEPES, 150mM NaCl, pH 7.4 (HBS), and ultracentrifuged for 30 minutes at 100 000g to sediment aggregates, leaving only single molecules in the supernatant. The supernatant was diluted to 0.1 μg/mL before introduction into the TIRFM chamber.
To study fibrin fibers in later stages of polymerization, we mixed platelet-poor plasma (PPP; preparation described in supplemental Methods) with fluorescently labeled fibrinogen (1 vol fluorescent fibrinogen to 17 vol PPP) with HBS containing thrombin and CaCl2 for a final concentration of 0.5 U/mL thrombin and 20mM CaCl2. The mixture was immediately introduced into the observation chamber.
To study the real-time polymerization of fibrin in plasma, PPP spiked with fluorescent fibrinogen (1 vol fluorescent fibrinogen to 10 vol PPP) was introduced into one side of the observation chamber and HBS containing 5 U/mL thrombin and 200mM CaCl2 was introduced into the other side to induce polymerization through diffusion at the interface of the 2 liquids.
Data processing was done with an ImageJ plug-in that measures the time evolution of total intensity of single molecules, by recording intensity in regions of interest centered automatically on the bright areas in the first image of a time-series stack and repeating the measurement throughout the stack (ImageJ Version 1.36b software). To measure total intensity of fibrin fibers, similar but larger regions of interest, between branch points of fibers, were selected manually when using the same plug-in. After background subtraction, bleaching correction was performed using the bleaching time constant of each dye (supplemental Methods). A new molecular calibration was developed to determine M, the number of molecules in each fiber (including fluorescent and unlabeled molecules; see supplemental Methods). Using M and considering the fiber structure of parallel rows1,13,14 we calculated the number of molecules in the cross-section of each fiber, c, and the corresponding diameter, D, of the hydrated fiber (see supplemental Methods).
Results and discussion
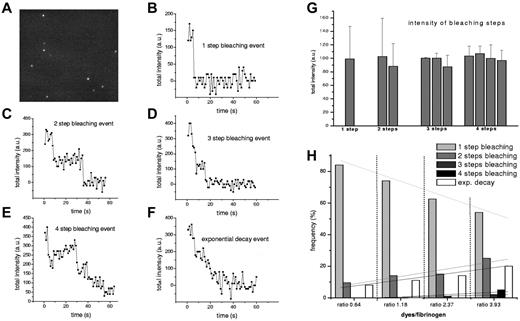
Individual fluorescently labeled fibrinogen molecules that attached to the glass chamber surface were visualized (Figure 1A). Because of the low fibrinogen concentration, prior removal of aggregates, and low probability of attachment to the surface, most fluorescent signals observed represented single molecules, as previously demonstrated (see supplemental Methods).12 Time sequences of fluorescence images showed that most of the signals bleached in 1 step (Figure 1B), some bleached in multiple steps (Figure 1C-E) or presented an exponential decay (Figure 1F). Bleaching in 1 step is the signature of a single molecule labeled with 1 fluorescent probe.10 Bleaching in several steps indicates the presence of several dyes attached to a fibrinogen molecule. The intensity of the bleaching steps was in the same range for all bleaching events (Figure 1G). An exponential-decay trace comes from the superposition of signals from several dyes that bleach over time, and their number can be calculated from the intensity analysis. With increasing bulk-labeling ratio, the number of 2-, 3-, and 4-step bleaching events increased slightly; however, the most frequent were the 1-step bleaching events, even at higher ratios (Figure 1H). Similar results were obtained with both dyes.
Bleaching of single fluorescently labeled fibrinogen molecules in TIRFM. (A) TIRFM image showing single fibrinogen molecules that adhered to the chamber glass surface from bulk solution. Time sequences of images like this showed the irreversible bleaching of the attached fluorophores in (B) 1 step, (C) 2 steps, (D) 3 steps, (E) 4 steps, or (F) showed sometimes an exponential decay. (G) Intensities of steps were in the same range for all individual bleaching events. Data plotted are from 1 single set of measurements and all the other datasets present similar values. Each data point represents a mean of at least 5 values and error bars are standard deviations. Because the magnitudes of the bleaching steps are all multiples of a single bleaching step, the number of bleaching steps accounts for the number of fluorophores attached, whereas the exponential decay curve accounts for a larger number of dyes attached to a fibrinogen molecule (number that can be determined from the intensity analysis). (H) Bleaching event distributions for several bulk-labeling ratios using tetramethyl-rhodamine–labeled fibrinogen show that by increasing the bulk-labeling ratio, the 1-step bleaching events decrease in favor of multiple steps events; however, 1-step bleaching events remain predominant. This distribution of active labeling we used for the molecular calibration. At least 90 bleaching events were collected for each ratio. The lines are linear fits of 1-step bleaching events, 2-step bleaching events, and so on. We used an Olympus Plan Apo 60×/1.45 oil TIRFM objective and an ORCA-ER Hamamatsu (Japan) camera. Images were acquired with National Instruments IMAQ Vision Builder 6 and image sequences were recorded at 1 frame/s. Resolution in the TIRFM image was 80 nm/pixel.
Bleaching of single fluorescently labeled fibrinogen molecules in TIRFM. (A) TIRFM image showing single fibrinogen molecules that adhered to the chamber glass surface from bulk solution. Time sequences of images like this showed the irreversible bleaching of the attached fluorophores in (B) 1 step, (C) 2 steps, (D) 3 steps, (E) 4 steps, or (F) showed sometimes an exponential decay. (G) Intensities of steps were in the same range for all individual bleaching events. Data plotted are from 1 single set of measurements and all the other datasets present similar values. Each data point represents a mean of at least 5 values and error bars are standard deviations. Because the magnitudes of the bleaching steps are all multiples of a single bleaching step, the number of bleaching steps accounts for the number of fluorophores attached, whereas the exponential decay curve accounts for a larger number of dyes attached to a fibrinogen molecule (number that can be determined from the intensity analysis). (H) Bleaching event distributions for several bulk-labeling ratios using tetramethyl-rhodamine–labeled fibrinogen show that by increasing the bulk-labeling ratio, the 1-step bleaching events decrease in favor of multiple steps events; however, 1-step bleaching events remain predominant. This distribution of active labeling we used for the molecular calibration. At least 90 bleaching events were collected for each ratio. The lines are linear fits of 1-step bleaching events, 2-step bleaching events, and so on. We used an Olympus Plan Apo 60×/1.45 oil TIRFM objective and an ORCA-ER Hamamatsu (Japan) camera. Images were acquired with National Instruments IMAQ Vision Builder 6 and image sequences were recorded at 1 frame/s. Resolution in the TIRFM image was 80 nm/pixel.
Using this molecular distribution of active fluorophores per fibrinogen molecule (Figure 1H) and the total intensity of a bleaching step, while taking into account the labeling probability, and considering the unlabeled plasma fibrinogen, a single-molecule calibration for the fluorescent fibrin fibers seen in TIRFM was developed (see supplemental Methods) and the total number of molecules in a fiber, including the “not seen” molecules, was determined.
Two different approaches were used to observe aspects of fibrin polymerization. Fluorescent fibrin fibers were observed in TIRFM after induction of polymerization in a vial by adding thrombin into recalcified citrated plasma and transferring the mixture to the chamber by flow (Figure 2A). The flow did not orient the fibers parallel to the flow direction, showing that within seconds, the network was already formed and stable. However, the flow did bend many of the fibers, indicating a deformable network.
Fluorescent fibrin fibers observed and characterized in TIRFM. (A) Typical TIRFM image of fluorescently labeled fibrin fibers formed by mixing thrombin into recalcified plasma and then insertion into the flow chamber. There was no preferential orientation of the fibers because of flow in the chamber, but most fibers were curved because of shear forces. (B) TIRFM observation chamber (2 clean glass coverslips on top of each other, separated laterally by double-stick tape to create a 75-μm thick chamber) allowed real-time observation of fibrin polymerization in TIRFM by introducing thrombin solution at one side of the chamber (green region) and allowing its diffusion into the region with fluorescent fibrinogen molecules (red region). (C) TIRFM images of the kinetics of growth of fibrin clot 1 molecule at a time. Growth of fibers continued for tens of minutes in the observation region, arbitrarily chosen in the center of the chamber close to the interface of thrombin and plasma. Frame 1 shows fluorescent fibrinogen in the central area of the chamber before introducing thrombin, and frames 2 and 3 were taken at 4.30 and 21 minutes after thrombin insertion. The sample was kept in the dark between frames 2 and 3 to avoid bleaching and thus show real lateral growth. (D-E) Graphs of fiber growth as a function of time, plotted as either number of molecules in the cross section of fiber, c, or hydrated fiber diameter, D. The 1-second time-point is the first moment of observation of the network after contact of thrombin with fibrinogen. According to the single-molecule calibration, lateral growth of fibers went up to thousands of molecules across (D), but some fiber regions reached an early stationary state (inset). Calculated diameters of the hydrated fibers (E) showed growth of up to 950 nm in 250 seconds for the largest fibers, and are in the same range as fiber diameters measured in unbleached fluorescence images, but more accurate. Regions of fibers analyzed were between branch points or intersections of fibers and had lengths from 2 to 10 μm. Image sequences were recorded at 1 frame/s. Resolution in all TIRFM images was 80 nm/pixel. Images presented are raw data with no bleaching correction performed for the frames in (C). Scale bar is 3 μm in panel A and 2.5 μm in panel C.
Fluorescent fibrin fibers observed and characterized in TIRFM. (A) Typical TIRFM image of fluorescently labeled fibrin fibers formed by mixing thrombin into recalcified plasma and then insertion into the flow chamber. There was no preferential orientation of the fibers because of flow in the chamber, but most fibers were curved because of shear forces. (B) TIRFM observation chamber (2 clean glass coverslips on top of each other, separated laterally by double-stick tape to create a 75-μm thick chamber) allowed real-time observation of fibrin polymerization in TIRFM by introducing thrombin solution at one side of the chamber (green region) and allowing its diffusion into the region with fluorescent fibrinogen molecules (red region). (C) TIRFM images of the kinetics of growth of fibrin clot 1 molecule at a time. Growth of fibers continued for tens of minutes in the observation region, arbitrarily chosen in the center of the chamber close to the interface of thrombin and plasma. Frame 1 shows fluorescent fibrinogen in the central area of the chamber before introducing thrombin, and frames 2 and 3 were taken at 4.30 and 21 minutes after thrombin insertion. The sample was kept in the dark between frames 2 and 3 to avoid bleaching and thus show real lateral growth. (D-E) Graphs of fiber growth as a function of time, plotted as either number of molecules in the cross section of fiber, c, or hydrated fiber diameter, D. The 1-second time-point is the first moment of observation of the network after contact of thrombin with fibrinogen. According to the single-molecule calibration, lateral growth of fibers went up to thousands of molecules across (D), but some fiber regions reached an early stationary state (inset). Calculated diameters of the hydrated fibers (E) showed growth of up to 950 nm in 250 seconds for the largest fibers, and are in the same range as fiber diameters measured in unbleached fluorescence images, but more accurate. Regions of fibers analyzed were between branch points or intersections of fibers and had lengths from 2 to 10 μm. Image sequences were recorded at 1 frame/s. Resolution in all TIRFM images was 80 nm/pixel. Images presented are raw data with no bleaching correction performed for the frames in (C). Scale bar is 3 μm in panel A and 2.5 μm in panel C.
To observe polymerization ab initio, fluorescently labeled fibrinogen in plasma was introduced at one side of the microfluidic observation chamber and thrombin was introduced at the other side to induce polymerization at the interface between the 2 liquids (Figure 2B). Because at the μm dimensions of the TIRFM chamber, liquids that make contact do not mix,15 polymerization was due exclusively to the diffusion of thrombin molecules into the fibrinogen region, and the rate of fiber growth was therefore diffusion dependent. As expected for the low concentration of thrombin reaching the fibrinogen region, the fibers were thick.16,17 Lateral growth of fibers continued for minutes (Figure 2C) and the single-molecule calibration showed that growth increases to thousands of molecules across (Figure 2D), but some fiber regions reach an early stationary state at lower values (Figure 2D inset). Fiber lateral growth is probably determined by the local kinetics of fibrinopeptide cleavage and oligomerization, as well as physical factors, such as the twisting of fibers.16,18 The calculated diameters of the hydrated fibers are variable along the length of the fibers and can grow to hundreds of nanometers (Figure 2E) being in the same range as those measured in unbleached fluorescent images.
In conclusion, we demonstrated that the dynamics of fibrin polymerization can be studied at the single-molecule level using TIRFM. Through analysis of bleaching individual fibrinogen molecules, we obtained the molecular distribution of fibrinogen labeling, which was used to obtain a single-molecule intensity calibration. We observed live growth kinetics with molecular accuracy, determined for the first time the number of molecules within individual fibrin fibers, and calculated their diameters, bridging in this way the gap from molecular to assembled structures. This new methodology can be used for further studies of the assembly of fibrin or other biopolymers.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Henry Shuman and Dr Yale Goldman for use of their TIRFM setup.
This work was supported by National Institutes of Health grants HL030954 and HL090774.
National Institutes of Health
Authorship
Contribution: A.H. designed the research, performed experiments, analyzed and discussed results, prepared the figures, and wrote the paper; K.C.G. and D.S. performed the fluorescence labeling of fibrinogen; and J.W.W. discussed results, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John W. Weisel, Dept of Cell and Developmental Biology, University of Pennsylvania Perelman School of Medicine, 421 Curie Blvd, 1054 BRB II/III, Philadelphia, PA 19104; e-mail: weisel@mail.med.upenn.edu.