Key Points
A novel fusion gene, OBFC2A/RARA, in variant acute promyelocytic leukemia.
In vitro all-trans retinoic acid sensitivity.
Abstract
Acute promyelocytic leukemia is characterized by the rearrangement of the retinoic acid receptor α (RARA) gene and its fusion with other genes. We report a novel case of variant acute promyelocytic leukemia with the karyotype der (2)t(2;17)(q32;q21). Array comparative genomic hybridization revealed distinct chromosome breakpoints within the RARA and oligonucleotide/oligosaccharide-binding fold containing 2A (OBFC2A) genes. Sequence analysis of the OBFC2A/RARA transcript showed that exon 5 of OBFC2A was fused with exon 3 of RARA through the same breakpoint as in previously described fusions of RARA. The single-stranded DNA binding protein encoded by OBFC2A is critical for genomic stability. Retention of the OB fold domain of OBFC2A in the fusion protein suggests the possibility of homodimerization. The leukemic cells from the patient showed neutrophilic differentiation in the in vitro all-trans retinoic acid assay. Mutation or rearrangement of the OBFC2A gene has not been previously reported in congenital or acquired disorders.
Introduction
Rearrangement of the retinoic acid receptor α (RARA) gene and fusion with the gene encoding promyelocytic leukemia protein (PML) has been reported as a cause of acute promyelocytic leukemia (APL).1,2 PML/RARA fusion causes reduced transcriptional activation, inhibition of myeloid differentiation, and APL.3 All-trans retinoic acid (ATRA) effectively reverses this and is used in induction therapy of APL. However, in APL variants RARA is fused with genes other than PML, such as promyelocytic leukemia zinc finger (PLZF),4 nucleophosmin (NPM1),5 nuclear mitotic apparatus (NUMA1),6 signal transducer and activator of transcription 5b (STAT5B),7 cAMP-dependent protein kinase type I-α regulatory subunit (PRKAR1A),8 FIP1-like 1 (FIP1L1),9 or BCL6 corepressor (BCOR).10 Unlike other forms, variant APL associated with STAT5B/RARA or PLZF/RARA are resistant to ATRA.7,11
This study reports a patient with variant APL because of a novel RARA gene rearrangement involving the oligonucleotide/oligosaccharide-binding fold containing 2A (OBFC2A) gene. The OBFC2A gene located at chromosome 2q32.3 encodes human single-stranded DNA binding protein 2 (hSSB2 or OBFC2A), which plays a role in DNA damage response and genomic stability.12,13 To our knowledge, this is the first report showing involvement of the OBFC2A gene in a human disease.
Methods
Case reports
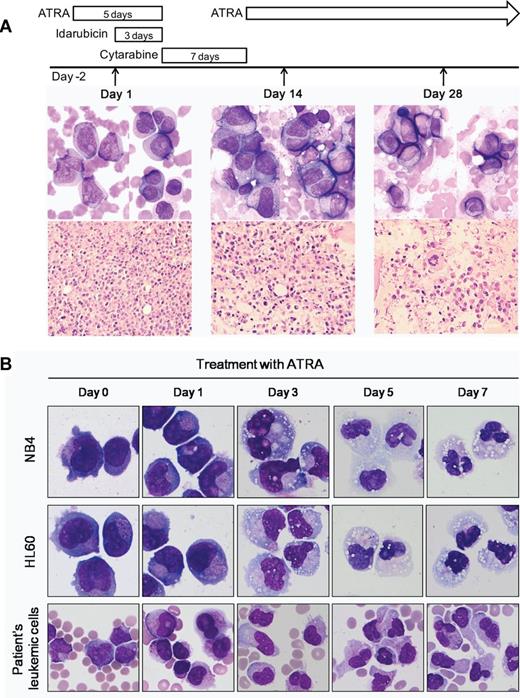
A 59-year-old man was transferred to our hospital with night sweats over the previous month, blood-tinged sputum, and petechia for 10 days. Laboratory investigations showed the following: Hb level, 7.8 g/dL; platelet count, 41 × 103/mm3; leukocyte count, 96.9 × 103/mm3 (61% blasts and abnormal promyelocytes; Figure 1A). Coagulation tests revealed the following: prothrombin time, 13.2 seconds (69.1%); partial thromboplastin time, 35.0 seconds; and fibrinogen 254 mg/dL. Although levels of d-dimer (39.97 μg/mL) and fibrin degradation products (136.7 μg/mL) were high, the patient did not show active bleeding. Because of clinical suspicion of APL, ATRA treatment (45 mg/m2 daily) was immediately initiated. Two days later, bone marrow (BM) examination was performed and idarubicin treatment was started. Morphologic analysis of the BM aspirate showed 69.2% microgranular abnormal promyelocytes (Figure 1A left panel). The immunophenotype by flow cytometric analysis was positive for CD13, CD33, CD45, CD65, and cMPO, and negative for CD34, HLA-DR, and B-cell and T-cell markers. Multiplex reverse transcription polymerase chain reaction (RT-PCR) assay (HemaVision) to amplify the fusion gene transcripts including PML/RARA was negative; hence, ATRA was discontinued. Induction therapy with idarubicin and cytarabine was started. After identification of the RARA rearrangement by fluorescence in situ hybridization (FISH), ATRA was restarted after a temporary discontinuance of 7 days and continued for approximately 70 days. On day 14, the patient showed the persistence of blasts and abnormal promyelocytes, but with more differentiated granulocytes (Figure 1A middle panel). He achieved complete remission on day 28, and 2 cycles of consolidation chemotherapy were administered, followed by allogenic stem-cell transplantation from an HLA-matched unrelated donor. One year after transplantation, he remains alive and in complete remission. The study was approved by the Institutional Review Board of the University of Ulsan College of Medicine and Asan Medical Center.
Morphologic features and in vitro ATRA sensitivity of the patient with clonal der(2)t(2;17)(q32;q21). (A) Day 1 (left panel): peripheral blood smear shows abnormal promyelocytes with bi-lobed or sharply indented nucleus and microgranulated cytoplasm. Some promyelocytes have Auer rods (Wright-Giemsa, ×1000). Core biopsy shows hypercellular bone marrow infiltrated by leukemic cells with high nucleocytoplasmic ratios and indented or irregular nuclear contours (H&E, ×400). Day 14 (middle panel): bone marrow touch preparation shows persistence of blasts and abnormal promyelocytes in addition to a few myelocytes or metamyelocytes (Wright-Giemsa, ×1000). Conversely, core biopsy reveals more differentiated granulocytes (H&E, ×400). Day 28 (right panel): bone marrow examination shows normocellular marrow with complete differentiation. (B) Leukemic cells of the patient show neutrophilic differentiation with 1μM ATRA treatment. ATRA induces differentiation of NB4 and HL60 cells. (Wright-Giemsa, ×1000). The image was acquired using a BX51 microscope (Olympus), ×1000 and ×400 lenses, and a DP25 digital camera (Olympus).
Morphologic features and in vitro ATRA sensitivity of the patient with clonal der(2)t(2;17)(q32;q21). (A) Day 1 (left panel): peripheral blood smear shows abnormal promyelocytes with bi-lobed or sharply indented nucleus and microgranulated cytoplasm. Some promyelocytes have Auer rods (Wright-Giemsa, ×1000). Core biopsy shows hypercellular bone marrow infiltrated by leukemic cells with high nucleocytoplasmic ratios and indented or irregular nuclear contours (H&E, ×400). Day 14 (middle panel): bone marrow touch preparation shows persistence of blasts and abnormal promyelocytes in addition to a few myelocytes or metamyelocytes (Wright-Giemsa, ×1000). Conversely, core biopsy reveals more differentiated granulocytes (H&E, ×400). Day 28 (right panel): bone marrow examination shows normocellular marrow with complete differentiation. (B) Leukemic cells of the patient show neutrophilic differentiation with 1μM ATRA treatment. ATRA induces differentiation of NB4 and HL60 cells. (Wright-Giemsa, ×1000). The image was acquired using a BX51 microscope (Olympus), ×1000 and ×400 lenses, and a DP25 digital camera (Olympus).
Chromosome analysis and FISH
A BM sample was processed after short-term (24 hours), unstimulated culture following standard procedures. Chromosomes were G-banded, and the karyotype was interpreted according to the International System for Human Cytogenetic Nomenclature 2009. FISH analysis was performed using a PML/RARA dual-color dual fusion translocation probe (Vysis) and a RARA dual-color break-apart probe (Vysis). All analyses were performed after obtaining written informed consent from the patient.
Array CGH analysis
High-resolution oligonucleotide array comparative genomic hybridization (CGH) was performed using the Whole Human Genome CGH microarray Kit 244K (Agilent Technologies; GEO accession no. GSE42984). Genomic DNA was labeled and hybridized to the array according to the manufacturer's protocol. Genomic imbalances were analyzed by the Genomic Workbench Standard Edition 5.0 (Agilent Technologies). Probes in the boundaries of copy number changes in chromosomes 2 and 17 were detected, and their position information was obtained from the manufacturer's sequence list for designing specific primers for fusion gene RT-PCR.
RT-PCR and sequencing for fusion transcript
Total RNA was extracted from BM aspirate at presentation and reverse transcribed into cDNA with random hexamers. OBFC2A/RARA mRNA was amplified with the following primers: 5′-CAGATTATCGAGGACAGCAG-3′ of OBFC2A and 5′-CACCATGTTCTTCTGGATGC-3′ of RARA. The fusion transcript was sequenced and compared with GenBank sequences.
In vitro ATRA treatment and morphologic assessment
Differentiation effects of ATRA were examined in NB4, HL60, and leukemic cells of the patient, according to the methodology used in a previous study.14
Results and discussion
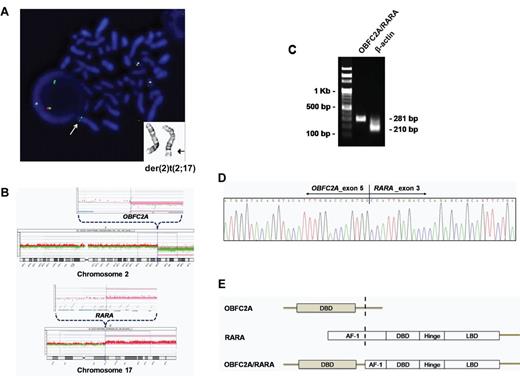
The karyotype of BM cells showed clonal der(2)t(2;17)(q32;q21) in all 20 metaphases (Figure 2A), of which 4 were subclones with an additional change of t(11;19)(q13;p13.1). FISH analysis showed an extra RARA signal with the PML/RARA probe and a 3′ RARA green signal with the RARA break-apart probe on der(2) (Figure 2A).
Identification of a novel OBFC2A/RARA fusion gene by FISH, array CGH, RT-PCR and direct sequencing. (A) Representative partial karyogram shows der(2)t(2;17)(q32;q21). FISH with the RARA break-apart probe reveals a 3′ RARA green signal on der(2), indicated by arrows. (B) Array CGH demonstrates that the breakpoints corresponding to a loss in 2q32.3-qter and a gain in 17q21.2-qter map to the OBFC2A and RARA genes. (C) The OBFC2A/RARA fusion transcript is amplified with an internal control (β-actin) by RT-PCR. (D) Partial nucleotide sequences surrounding the OBF2CA/RARA fusion region reveal the junction of OBFC2A exon 5 and RARA exon 3. (E) Schematic diagram represents the OBFC2A, RARA, and OBF2CA/RARA fusion proteins, including the activation function-1 (AF-1), DNA-binding domain (DBD), hinge, and ligand-binding domain (LBD).
Identification of a novel OBFC2A/RARA fusion gene by FISH, array CGH, RT-PCR and direct sequencing. (A) Representative partial karyogram shows der(2)t(2;17)(q32;q21). FISH with the RARA break-apart probe reveals a 3′ RARA green signal on der(2), indicated by arrows. (B) Array CGH demonstrates that the breakpoints corresponding to a loss in 2q32.3-qter and a gain in 17q21.2-qter map to the OBFC2A and RARA genes. (C) The OBFC2A/RARA fusion transcript is amplified with an internal control (β-actin) by RT-PCR. (D) Partial nucleotide sequences surrounding the OBF2CA/RARA fusion region reveal the junction of OBFC2A exon 5 and RARA exon 3. (E) Schematic diagram represents the OBFC2A, RARA, and OBF2CA/RARA fusion proteins, including the activation function-1 (AF-1), DNA-binding domain (DBD), hinge, and ligand-binding domain (LBD).
Array CGH analysis to identify the exact region of rearrangement defined a loss in 2q32.3-qter and a gain in 17q21.2-qter. The breakpoints were located within the OBFC2A gene in 2q32.3 and the RARA gene in 17q21.2 (Figure 2B). To confirm the presence of OBFC2A/RARA fusion transcript, we performed RT-PCR using primers specific for OBFC2A and RARA. The predicted 281-bp product was specifically amplified from the patient's cDNA (Figure 2C). Direct sequence analysis showed that the RARA portion of the transcript started in exon 3 and was fused in-frame to exon 5 of OBFC2A (Figure 2D). As a result, the OBFC2A 5′-region encoding the DNA-binding domain (DBD) was fused to the 3′-region of RARA, which includes the DBD and ligand-binding domain (LBD; Figure 2E). The OBFC2A/RARA fusion gene is predicted to encode a 551-amino acid protein.
The OBFC2A gene consists of 7 exons spanning 10.4 kb and ubiquitously encodes a protein component of the heterotrimeric complex SOSS (sensor of single-stranded DNA). SOSS complexes maintain genome stability via DNA damage-response pathways, particularly homologous recombination-dependent repair of DNA double-strand breaks.12,13 It is noteworthy that PML also supports DNA double-strand break repair by homologous recombination.15 The OBF2CA protein is characterized by an N-terminal oligonucleotide/oligosaccharide-binding fold (OB fold) that binds to single-stranded DNA substrate, followed by an unstructured proline/glycine-rich tail region.16 Most of this tail region is deleted in the fusion protein, but the OB fold is retained, providing the possibility of dimerization of the OBFC2A/RARA fusion protein through the OB fold.17
RARA functions by binding as a heterodimer with retinoid X receptor (RXR) to retinoic acid response elements (RARE).3 This RARA-RXR complex is required for the promyelocyte differentiation process. Binding of retinoic acid (RA) to RAR results in a conformational change in the heterodimer, allowing release of co-repressors, recruitment of co-activators, and consequently, gene expression. RARA fusion proteins strongly bind to co-repressors and requires a pharmacologic dose of RA to release co-repressors and recruit co-activators. RA degrades the PML-RARA fusion protein through proteosome-mediated pathways and caspases.18,19 As in all other RARA fusion proteins, OBFC2A/RARA contains the DBD and LBD domains of RARA.4-9,20 The OBFC2A/RARA fusion protein is expected to produce the same biologic effects as those mediated by PML/RARA. It might be a dominant negative mutant for OBFC2A and promote tumor formation. Variant APL with OBFC2A/RARA fusion gene appeared to be sensitive to ATRA by in vitro assay (Figure 1B), although on day 14, there was early morphologic features of differentiation in the bone marrow because of the discontinuation of ATRA for 7 days.
To summarize, we report a patient with a novel OBFC2A/RARA fusion gene that gives rise to ATRA-sensitive variant APL. Additional studies on the OBFC2A/RARA fusion are required to understand its mechanism of leukemogenesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank Jee-Young Lee for performing cytogenetic analysis and Professor Hyun-Sook Chi for providing the patient sample.
Authorship
Contribution: D.W. analyzed data and wrote the paper; S.Y.S. performed FISH experiments; C.-J.P., S.J., and H.-S.C. performed morphologic analysis; K.-H.L. was involved in the management of the patient and provided clinical data; J.-O.L. performed array CGH and molecular studies; and E.-J.S. designed the research, interpreted data, and was responsible for final approval of the draft. All authors reviewed the manuscript and contributed to the final draft.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eul-Ju Seo, MD, PhD, Department of Laboratory Medicine, Asan Medical Center, 88 Olympic-ro 43-gil, Songpa-gu, Seoul 138-736, Korea; e-mail: ejseo@amc.seoul.kr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal