Key Points
The SALL4/NuRD/PTEN pathway is important for acute myeloid leukemogenesis.
Targeting AML can be achieved by blocking the interaction between transcription factor SALL4 and the epigenetic NuRD complex.
Abstract
An exciting recent approach to targeting transcription factors in cancer is to block formation of oncogenic complexes. We investigated whether interfering with the interaction of the transcription factor SALL4, which is critical for leukemic cell survival, and its epigenetic partner complex represents a novel therapeutic approach. The mechanism of SALL4 in promoting leukemogenesis is at least in part mediated by its repression of the tumor suppressor phosphatase and tensin homolog deleted on chromosome 10 (PTEN) through its interaction with a histone deacetylase (HDAC) complex. In this study, we demonstrate that a peptide can compete with SALL4 in interacting with the HDAC complex and reverse its effect on PTEN repression. Treating SALL4-expressing malignant cells with this peptide leads to cell death that can be rescued by a PTEN inhibitor. The antileukemic effect of this peptide can be confirmed on primary human leukemia cells in culture and in vivo, and is identical to that of down-regulation of SALL4 in these cells using an RNAi approach. In summary, our results demonstrate a novel peptide that can block the specific interaction between SALL4 and its epigenetic HDAC complex in regulating its target gene, PTEN. Furthermore, targeting SALL4 with this approach could be an innovative approach in treating leukemia.
Introduction
Members of the SAL gene family belong to a group of C2H2 zinc finger transcription factors characterized by multiple zinc finger domains present in the protein.1,2 Sal is a nonclustered region-specific homeobox gene that plays an essential role in Drosophila, and Sal-related genes have been isolated from Caenorhabditis elegans,3 fish,4 frogs (Xenopus),5,6 mice,7 and humans.2 Each of these homologs is expressed during embryonic development and in a more limited set of specific adult tissues. Murine Sall4 also plays a crucial role in development. Sall4-null mice die at embryonic day 6.5.8-11 We have shown that murine Sall4 is essential for the maintenance of pluripotency and self-renewal properties of embryonic stem cells by interacting with 2 other key regulators, Nanog and Oct4.11,12
After birth, SALL4 expression is down-regulated and absent in most adult tissues. However, SALL4 is expressed in various cancers, including a subset (30%) of solid tumors such as breast cancer,13 ovarian cancer,14 gastric cancer,15 Wilms tumor,16 and germ cell tumors,14,17-21 as well as in leukemias—most notably, almost all cases of human acute myeloid leukemia (AML)22 and approximately 75% of B-cell acute lymphoblastic lymphoma/leukemia.23 We have also reported that SALL4 is enriched in the “side-population” of the leukemia and solid tumor cells. This side population is implicated in drug resistance and cancer initiation and is used to isolate cancer initiation cells.24 SALL4 expression is also correlated with worse prognosis in AML patients.24
We have also shown that SALL4 has functional roles in leukemogenesis. Transgenic mice expressing SALL4 develop myelodysplastic syndrome and AML.22 Loss-of-function studies have demonstrated that SALL4 is a key regulator of leukemic cell survival, and down-regulation of SALL4 led to significant apoptosis of leukemic cells in a cell line model.25 We have recently characterized a “SALL4 leukemic initiation signature” in the transgenic murine model and found that SALL4 mainly acts as a repressor by interacting with the nucleosome remodeling and deacetylase (NuRD) complex that comprises HDAC1 and HDAC2. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN), one of the factors that are essential for the self-renewal of leukemia stem cells,26 is repressed by SALL4 through the NuRD, a HDAC complex.27
In the present study, we tested the hypothesis that blocking the SALL4/HDAC interaction would lead to a biologic effect on cancer cells, particularly in AML cells. We first demonstrated that the HDAC-recruiting region of SALL4 contains 12 amino acids. We then showed that this 12–amino acid peptide promotes growth inhibition of SALL4-expressing leukemic cells, similar to what is observed after treatment with a classic HDAC inhibitor, trichostatin A (TSA) and after down-regulation of SALL4 by shRNA. The antitumor effect of this peptide can be rescued by a PTEN inhibitor. These studies demonstrate a novel therapeutic strategy for inhibition of the growth of tumor cells dependent on SALL4 pathways.
Methods
Peptide synthesis, binding affinity, and delivery assays
Peptides were synthesized by Biosynthesis Inc using standard solid-phase peptide synthesis chemistry and purified by the manufacturer typically to 95% purity. Peptides were dissolved in deionized H2O to concentration of 20mM, diluted in sterile PBS to a final concentration of 2mM and aliquoted and stored at −80°C. For experiments in which peptide was added to nuclear extracts, a 20mM concentration of peptide was added directly to 100 μL of nuclear extract from 1 × 106 cells to final concentration of 0.5, 1.0, 1.5, or 2.0mM, respectively. For delivery of peptide into adherent cell lines, cells were grown in 6-well plates (35-mm dish) to 50% confluence and various concentrations of peptides were added. For each treatment with Pep-1 peptide carrier (Chariot reagent; Active Motif), 10 μL of 2mM diluted peptide was mixed with 10 μL of 10-fold diluted Pep-1 for 30 minutes on ice. Medium was removed, followed by addition of peptides and Pep-1 mixture to the cells along with 400 μL of serum-free medium. The cells were incubated further for 1 hour at 37°C, after which time 500 μL of complete medium (with 10% serum) was added. Alternatively, for nonadherent cells such as HL-60, 20mM peptide diluted in 10 μL of PBS was mixed with 10 μL of diluted Pep-1, incubated for 30 minutes on ice, and added directly to the cell suspension (1 mL of 3 × 105 cells). Primary AML cells were treated the same way as HL-60 by scaling up the peptide and carrier reagent accordingly.
Culture of primary AML samples
Three frozen primary AML patient samples (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were obtained from Brigham and Women's Hospital (Boston, MA) under institutional review board–approved protocol number 2011-P-000096/1. This study was conducted in accordance with the Declaration of Helsinki. Culture conditions were adapted from a previously published protocol.28-31 In brief, after thawing, the frozen AML samples were incubated in RPMI 1640 medium without serum for 1-3 hours and DNA fragments from dead cells were removed by washing. After 3 washes with the medium, 1 × 106 cells per well of a 12-well plate were maintained in 1 mL of serum-free medium (StemSpan-H3000; StemCell Technologies) supplied with StemSpan CC100 cytokine cocktail (StemCell Technologies) that, based on our previous experience, supports 40%-50% viability at 72 hours after thaw culturing. These cells were then used for the down-regulation of SALL4 and peptide treatment experiments.
Xenotransplantation
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory) were bred and maintained in the Children's Hospital Boston animal facility. All animal work was approved by and done according to the guidelines of the institutional animal care and use committee under protocol 10-10-1832. Human primary AML cells exposed to various peptides or carrier only (1.0 × 106 cells per mouse) or transduced with SALL4-shRNA or control lentivirus (1.5 × 106 cell per mouse) were transplanted into 10- to 12-week-old mice, which received 135 cGy of sublethal irradiation 2-4 hours before the injection via the dorsal tail vein. Mice were euthanized when they became ill or at 78 days after transplantation. BM was removed from the 2 femurs by flushing with RPMI 1640 medium, spleen cells were abstained by mincing and filtering through a cell strainer, and peripheral blood was collected from the hearts. These samples were subsequently subjected to flow cytometry analysis using FITC-conjugated anti–human CD45 antibody and APC-conjugated anti–mouse CD45 antibody (eBiosciences). The percentage of human CD45+ cells was calculated as follows: % human CD45+ cells = no. human CD45+ cells/(no. human CD45+ cells + no. mouse CD45+ cells) × 100. In addition, both the Mantel-Cox and Gehan-Breslow-Wilcoxon tests were used for survival analyses.
Results
A peptide derived from the aminoterminal 12–amino acid sequence of SALL4 interacts with the HDAC complex
We have shown previously that SALL4 interacts with NuRD27 and others have suggested that another SALL gene family member, SALL1, can recruit the NuRD complex through interaction with a conserved 12–amino acid sequence at its N-terminus.32-34 Because the N-termini of SALL1 and SALL4 are almost identical, we hypothesized that the N-terminus of SALL4 is involved in the recruitment of HDAC/NuRD (in this manuscript we refer to this 12–amino acid peptide at the N-terminus of SALL4 as wild-type [wt]). It has been shown by others that mutating amino acids 3-5 of this 12–amino acid wt peptide abrogates its binding to the NuRD complex. Among these 3 amino acids, mutation of residue 5 (Lys) alone abolishes the NuRD/HDAC interaction to the greatest extent.33,35,36 Therefore, we mutated residue 5, converting Lys to Ala in the context of the 12–amino acid wt peptide to act as a negative control. A second negative control, scrambled (scr) peptide, was designed with the same 12 amino acids as that of the wt peptide but in an scr sequence. Designing the scr peptide in this manner can maintain the overall net charge of this peptide, which affects cellular uptake of the peptide (Figure 1A).
A peptide derived from the aminoterminus of SALL4 can interact with the NuRD complex components, HDAC1/HDAC2. (A) The top diagram compares the structures of the 2 SALL4 isoforms, SALL4A and SALL4B, demonstrating the conserved aminoterminus. Below are the amino acid sequences of the peptides used in this study. In the mut peptide, Lys 5 is replaced with an alanine residue. (B) The wt peptide associates with HDAC1 and HDAC2. Nuclear extracts from 1 × 106 SNU-398 cells were incubated with 20μM FITC-labeled wt, mut, or scr peptides and then immunoprecipitated with anti-FITC or nonimmune IgG, followed by Western blotting with anti-HDAC1 (top panel) or anti-HDAC2 (bottom panel) antibodies. In the first lane, marked “Input,” 10% of the extracts were subjected to Western blot analysis without immunoprecipitation. (C) The wt peptide associates with HDAC activity. Nuclear extracts from 1 × 106 SNU-398 cells were incubated with 20μM FITC-labeled wt, mut, or scr peptides. Extracts were subsequently immunoprecipitated with anti-FITC antibody and the complexes pulled down were subjected to an HDAC activity assay as described in the Methods. TSA was added at a concentration of 100nM. Input is the sample in which 10% of the extract was assayed for HDAC activity without the immunoprecipitation step. (D) The wt peptide binds with high affinity to protein complexes in SNU-398 nuclear extracts. Competitive binding of nonlabeled wt or scr peptides was measured by fluorescent polarization. A 1μM concentration of FITC-labeled wt peptide was added to nuclear extracts from 2 × 107 SNU-398 cells, along with increasing concentrations of nonlabeled scr or wt peptide. The fluorescent polarization (y axis) is decreased if the FITC-labeled peptide is displaced from a larger complex.
A peptide derived from the aminoterminus of SALL4 can interact with the NuRD complex components, HDAC1/HDAC2. (A) The top diagram compares the structures of the 2 SALL4 isoforms, SALL4A and SALL4B, demonstrating the conserved aminoterminus. Below are the amino acid sequences of the peptides used in this study. In the mut peptide, Lys 5 is replaced with an alanine residue. (B) The wt peptide associates with HDAC1 and HDAC2. Nuclear extracts from 1 × 106 SNU-398 cells were incubated with 20μM FITC-labeled wt, mut, or scr peptides and then immunoprecipitated with anti-FITC or nonimmune IgG, followed by Western blotting with anti-HDAC1 (top panel) or anti-HDAC2 (bottom panel) antibodies. In the first lane, marked “Input,” 10% of the extracts were subjected to Western blot analysis without immunoprecipitation. (C) The wt peptide associates with HDAC activity. Nuclear extracts from 1 × 106 SNU-398 cells were incubated with 20μM FITC-labeled wt, mut, or scr peptides. Extracts were subsequently immunoprecipitated with anti-FITC antibody and the complexes pulled down were subjected to an HDAC activity assay as described in the Methods. TSA was added at a concentration of 100nM. Input is the sample in which 10% of the extract was assayed for HDAC activity without the immunoprecipitation step. (D) The wt peptide binds with high affinity to protein complexes in SNU-398 nuclear extracts. Competitive binding of nonlabeled wt or scr peptides was measured by fluorescent polarization. A 1μM concentration of FITC-labeled wt peptide was added to nuclear extracts from 2 × 107 SNU-398 cells, along with increasing concentrations of nonlabeled scr or wt peptide. The fluorescent polarization (y axis) is decreased if the FITC-labeled peptide is displaced from a larger complex.
We first evaluated whether the wt peptide or the mutant (mut) or scr controls could bind to HDAC1 and HDAC2, which comprise part of the NuRD complex.37 The hepatocellular carcinoma (HCC) cell line SNU-398 expresses an abundant amount of SALL4 and was chosen for the following experiments. Pull-down by an anti-FITC antibody on nuclear extracts from SNU-398 cells pretreated with FITC-labeled wt, mut, or scr peptides revealed that HDAC1 and HDAC2 could be easily detected in nuclear complexes obtained from the cells treated with wt peptide. Much reduced HDAC binding to the mut peptide was observed and binding to the scr peptide was undetectable (Figure 1B). We further hypothesized that if the peptide interacted with HDAC1 and/or HDAC2, then the peptide pull-down could exhibit HDAC activity. To test this possibility, we measured the histone deacetylase activity of the peptide pull-down complex using a fluorescent assay kit (Figure 1C). The active enzymatic activity of the HDACs tested by this assay corresponded to the amount of the HDAC proteins in the sample. Although the wt peptide pull-down had the most HDAC activity and the mut showed some residual activity, the scr peptide, which did not bind to HDAC, exhibited no HDAC activity in the pull-down, comparable to that observed with the IgG control.
We next tested the binding affinity of the wt and scr peptides to the HDAC complex. For this, we used fluorescent polarization (FP), a technology based on molecular movement and rotation in which fluorescent molecules are excited with polarized light. If the fluorescent molecules are part of a large complex, they rotate less and the emitted light has high polarity. In contrast, smaller molecules rotate faster and have low polarity (“depolarized”). FP has long been a valuable biophysical research tool for investigating protein-protein or peptide-protein interactions at the molecular level. When a fluorescence-labeled peptide binds to a target protein or a complex, it becomes part of the large molecular complex, is less mobile, and has high polarization. However, an excessive amount of unlabeled peptide added to this mixture will displace the labeled peptide and the free labeled peptide will have low polarization. Using this approach, FITC-labeled wt or scr peptides were first mixed with nuclear extracts from SNU-398 cells. Serial dilutions of unlabeled wt or scr peptides were then added as competitors, respectively, and measured by FP assay. Although the wt peptide had a half-maximal inhibitory concentration of 5μM, that for the scr peptide could not be calculated because of the lack of binding activity (Figure 1D and supplemental Figure 1). In summary, the 12–amino acid wt peptide derived from the aminoterminus of SALL4 can bind to an active HDAC complex.
The wt peptide competes with endogenous SALL4 to block interaction with HDAC and repression of PTEN
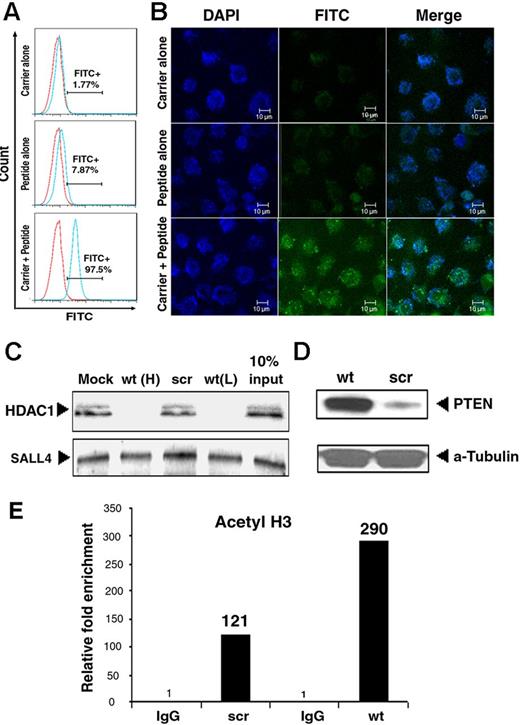
We next tested the cellular uptake and localization of these peptides in various types of cells using FITC-labeled wt or scr peptides. Despite the net positive charge of these peptides, their penetration into cells is cell-type specific. Some cell lines, such as HL-60 (an AML cell line), allowed peptide entry without any modification or facilitator, whereas others such as SNU-398 were not penetrable at all. To achieve a similar level of peptide delivery across a range of cell types, we used a 21–amino acid residue peptide carrier, Pep-1.38 The overall success rate of peptide uptake by the cells was more than 95% with Pep-1, as evaluated by flow cytometry (Figure 2A). In addition, confocal microscopic studies confirmed the uptake of these peptides throughout the cell, including nuclei (as evidenced by the DAPI overlay in Figure 2B). We further confirmed that Pep-1 could facilitate peptide entry into all cell types at high levels (supplemental Table 2) and Pep-1 was therefore used in subsequent studies.
The wt peptide blocks SALL4 repression of PTEN. (A) Flow cytometric analysis on SNU-398 cells treated with Pep-1 carrier peptide alone (top panel), wt peptide alone (middle panel), or Pep-1 carrier + peptide (bottom panel). More than 90% of cells showed FITC-labeled peptide uptake after the use of Pep-1 carrier. (B) Confocal images of the distribution of FITC-labeled peptide in SNU-398 cells treated as shown in panel A. (C) The endogenous SALL4 interaction with HDAC is blocked by wt (wt(H): 20μM; wt(L): 10μM), but not scr peptide. SALL4 was immunoprecipitated from SNU-398 nuclear extracts (1 × 106 cells) pretreated with wt or scr peptides as indicated using an anti-SALL4 antibody. Immunoprecipitates were analyzed by Western blot using HDAC1 (top panel) or SALL4 (bottom panel) antibodies. The interaction was completely abrogated by pretreatment with 20μM wt peptide (lane 2) but not scr peptide (lane 3). An equal amount of SALL4 protein was present in all samples. In lane 4 (10% input), 10% of the amount of nuclear extract used in lanes 1 through 3 was subjected to Western blot analysis without immunoprecipitation. (D) Western blot showed increased PTEN expression after wt peptide treatment compared with scr. α-tubulin was used as a loading control. (E) ChIP-quantitative RT-PCR showed increased H3 acetylation markers at the PTEN promoter in SNU-398 cells on wt peptide treatment. Enrichment with control IgG was set at 1.
The wt peptide blocks SALL4 repression of PTEN. (A) Flow cytometric analysis on SNU-398 cells treated with Pep-1 carrier peptide alone (top panel), wt peptide alone (middle panel), or Pep-1 carrier + peptide (bottom panel). More than 90% of cells showed FITC-labeled peptide uptake after the use of Pep-1 carrier. (B) Confocal images of the distribution of FITC-labeled peptide in SNU-398 cells treated as shown in panel A. (C) The endogenous SALL4 interaction with HDAC is blocked by wt (wt(H): 20μM; wt(L): 10μM), but not scr peptide. SALL4 was immunoprecipitated from SNU-398 nuclear extracts (1 × 106 cells) pretreated with wt or scr peptides as indicated using an anti-SALL4 antibody. Immunoprecipitates were analyzed by Western blot using HDAC1 (top panel) or SALL4 (bottom panel) antibodies. The interaction was completely abrogated by pretreatment with 20μM wt peptide (lane 2) but not scr peptide (lane 3). An equal amount of SALL4 protein was present in all samples. In lane 4 (10% input), 10% of the amount of nuclear extract used in lanes 1 through 3 was subjected to Western blot analysis without immunoprecipitation. (D) Western blot showed increased PTEN expression after wt peptide treatment compared with scr. α-tubulin was used as a loading control. (E) ChIP-quantitative RT-PCR showed increased H3 acetylation markers at the PTEN promoter in SNU-398 cells on wt peptide treatment. Enrichment with control IgG was set at 1.
We next investigated whether wt peptide could compete for the interaction of endogenous SALL4 with HDAC1. SNU-398 cells were treated with wt or scr peptides along with the peptide carrier Pep-1. Binding to HDAC1 by endogenous SALL4 was completely abrogated when cells were treated with the wt, but not the scr, peptide (Figure 2C). This result demonstrated that wt peptide can compete with SALL4 in recruiting HDACs.
We then investigated whether this peptide could reverse the repressive effect of SALL4 on its target genes. We focused on PTEN because it is implicated in SALL4-mediated leukemogenesis.25,27 In addition, we have previously reported that SALL4 could specifically bind to the PTEN promoter region in a ChIP assay.27 When SALL4 was overexpressed in 293T cells, we observed an enrichment of SALL4 and decreased H3 acetylation at this binding site, which was correlated with decreased PTEN RNA expression.27 Therefore, we hypothesized that SALL4 could recruit the HDAC/NuRD complex to repress PTEN and that the wt peptide would compete with endogenous SALL4 in recruiting the HDAC/NuRD complex to the PTEN promoter and release the repression effect of SALL4 on PTEN. Furthermore, this model would predict that wt peptide treatment of SALL4-expressing cells would lead to increased epigenetic markers of gene activation, such as H3 acetylation, at the PTEN promoter–binding site, as well as an increase in PTEN expression. To test this possibility, SNU-398 cells were treated with scr or wt peptide and subsequently evaluated for PTEN expression and H3 acetylation at the SALL4-binding site at the PTEN promoter region. The wt peptide-treated SNU-398 cells demonstrated increased PTEN protein (Figure 2D) and increased H3 acetylation on the PTEN promoter region (Figure 2E), which was not observed with scr peptide–treated SNU-398 cells (Figure 2D-E). Similar results were also observed when THP-1 was used (supplemental Figure 2). Therefore, our results are compatible with the model that SALL4 recruits HDACs to repress PTEN expression and that blocking the SALL4-HDAC interaction with the wt peptide results in increased expression of PTEN.
The wt peptide exerts an antiproliferative effect on SALL4-expressing cancer cells
Multiple leukemic and solid tumor cancer cell lines with high or low/no SALL4 expression (Figure 3A) were chosen to test the biologic effect(s) of the wt peptide. The wt or scr peptide was delivered into tumor cells at various concentrations and their effects on cells were evaluated 72 hours after treatment using the MTS cell viability assay. Because we demonstrated that the wt peptide could compete with SALL4 in the recruitment of the HDAC-containing NuRD complex and could potentially work as an HDAC inhibitor, we used the HDAC inhibitor TSA as a positive control in the same group of experiments. All SALL4-expressing leukemic or solid tumor cell lines, including HL-60, THP1, HuH-7, and SNU-398, exhibited decreased cell viability when treated with wt peptide but remained unaffected by treatment with scr peptide (Figure 3B). TSA treatment led to a similar antiproliferative effect on these cancer cells. The effect of wt peptide is SALL4 specific, because it had no effect on KBM5, an AML cell line with undetectable endogenous SALL4 expression. We were surprised to see that the wt peptide had no effect on a high SALL4-expressing endometrioid cancer cell line AN3CA. This prompted us to investigate whether PTEN, a gene repressed by SALL4 through its interaction with NuRD,27 is present in this cell line. According to our model, the function of wt peptide requires both the expression of SALL4 and intact SALL4 downstream target genes such as PTEN. In tumor cells with PTEN deletion, even though wt peptide could compete with endogenous SALL4 in interacting with the HDAC complex, we might not observe an antiproliferative effect. Therefore, the expression of PTEN was evaluated in AN3CA cells along with other cell lines. Although the PTEN protein was present in all of the other cell lines, it was absent in AN3CA cells (Figure 3C), which is consistent with the reported deletion mutation of the gene in this cell line.39,40 These results suggest that the action of the peptide requires expression of SALL4 and that in cells such as KBM5 lacking SALL4, it is not effective. Furthermore, the antiproliferative effect also requires certain downstream targets such as PTEN.
The wt peptide reduces tumor cell viability in a SALL4/PTEN-dependent manner. (A) SALL4 expression in various tumor cell lines. SALL4 RNA expression was measured by quantitative RT-PCR and normalized to GAPDH. Expression of SALL4 in KBM5 cells was set as 1. (B) Reduced viability of tumor cells treated with wt peptide and TSA at various concentrations was observed in SALL4-expressing THP1, HL-60, SNU-398, and HuH-7 cell lines, but not in non-SALL4–expressing KBM5 cells at 72 hours. Surprisingly, AN3CA, a uterine cancer cell line with high SALL4 expression did not respond to wt peptide treatment. Cell viability (y axis) represents the relative result of the MTS assay. The value for each cell line with Pep-1 treatment alone was set as 1 (n = 3). *P < .05. (C) Western blot on endogenous PTEN protein expression in various untreated cancer cell lines. AN3CA has no detectable endogenous PTEN. (D) Peptide treatment can affect the PTEN/AKT pathway. Shown is a Western blot of the protein expression levels of PTEN, pAKT, and total AKT after peptide treatments. (E) A PTEN inhibitor (SF1670) can reverse wt peptide effects on viability of THP1 (SALL4+) cells, but has no effect on KBM5 (SALL4−) cells. The graph shows the results of MTS analysis of cells treated with wt peptide, scr peptide, TSA (100nM), or wt peptide + PTEN inhibitor SF1670 (wt + inh; n = 3). *P < .05.
The wt peptide reduces tumor cell viability in a SALL4/PTEN-dependent manner. (A) SALL4 expression in various tumor cell lines. SALL4 RNA expression was measured by quantitative RT-PCR and normalized to GAPDH. Expression of SALL4 in KBM5 cells was set as 1. (B) Reduced viability of tumor cells treated with wt peptide and TSA at various concentrations was observed in SALL4-expressing THP1, HL-60, SNU-398, and HuH-7 cell lines, but not in non-SALL4–expressing KBM5 cells at 72 hours. Surprisingly, AN3CA, a uterine cancer cell line with high SALL4 expression did not respond to wt peptide treatment. Cell viability (y axis) represents the relative result of the MTS assay. The value for each cell line with Pep-1 treatment alone was set as 1 (n = 3). *P < .05. (C) Western blot on endogenous PTEN protein expression in various untreated cancer cell lines. AN3CA has no detectable endogenous PTEN. (D) Peptide treatment can affect the PTEN/AKT pathway. Shown is a Western blot of the protein expression levels of PTEN, pAKT, and total AKT after peptide treatments. (E) A PTEN inhibitor (SF1670) can reverse wt peptide effects on viability of THP1 (SALL4+) cells, but has no effect on KBM5 (SALL4−) cells. The graph shows the results of MTS analysis of cells treated with wt peptide, scr peptide, TSA (100nM), or wt peptide + PTEN inhibitor SF1670 (wt + inh; n = 3). *P < .05.
The wt peptide affects both PTEN and its downstream target, pAKT
To further evaluate the specificity of the SALL4/PTEN pathway, we next investigated whether wt peptide treatment could affect the expression of PTEN and its downstream target, phosphorylated AKT (pAKT). Indeed, we observed increased PTEN expression after wt peptide or TSA treatment of SALL4-expressing THP1 cells, whereas the corresponding levels of pAKT on Ser 473 were significantly reduced. No effect on the PTEN/pAKT pathway after scr peptide treatment was observed in THP1 cells (Figure 3D left panel). In contrast, treatment of the non-SALL4–expressing KBM5 cells with wt peptide had little or no effect on the levels of PTEN and pAKT in this cell line (Figure 3D right panel) compared with scr peptide treatment. In addition, wt peptide treatment had no effect on endogenous SALL4 expression (supplemental Figure 3).
To further investigate whether the effect of wt peptide on tumor cells is related to PTEN, we used a PTEN inhibitor (SF1670).41 This PTEN inhibitor does not affect the expression level of PTEN, but can reverse the effect of PTEN on dephosphorylation of its downstream target, pAKT.41 Although SALL4-expressing THP1 cells treated with wt peptide alone showed decreased cell viability, co-treatment with this PTEN inhibitor restored the cell viability to the baseline levels, similar to what happened after scr peptide treatment (Figure 3E left panel). As a control for the specificity of SALL4, we also treated KBM5, a non-SALL4–expressing cell line, with a PTEN inhibitor. Neither wt or scr peptide treatment showed any change in cell viability, and co-treatment of this PTEN inhibitor with wt peptide in this cell line also had no effect (Figure 3E right panel). Results similar to that observed in THP1 were seen in HCC cell lines such as SNU-398 cells (high SALL4 expression), whereas SNU-387 cells (with undetectable SALL4 expression) demonstrated a pattern similar to KBM5 cells (data not shown).
In summary, the antiproliferative effect of this wt peptide was only observed in SALL4-expressing tumor cells with an intact PTEN gene and can be rescued with a PTEN inhibitor. Combining our observations on the biologic effects of the peptide treatment and the expression of PTEN and pAKT in these cells, we conclude that the role of the wt peptide in promoting tumor cell death is correlated with its effect on the expression levels of PTEN and pAKT, suggesting that it is working, at least in part, specifically through the PTEN/AKT pathway.
Treatment of primary AML cells with the SALL4 peptide leads to impaired leukemic engraftment in vivo similar to that of down-regulation of SALL4
We have shown previously that SALL4 is critical for leukemic cell survival in the AML cell line NB4.25 In the present study, we investigated whether this is also the case for primary human AML samples. Three SALL4/PTEN-expressing AML samples were selected for experiments in which cells were first tested under culture conditions that maintain/promote the viability of AML cells.28-31 The same SALL4-specific shRNA that was validated in our previous studies12,25,42 was used in these AML patient samples (Figure 4A). Forty-eight hours after transduction, we determined the viability of cells by flow cytometry after staining with annexin V and propidium iodide (PI). Live cells are annexin V− and PI−, the double-negative population. When the living cell percentage was normalized to 100% in the control scr shRNA-treated group, decreased viability was observed after reduction of SALL4 expression in primary AML cells in culture (SALL4 shRNA1: 80.9% ± 2.75%, n = 3, P = .0002; SALL4 shRNA2: 87.3% ± 4.59%, n = 3, P = .0008; Figure 4B). Furthermore, in a xenotransplantation model, we observed increased survival of leukemic cell recipient NSG mice after knocking down SALL4 expression in the primary AML samples.
Down-regulation of SALL4 by shRNA leads to decreased cell viability of primary human AML cells in culture and reduced leukemic development in vivo. (A) Down-regulation of SALL4 RNA in primary AML cells using shRNA. Control scr shRNA-infected cells and SALL4 shRNA-infected cells (shRNA1 and shRNA2) were analyzed 48 hours after transduction. The expression of SALL4 RNA in AML cells infected with SALL4 shRNA-expressing retroviruses was reduced to 35% of those infected with scr pRS control vectors, evaluated by quantitative RT-PCR after normalized to GAPDH (n = 3 biologic samples). Error bars indicate SD. (B) Increased apoptosis and cell death were observed via flow cytometry analysis of annexin V/PI staining in AML cells on SALL4 knock-down (shRNA1 and shRNA2). Data were derived from 3 independent experiments. Viable cells were defined as the double-negative (annexin V−/PI−) population. The viability was set as 100 for the control group. (C) Xenotransplantation showed increased survival of mice receiving SALL4-reduced leukemic cells. A total of 1.5 million primary human AML cells were transduced as described in the Methods and cultured for 48 hours before transplantation via tail vein injection. Although the median survival of recipient mice with control scr shRNA retrovirus-transduced primary human AML cells (n = 7) was 33 days, the median survival of recipient mice with SALL4 shRNA retrovirus-infected primary human AML cells (n = 6) was 109 days. The log-rank (Mantel-Cox) P = .03 and the Gehan-Breslow-Wilcoxon P = .01. (D-F) Leukemia development in xenotransplant recipient mice. AML is defined as a blast count more than 20% in the peripheral blood and/or BM with multiple organ involvements observed in recipient mice. Blasts were present in liver (D, 200×), spleen (E, 200×), and BM (F, 200×) as assessed by H&E staining. (G-H) AML cells in xenograft recipients were human CD45+. Flow cytometry was performed on BM from recipients after transduction with control scr (G) or SALL4 shRNA (H) retrovirus. The red line represents isotype control and the blue line represents anti–human CD45 antibody.
Down-regulation of SALL4 by shRNA leads to decreased cell viability of primary human AML cells in culture and reduced leukemic development in vivo. (A) Down-regulation of SALL4 RNA in primary AML cells using shRNA. Control scr shRNA-infected cells and SALL4 shRNA-infected cells (shRNA1 and shRNA2) were analyzed 48 hours after transduction. The expression of SALL4 RNA in AML cells infected with SALL4 shRNA-expressing retroviruses was reduced to 35% of those infected with scr pRS control vectors, evaluated by quantitative RT-PCR after normalized to GAPDH (n = 3 biologic samples). Error bars indicate SD. (B) Increased apoptosis and cell death were observed via flow cytometry analysis of annexin V/PI staining in AML cells on SALL4 knock-down (shRNA1 and shRNA2). Data were derived from 3 independent experiments. Viable cells were defined as the double-negative (annexin V−/PI−) population. The viability was set as 100 for the control group. (C) Xenotransplantation showed increased survival of mice receiving SALL4-reduced leukemic cells. A total of 1.5 million primary human AML cells were transduced as described in the Methods and cultured for 48 hours before transplantation via tail vein injection. Although the median survival of recipient mice with control scr shRNA retrovirus-transduced primary human AML cells (n = 7) was 33 days, the median survival of recipient mice with SALL4 shRNA retrovirus-infected primary human AML cells (n = 6) was 109 days. The log-rank (Mantel-Cox) P = .03 and the Gehan-Breslow-Wilcoxon P = .01. (D-F) Leukemia development in xenotransplant recipient mice. AML is defined as a blast count more than 20% in the peripheral blood and/or BM with multiple organ involvements observed in recipient mice. Blasts were present in liver (D, 200×), spleen (E, 200×), and BM (F, 200×) as assessed by H&E staining. (G-H) AML cells in xenograft recipients were human CD45+. Flow cytometry was performed on BM from recipients after transduction with control scr (G) or SALL4 shRNA (H) retrovirus. The red line represents isotype control and the blue line represents anti–human CD45 antibody.
To ensure that similar viable leukemic cells were given to the recipient mice, early time point engraftment was measured, which showed a similar human leukemic cell engraftment between the control-treated and SALL4 knock-down recipients at 24 hours after transplantation. However, an increased engraftment in the control-treated recipient group compared with the SALL4 knockdown recipient group was observed at 48 hours after transplantation (supplemental Figure 4). In the long-term follow-up study, all recipients receiving AML cells transduced with scr control shRNA (n = 7) died within 3 months, whereas only 2 of 6 mice receiving SALL4 shRNA-treated primary AML died during the study period (Figure 4C and supplemental Table 3).
Although the median survival of scr shRNA control recipient mice (n = 7) was 33 days, the median survival of SALL4 shRNA recipient mice (n = 6) was 109 days (P = .01). The transplanted leukemia disease was characterized by immature blasts with human CD45 expression in the peripheral blood, BM, and tissues such as the liver and spleen (Figure 4D-G). In contrast, nonleukemic SALL4 shRNA recipient mice showed less than 0.5% human CD45+ cells (Figure 4H) in the BM or spleen samples.
We next investigated whether we could achieve a therapeutic effect using the wt peptide in primary human AML samples. The same SALL4/PTEN-expressing primary AML samples were selected for peptide treatment experiments. Leukemic cells were subjected to 3 treatment regimens: peptide (wt or scr), peptide + PTEN inhibitor, or HDAC inhibitor (TSA). To assist peptide delivery into cells, a carrier agent, Pep-1, was used for all the experiments and control-treated cells were given Pep-1 alone (mock). At 96 hours from the commencement of first treatment dose, cellular viability was determined by flow cytometry after staining with annexin V and PI. We observed a significant decrease in the cell viability of primary AML cells after wt peptide treatment, which could be rescued by the PTEN inhibitor SF1670 (Figure 2A). Cells treated with either scr peptide or the Pep-1 peptide carrier (Mock) had more than 97% cell viability, which was similar to that of nontreated cells at the end of day 4. However, we observed only 55.3% ± 12.02% cell viability on wt peptide treatments (n = 3, P = .006) that could in part be rescued by the PTEN inhibitor SF1670, which restored cell viability back to 78.3% ± 11.7% (n = 3). Similar to wt peptide treatment, TSA also induced cell death, with a cell viability of 54.7% ± 21.6% observed at day 4 after treatment (n = 3, P = .041; Figure 5A).
Treatment of SALL4 peptide in primary human AML cells induces reduced cell viability in culture and impaired leukemic engraftment in vivo. (A) wt peptide treatment reduces cell viability of human AML cells in culture and this effect can be reversed in part by the PTEN inhibitor SF1670. A total of 1 × 106 AML cells were left untreated (None); treated with Pep-1 alone (Mock); treated with 20μM wt or scr peptide; treated with wt peptide + 400nM PTEN inhibitor SF1670 (wt + inh); or treated with 100nM TSA for 48 hours, as described in “Methods.” Cell viability of untreated primary AML cells is set as 100 (n = 3 biologic samples). Error bars indicate SD. (B) Schematic diagram showing the steps and time course of the peptide treatment in the xenotransplantation assay. One million primary human leukemic cells were treated twice with peptides at a 24-hour interval, followed by transplantation into sublethally irradiated NSG mice. Mice were killed when they became ill. (C) Wright-Giemsa staining of a cytospin preparation of BM from scr- or carrier alone–treated mice shows marked expansion of immature blasts, which is not present in the wt-treated recipient BM (left panel) Scale bar indicates 10 μm. The scr- or Pep-1 carrier alone–treated mice also have enlarged spleens (middle panel) and kidneys (right panel), whereas the wt-treated recipient mice show normal spleens and kidneys. (D) Histology section of the kidney (arrow) shows effacement of normal architecture by leukemic infiltration. Scale bar indicates 20 μm. (E-F) SALL4 wt peptide treatment significantly impaired human AML cell engraftments in NSG mice analyzed at 78 days after transplantation. The percentage of human CD45+ cells engrafted in the BM, spleen, and peripheral blood was determined by flow cytometry. Representative FACS results from wt, scr, and Pep-1 carrier–only (mock) treatments are shown in panel E and a statistical summary is shown in panel F (n = 5 mice per group). P < .01 in BM; P < .05 in the spleen and peripheral blood by ANOVA with the Tukey multiple comparison test.
Treatment of SALL4 peptide in primary human AML cells induces reduced cell viability in culture and impaired leukemic engraftment in vivo. (A) wt peptide treatment reduces cell viability of human AML cells in culture and this effect can be reversed in part by the PTEN inhibitor SF1670. A total of 1 × 106 AML cells were left untreated (None); treated with Pep-1 alone (Mock); treated with 20μM wt or scr peptide; treated with wt peptide + 400nM PTEN inhibitor SF1670 (wt + inh); or treated with 100nM TSA for 48 hours, as described in “Methods.” Cell viability of untreated primary AML cells is set as 100 (n = 3 biologic samples). Error bars indicate SD. (B) Schematic diagram showing the steps and time course of the peptide treatment in the xenotransplantation assay. One million primary human leukemic cells were treated twice with peptides at a 24-hour interval, followed by transplantation into sublethally irradiated NSG mice. Mice were killed when they became ill. (C) Wright-Giemsa staining of a cytospin preparation of BM from scr- or carrier alone–treated mice shows marked expansion of immature blasts, which is not present in the wt-treated recipient BM (left panel) Scale bar indicates 10 μm. The scr- or Pep-1 carrier alone–treated mice also have enlarged spleens (middle panel) and kidneys (right panel), whereas the wt-treated recipient mice show normal spleens and kidneys. (D) Histology section of the kidney (arrow) shows effacement of normal architecture by leukemic infiltration. Scale bar indicates 20 μm. (E-F) SALL4 wt peptide treatment significantly impaired human AML cell engraftments in NSG mice analyzed at 78 days after transplantation. The percentage of human CD45+ cells engrafted in the BM, spleen, and peripheral blood was determined by flow cytometry. Representative FACS results from wt, scr, and Pep-1 carrier–only (mock) treatments are shown in panel E and a statistical summary is shown in panel F (n = 5 mice per group). P < .01 in BM; P < .05 in the spleen and peripheral blood by ANOVA with the Tukey multiple comparison test.
We also tested the ability of peptide treatment to inhibit the development of AML in vivo using xenografts. Consistent with our cell-culture observations, we observed reduced engraftment of human AML cells treated with wt peptides after xenotransplantation. Primary human AML cells treated with Pep-1 only or Pep-1 + wt or scr peptide were transplanted into sublethally irradiated NSG mice by tail vein injection. To ensure that similar viable leukemic cells were given to the recipient mice, early time point engraftment was measured and showed a similar human leukemic cell engraftment between the scr and wt peptide–treated recipients at 24 hours after transplantation. However, at 2 weeks after transplantation, the engraftment from scr peptide–treated recipients was statistically higher than that from wt peptide–treated recipients (supplemental Figure 5). A long-term follow-up study was also conducted. Eleven weeks after transplantation, mice that had received Pep-1-alone or scr peptide became moribund and all mice were euthanized for analysis (Figure 5B). The recipient mice from both the Pep-1 alone- or Pep-1 + scr peptide–treated group had blasts on BM cytospin, splenomegaly, and leukemic infiltration of the kidneys (Figure 5C-D). In addition, we evaluated the engraftment of human CD45+ cells by flow cytometry (Figure 5E). Although the average engraftment of Pep-1 alone- or Pep-1 + scr peptide–treated cells was greater than 74% in the BM, the average engraftment of the Pep-1 + wt peptide–treated group was only 0.5%. Similar differences were observed in the spleen and peripheral blood (Figure 5F). These results demonstrate that blocking the SALL4 interaction with HDAC/NuRD inhibits the leukemic properties of human AML cells both in culture and in xenograft models, similar to what we observed after down-regulation of SALL4 in these cells.
Discussion
Approximately 13 000 new cases and 9000 deaths from AML were estimated to occur in the United States in 2010. This is a disease in which standard chemotherapy has not changed in more than 25 years and survival remains extremely poor. To develop effective therapeutics, it is important to understand the mechanism(s) driving the development of AML and translate that knowledge into more targeted and effective therapy. Transcription factors play a key role in tumor development, including leukemogenesis, and some of these transcription factors are being used as diagnostic and prognostic markers in cancers. Targeting transcription factors in cancer through blocking of oncogenic complexes formation has been an exciting recent approach.43-46
HDAC inhibitors have been used in various cancer treatments with variable results. The best therapeutic outcomes to date are observed in hematologic malignancies. Most of the existing HDAC inhibitors target the enzymatic activities of HDACs, which are in general nonspecific and indiscriminately reexpress silenced genes in normal and tumor cells. The antineoplastic effects and functional mechanisms of HDAC inhibitors are likely tissue specific and context dependent, depending on transcription factors. Targeting the transcription factor(s) that recruits HDACs in cancers could potentially achieve more specific therapeutic effects.
The embryonic stem cell gene SALL4 encodes a zinc finger transcription factor. Its expression is down-regulated during development and it is absent in most adult tissues, but it is aberrantly reexpressed in cancer cells, including AML. Knocking down the SALL4 gene by shRNA in leukemia and solid tumors leads to cell death and growth inhibition both in vitro and in vivo.13,25,42,47,48 The unique expression pattern of SALL4 and its essential functional role for cancer cell survival makes it an ideal candidate for targeting cancer cells.
The important role of transcription factor SALL4 in leukemic stem or initiating cells is supported by its interactions with several key players in the self-renewal of embryonic stem cells and leukemic stem or initiating cells, particularly by repression of PTEN expression. We have discovered that SALL4 acts mainly as a repressor by interacting with an epigenetic HDAC/NuRD complex and that the oncogenic role of SALL4 in cancer development is in part through its repressive function on the tumor suppressor PTEN that occurs by recruiting the HDAC/NuRD complex. In the present study, we investigated whether blocking the SALL4 interaction with the HDAC/NuRD complex could have a biologic effect in cancer cells, particularly AML cells. We hypothesized that HDAC inhibitors (either by targeting the enzymatic activities of HDAC and/or disrupting the interaction between the HDAC and its transcription factor recruiter) can impair the repressor function of SALL4 and negatively affect the self-renewal and survival of leukemic cells by reactivating the expression of PTEN (Figure 6). In the present study, we present data that support this hypothesis. First, we have demonstrated that the SALL4 HDAC/NuRD–recruiting region is located at the N-terminus of SALL4 and contains 12 critical amino acids. Furthermore, we have shown that this 12–amino acid peptide has demonstrated growth inhibition of leukemic cells similar to that of classic HDAC inhibitors such as TSA and to down-regulation of the SALL4 gene by shRNA. This supports our proposal that some of the therapeutic effects of HDAC inhibitors on hematologic malignancies are probably mediated by the specific effect(s) of transcription factor SALL4. These studies not only lead to a novel approach in treating AML, but also provide new understanding of the mechanisms of action of HDAC inhibitors in AML.
Working model of the novel peptide or a small molecule that targets the interaction between the HDAC complex and its transcription factor recruiter, SALL4. (A) Top panel: SALL4 represses its downstream targets by recruiting an HDAC complex, NuRD, to specific promoter regions such as the PTEN promoter, resulting in histone deacetylation, a more compact chromatin structure, and transcription repression. Bottom panel: the wt peptide (or a small molecule) competes with SALL4 in interacting with NuRD. The repression of PTEN by SALL4 is therefore lost and the PTEN expression is up-regulated, leading to tumor growth inhibition. (B) In some tumor cells (such as the human endometrioid cancer line AN3CA), PTEN is deleted, so the disruption of the SALL4/HDAC complex does not affect cell growth. (C) Some tumors (such as KBM5) do not express SALL4, in which case PTEN will be regulated independently of SALL4 and the wt peptide will not have any effects on cell growth.
Working model of the novel peptide or a small molecule that targets the interaction between the HDAC complex and its transcription factor recruiter, SALL4. (A) Top panel: SALL4 represses its downstream targets by recruiting an HDAC complex, NuRD, to specific promoter regions such as the PTEN promoter, resulting in histone deacetylation, a more compact chromatin structure, and transcription repression. Bottom panel: the wt peptide (or a small molecule) competes with SALL4 in interacting with NuRD. The repression of PTEN by SALL4 is therefore lost and the PTEN expression is up-regulated, leading to tumor growth inhibition. (B) In some tumor cells (such as the human endometrioid cancer line AN3CA), PTEN is deleted, so the disruption of the SALL4/HDAC complex does not affect cell growth. (C) Some tumors (such as KBM5) do not express SALL4, in which case PTEN will be regulated independently of SALL4 and the wt peptide will not have any effects on cell growth.
Although PTEN is being used here as a “proof-of-concept” example for our proposed peptide action model, we expect that other SALL4 target genes may be affected by the peptide as well. We are in the process of performing global expression profiling after the peptide treatment in SALL4-expressing cancers, including AML. The expression of SALL4 activated genes such as c-myc are not affected by peptide treatment (data not shown).
To determine whether targeting SALL4 with PTEN can be used in treating solid tumors, we treated the HCC cell line with wt and control scr and mut peptides. We found that, similar to our observations in AML, treating SALL4-expressing HCC cell lines can lead to cell death in culture and decreased tumorigenesis in murine xenograft models, identical to the phenotype generated by loss-of-function of SALL4 using an shRNA approach (K.J.Y. and L.C, manuscript under review). These findings suggest targeting SALL4 can also be used as an innovative approach in treating solid tumors.
Although peptide shows potential for anticancer usage, we are concerned about its potential toxic effects on normal cells, particular hematopoietic stem/progenitor cells. In the present study, we treated the selected normal CD34+ cells with this peptide or its controls, cultured these cells under ex vivo culture conditions for 7-8 days, and then transplanted them into sublethally irradiated NSG recipient mice. Thus far, we have not observed any cytotoxicities such as apoptosis/death of the CD34+ cells during ex vivo culture or any negative impact on in vivo engraftment after peptide treatment (H.T. and L.C., manuscript in preparation). The long-term in vivo effect of this peptide on additional normal tissues is still under study.
The results of the present study demonstrate that PTEN can be used as a cell type–specific, SALL4 gene–specific HDAC inhibitor and/or as a SALL4/PTEN modulator in cancer treatment. Future studies will focus on optimizing peptide design and the development of small-molecule peptide mimetics that can specifically block/compete the interaction between SALL4 and the HDAC complex in the treatment of leukemia and other malignancies. We believe that this novel approach of targeting the interaction between a transcription factor (SALL4) and its epigenetic complex (HDAC/NuRD) will serve as a paradigm for targeting other transcription factors in other malignancies, both in solid tumors and leukemia, and thus provide a new direction in targeted cancer therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Joline Lim for assistance in the preparation of the manuscript.
This work was supported in part by the National Institutes of Health (grant PO1 DK080665 to D.G.T. and grants RO1HL092437 and PO1HL095489 to L.C.).
National Institutes of Health
Authorship
Contribution: C.G., T.D., K.J.Y., and H.T. performed the experiments and wrote the manuscript; K.J.Y. contributed to the design of the experiments; H.R.L. provided critical reagents; J.E.B. designed the experiments; H.-w.J. performed the experiments; and D.G.T. and L.C. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Li Chai, Department of Pathology, Joint Program in Transfusion Medicine, Brigham and Women's Hospital, Harvard Medical School, 75 Francis St, Boston, MA 02115; e-mail: lchai@partners.org.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal