Key Points
We demonstrate a critical role for histone deacetylase 1 and 2 (HDAC1/2) in T-cell development and the maintenance of genomic stability.
Abstract
Histone deacetylase 1 and 2 (HDAC1/2) regulate chromatin structure as the catalytic core of the Sin3A, NuRD and CoREST co-repressor complexes. To better understand the key pathways regulated by HDAC1/2 in the adaptive immune system and inform their exploitation as drug targets, we have generated mice with a T-cell specific deletion. Loss of either HDAC1 or HDAC2 alone has little effect, while dual inactivation results in a 5-fold reduction in thymocyte cellularity, accompanied by developmental arrest at the double-negative to double-positive transition. Transcriptome analysis revealed 892 mis-regulated genes in Hdac1/2 knock-out thymocytes, including down-regulation of LAT, Themis and Itk, key components of the T-cell receptor (TCR) signaling pathway. Down-regulation of these genes suggests a model in which HDAC1/2 deficiency results in defective propagation of TCR signaling, thus blocking development. Furthermore, mice with reduced HDAC1/2 activity (Hdac1 deleted and a single Hdac2 allele) develop a lethal pathology by 3-months of age, caused by neoplastic transformation of immature T cells in the thymus. Tumor cells become aneuploid, express increased levels of c-Myc and show elevated levels of the DNA damage marker, γH2AX. These data demonstrate a crucial role for HDAC1/2 in T-cell development and the maintenance of genomic stability.
Introduction
Histone deacetylase (HDAC) 1 and 2 are sister proteins (∼ 80% identical), functionally redundant in many cell types,1-3 which are recruited together into 3 main transcriptional complexes: Sin3A,4 NuRD5,6 and CoREST.7,8 Classically, HDAC1 and 2 (HDAC1/2) function has been viewed in the context of transcriptional repression, because deacetylation of histone tails results in the tightening of nucleosomal arrays.9,10 However, genome-wide mapping of HDAC1 (or Rpd3) binding sites in human cells11 and yeast,12 reveals a positive correlation with gene activity, suggesting a role for HDACs in the cyclical acetylation of histones within the vicinity of active promoters.13 HDAC1/2 may thus have roles in both gene activation and repression.
In the clinic, the HDAC inhibitor SAHA is used to treat patients with cutaneous T-cell lymphoma.14 Therefore a greater understanding of the key pathways regulated by HDAC1/2 activity in T-lymphocyte development will help inform their use as drug targets. Moreover, as a dispensable cell type in a standard pathogen free environment, it is an excellent model to study essential genes, such as HDAC1/2. T-cell development, from immature double negative (DN), to double positive (DP) intermediates and mature (but naive) CD4 single positive (CD4SP, helper) and CD8 single positive (CD8SP, cytotoxic) populations is regulated by a number of transcription factors and chromatin modifying complexes.15,16 Runx1 and Runx3 have nonredundant roles in the regulation of CD4 expression.17-19 The zinc-finger transcriptional regulator, Gata3, is crucial for the development of the earliest T-cell progenitors.20 ThPOK expression is necessary to direct CD4 expression in MHC II restricted cells and is also sufficient to redirect class-I MHC T cells into the CD4 T-helper lineage.21 Individual HDACs have also been implicated in T-lymphocyte function. HDAC7 regulates cell survival and TCR signaling in DP thymocytes,22 HDAC6 and HDAC9 regulate the activity of T-regulatory cells,23,24 while HDAC11 directly controls IL-10 expression levels in antigen presenting cells.25
Deletion of either Sin3A26 or Mi2β,27 central components of the HDAC1/2 containing Sin3A and NuRD complexes, respectively, perturbs thymopoiesis. Loss of Sin3A causes a deficiency in the DN to DP transition and a significant reduction in the number of CD8SP cells. Mi2β also plays a role in DN to DP transition, and in addition is required for transcriptional activation of the Cd4 gene. These 2 studies clearly implicate HDAC1/2 in T-cell development. However, deletion of Hdac1 alone at the double positive stage of development produced a relatively mild phenotype28 ; suggesting that, as in many other tissue systems, deletion of both Hdac1 and Hdac2 is required to observe a more substantial phenotypic effect.1-3 We demonstrate here that double knock-out of Hdac1/2 results in a substantial block in T-cell development with a dramatic reduction in thymocyte cellularity and a failure to undergo the DN to DP transition. Furthermore, Hdac1/2 haploinsufficiency causes a lethal pathology at approximately 12-15 weeks of age, caused by neoplastic transformation of immature T cells in the thymus. Tumor cells are aneuploid, show amplification of c-Myc protein, increased levels of global histone acetylation and γH2AX, a marker of DNA damage. These data reveal critical roles for HDAC1/2 in T-cell development and the maintenance of genomic stability.
Methods
Generation of T-cell specific Hdac1 and Hdac2 knock-out mice
Mouse work was performed according to the regulations specified by the A(SP)A act 1986, under the Home Office project license PPL 80/2115, belonging to S.M.C. Hdac1 and Hdac2 conditional knock-out alleles were generated using the targeting constructs previously described by Dovey et al,29 to target Hdac1 and Hdac2 loci in AB1.1;129S5 mouse embryonic stem cells using standard gene targeting methods. See supplemental Figure 1 for detailed targeting strategy (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Hdac1Lox/Lox and Hdac2Lox/Lox conditional knock-out mice, in which exon 2 has been flanked by LoxP sites, were inter-crossed with the transgenic Lck-Cre line30 to generate a T-cell specific Hdac1/2 deletion. These mice were further bred to the transgenic OT-II (TgOT-II-TCR) mice31 to produce MHC class II restricted T cells. Genotyping primers are provided in supplemental Table 2.
Isolation and cytometric flow analysis of lymphoid cells
Cells from thymus, spleen, and lymph nodes were collected in ice-chilled cell collection buffer (PBS, 3% fetal calf serum) and single-cell suspensions generated. Cells were suspended in red blood cell lysis buffer (0.16M NH4Cl, 0.01M KHCO3), incubated at room temperature for 5 minutes, pelleted, and resuspended in cell collection buffer. Cells for FACS were stained using the antibodies described in supplemental Table 2. Cells were analyzed using a BD FACSCanto II flow cytometer (BD Biosciences). FCS 3.0 files generated by FACSDiva software (BD Biosciences) were exported and analyzed using VenturiOne (Applied Cytometry). Samples for mRNA extraction and whole genome expression profiling were stained and collected into cold PBS using a high-speed MoFLo Legacy cell sorter (Beckman Coulter).
Protein and enzymatic analysis
To confirm loss of HDAC1 and HDAC2, cells were lysed with hypotonic buffer: 10mM HEPES pH7.9, 10mM KCl, 0.1mM EDTA and 10mM DTT. After centrifugation at 1500g for 5 minutes nuclear extracts were prepared from pellets with: 20mM HEPES 7.9, 0.4M NaCl, 1mM EDTA, and 25% glycerol. Ten μg of nuclear extract was separated using SDS-PAGE and membranes probed with the appropriate antibodies (supplemental Table 1). Acid extraction of histones was performed as previously described.29 Five μg of extract was loaded in each lane and membranes probed using a panel of antibodies raised against a number of histone modifications. Membranes were scanned using the Odyssey Infrared imaging system and quantification of proteins achieved using the appropriate IRDye conjugated secondary antibodies (LiCOR Biosciences).
For immunoprecipitation, 70 μg of nuclear extract was incubated overnight at 4°C with antibody-coated protein-G agarose beads (GE Life Sciences). After 4 washes in nuclear extract buffer, beads were split into 2 aliquots. One aliquot was used to assess the deacetylase activity of the immunoprecipitates, while the second was resolved by SDS-PAGE and probed with antibodies raised against known components of the immunoprecipitated complexes.
Microarray and comparative genomic hybridization analysis
Comparative gene expression profiles of stage-matched (TCRβlow/CD5low) pre-lymphomic HD1Δ2/Δ2; HD2WT/Δ2 and wild-type thymocytes were compared using an Illumina MouseWG-6 v2 Expression BeadChip platform (Illumina). Total RNA was isolated using a standard Trizol (Invitrogen) protocol and Phase Lock Gel Heavy Tubes (5 Prime). Quality control of total mRNA was performed using a 2100 Bioanalyser (Agilent). Only samples that had an RNA integrity number (RIN) of 8.6 or higher were selected for processing and array hybridization. Processing of samples followed the manufacturer's instructions. Detection P values less than .01 were used to filter all data. Significant differential expression between sample sets was defined as probes that exhibited a robust fold change of ≥ 2 (Fc ≥ 2) with an adjusted P value of less than .005. An analysis of functionally related gene groups among deregulated genes and chromosomal distribution of deregulated genes was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7.32
DNA copy number variation in diseased thymocytes was assessed using the Mouse Genome Comparative Genomic Hybridization 244K Microarray (Agilent Technologies) according to the manufacturer's protocol. DNA was labeled with Cy3 or Cy5 according to BioPrime aCGH genomic labeling protocol (Invitrogen) and cleaned using purelink PCR purification kit (Invitrogen). Slides were hybridized for 48 hours, washed, and scanned with an Agilent microarray scanner. Raw data were then extracted using Agilent Feature Extraction. All following data analysis was performed in R using Bioconductor free packages (http://www.bioconductor.org), with background corrected then normalized using the loess algorithm. (Array comparative genomic hybridization experiments: accession number E-MTAB-1433; global transcriptional profiling in TCRβ/CD5Low cells: accession number E-MTAB-1432.)
Results
HDAC1/2 are required for normal T-cell development
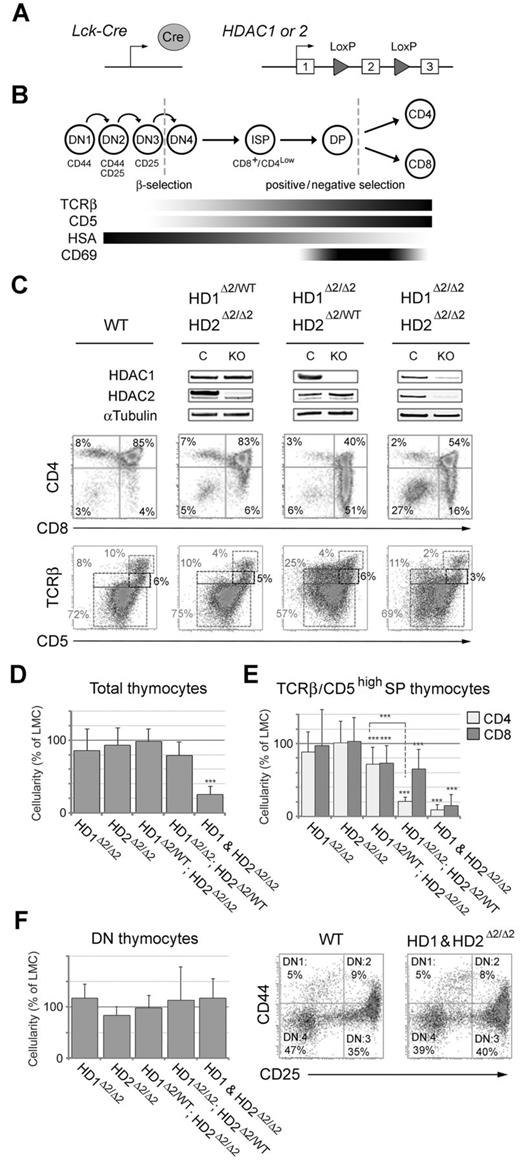
To investigate the role of HDAC1/2 in T-cell development we bred Hdac1Lox/Lox and Hdac2Lox/Lox conditional knock-out mice, in which exon 2 was flanked by LoxP sites (Figure 1A and supplemental Figure 1), to the transgenic Lck-Cre line.30 Southern blot and PCR analysis of DNA from Hdac1Lox/Lox; Lck-Cre and Hdac2Lox/Lox; Lck-Cre T cells revealed that Cre-mediated deletion of exon 2 was > 99% efficient, disrupting the ORF of both Hdac1 and Hdac2 such that a premature STOP codon is incorporated into exon 3, with a subsequent loss of protein (supplemental Figure 2). Hdac1Lox/Lox; Lck-Cre and Hdac2Lox/Lox; Lck-Cre mice (henceforward referred to as Hdac1Δ2/Δ2 and Hdac2Δ2/Δ2) were then inter-crossed to generate all genotypic combinations: single (Hdac1Δ2/Δ2, or Hdac2Δ2/Δ2), compound (Hdac1Δ2/Δ2; Hdac2Δ2/WT, or Hdac1Δ2/WT; Hdac2Δ2/Δ2) and double (Hdac1&2Δ2/Δ2) T-cell specific knock-outs. T-cell development can be delineated into a series of discrete steps by measuring the expression of the T-cell receptor (TCR), its co-receptors CD4 and CD8, and CD5 using flow cytometric analysis (Figure 1B). The Lck-Cre transgene is active early, during the double-negative (DN) stage of thymopoiesis, allowing us to monitor HDAC1/2 function during each of these critical stages.
Analysis of thymocytes lacking HDAC1/2 isolated from neonatal mice. (A) Schematic diagram of the model system used. Cre expression is driven from the proximal promoter of the T-cell specific tyrosine kinase, Lck. Both Hdac1 and Hdac2 are inactivated by deletion of exon 2, which is flanked by LoxP sites (triangles). (B) Schematic diagram illustrating the key steps of intra-thymic T-cell development. Early lymphoid progenitor cells enter the thymus from the bone marrow as double negative (DN) cells, the most immature cells of the thymus and exit to the periphery as either CD4SP (helper) MHC class II–restricted, or CD8SP (cytotoxic) MHC class I–restricted T cells. DN1, DN2, DN3, and DN4 stages of DN cell differentiation are distinguished by the relative expression levels of CD44 and CD25. Likewise, stages of DN to CD4+ or CD8+ T-cell differentiation are distinguished by the relative expression levels of CD4 and CD8. Bottom panel: Expression of other cell-surface markers, in combination with the expression of CD4/8, used to distinguish compartments of intra-thymic development of T cells of the TCRα/β lineage. (C) Thymocytes of the indicated genotype were isolated from neonatal mice (1-2 weeks old) and used either to make nuclear extract for Western blotting (top panel), or for 2 color FACS analysis (bottom panels). Percentages represent the mean where n > 6 for each genotype. (D-E) Comparative thymocyte cellularity compared with litter-mate controls from the genotypes indicated (***P < .001, paired t test). (F) Analysis of double negative (DN) thymocytes. Comparative thymocyte cellularity (left panel), and 2-color FACS analysis (right panel) for thymocytes of the indicated genotype.
Analysis of thymocytes lacking HDAC1/2 isolated from neonatal mice. (A) Schematic diagram of the model system used. Cre expression is driven from the proximal promoter of the T-cell specific tyrosine kinase, Lck. Both Hdac1 and Hdac2 are inactivated by deletion of exon 2, which is flanked by LoxP sites (triangles). (B) Schematic diagram illustrating the key steps of intra-thymic T-cell development. Early lymphoid progenitor cells enter the thymus from the bone marrow as double negative (DN) cells, the most immature cells of the thymus and exit to the periphery as either CD4SP (helper) MHC class II–restricted, or CD8SP (cytotoxic) MHC class I–restricted T cells. DN1, DN2, DN3, and DN4 stages of DN cell differentiation are distinguished by the relative expression levels of CD44 and CD25. Likewise, stages of DN to CD4+ or CD8+ T-cell differentiation are distinguished by the relative expression levels of CD4 and CD8. Bottom panel: Expression of other cell-surface markers, in combination with the expression of CD4/8, used to distinguish compartments of intra-thymic development of T cells of the TCRα/β lineage. (C) Thymocytes of the indicated genotype were isolated from neonatal mice (1-2 weeks old) and used either to make nuclear extract for Western blotting (top panel), or for 2 color FACS analysis (bottom panels). Percentages represent the mean where n > 6 for each genotype. (D-E) Comparative thymocyte cellularity compared with litter-mate controls from the genotypes indicated (***P < .001, paired t test). (F) Analysis of double negative (DN) thymocytes. Comparative thymocyte cellularity (left panel), and 2-color FACS analysis (right panel) for thymocytes of the indicated genotype.
We began by analyzing Hdac1/2 deleted T cells from mice at 6-8 weeks of age (supplemental Figure 3). Loss of either HDAC1, or HDAC2 alone produced no discernible phenotype with regards to the proportions of DN (CD4−CD8−), double positive (DP, CD4+CD8+), CD4 single-positive (CD4SP, CD4+CD8−) and CD8 single-positive (CD8SP, CD4−CD8+) T cells (supplemental Figure 3B). Mice with a compound heterozygous/homozygous genotype, in which only a single copy of Hdac1 remains (Hdac1Δ2/WT; Hdac2Δ2/Δ2), also had a phenotype comparable to wild-type. However, in both Hdac1Δ2/Δ2; Hdac2Δ2/WT (bearing only a single copy of Hdac2) and Hdac1&2Δ2/Δ2 mice, we observed a significant increase in CD4Low/CD8High cells, including a 4-fold increase in the percentage of immature single positive cells compared with wild type (ISP, CD4Int/CD8+ − 4% versus 18% and 17% respectively), indicating a block in development. Despite a strong phenotypic effect, we consistently observed near wild-type levels of HDAC2 protein in double knock-out mice (supplemental Figure 3A arrow), presumably because of selective pressure over time, because of the essential requirement for HDAC1/2 during T-cell development. In an attempt to overcome this we switched our analysis to the neonatal stage (10-14 days), because at earlier time points we observed a more robust, simultaneous, loss of HDAC1 and HDAC2 protein (Figure 1C). Neonatal Hdac1Δ2/Δ2; Hdac2Δ2/WT mice again display a significant increase in CD4low/CD8high cells (Figure 1C, 4% to 51%). We also observed a 2-fold reduction in the percentage of mature TCRβhigh/CD5high thymocytes (10% to 4%) and a 3-fold increase in TCRβInt/CD5Low cells (8% to 25%), identifying this population as developmentally immature. The absolute number of CD4Low/CD8High cells is increased 10-fold, while DP cells are reduced by 2.6-fold, indicating a block in DN to DP transition because of Hdac1/2 haploinsufficiency (supplemental Figure 4A). This also leads to increased levels of apoptosis in CD4Low/CD8High and DP cells in both neonates and 6-week old mice (supplemental Figure 4B).
Uniquely among all the genotypes tested, Hdac1&2Δ2/Δ2 mice had a marked 5-fold reduction in thymocyte cellularity (Figure 1D), with decreases in the number of mature CD4SP and CD8SP cells (Figure 1E). This reduction is likely related to the increased percentage of DN cells in Hdac1&2Δ2/Δ2 mice (Figure 1C, 3% to 27%). However, the absolute number of DN cells was similar in mice of all genotypes (Figure 1F left panel), as were the relative populations of DN1, DN2, DN3 and DN4 cells (adjudged by CD25/CD44 staining, right panel). The DN to DP transition involves a proliferative burst, which may be impaired because of a generic requirement for HDAC1/2 during cell cycle progression, similar to that observed in other Hdac1/2 knock-out systems.1,33,34
HDAC1/2 deficient T cells exhibit positive selection defects and reduced CD4 lineage commitment
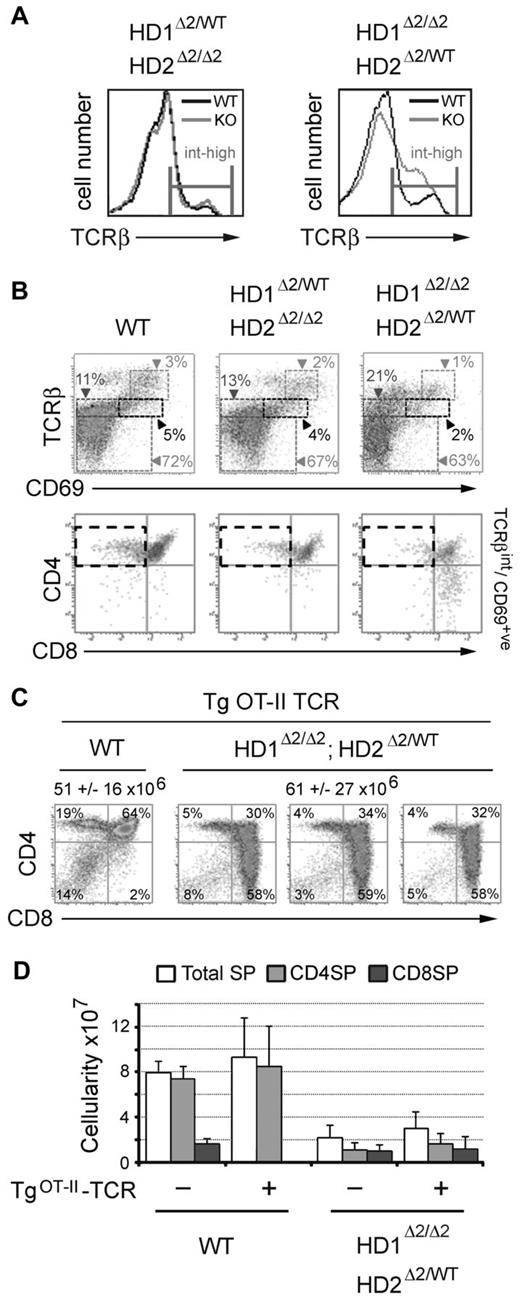
To prevent defective proliferation from confounding analysis of the developmental phenotypes, we focused our analysis on the 2 compound heterozygote/homozygote genotypes, Hdac1Δ2/WT; Hdac2Δ2/Δ2 and Hdac1Δ2/Δ2; Hdac2Δ2/WT, in comparison with wild-type mice. Staining for TCRβ expression reveals a significant increase in developmentally immature cells in Hdac1Δ2/Δ2; Hdac2Δ2/WT mice compared with Hdac1Δ2/WT; Hdac2Δ2/Δ2 and wild-type controls, at a level of TCRβ consistent with DP cells (Figure 2A right panel). To progress from the DP intermediate stage to CD4SP (helper), or CD8SP (cytotoxic) lineages, thymocytes must undergo positive selection mediated by TCR/MHC interactions, or die from neglect. Staining for CD69, which is transiently expressed in cells undergoing positive selection, in conjunction with TCRβ staining, permits fractionation of individual developmental subpopulations, namely: immature (TCRβlow/CD69low), preselection (TCRβint/CD69low), thymocytes undergoing selection (TCRβint/CD69int), and postselection (TCRβhigh/CD69high) cells. The proportion of positively selected thymocytes is reduced 2-fold, and postselection thymocytes 3-fold (Figure 2B), in cells with a single copy of Hdac2 (Hdac1Δ2/Δ2; Hdac2Δ2/WT). Gating on TCRβint/CD69int cells undergoing positive selection, we observed very few CD4high/CD8low cells, which are primed for terminal differentiation into CD4 or CD8 SP thymocytes (Figure 2B bottom panels). Hdac1Δ2/WT; Hdac2Δ2/Δ2 mice (single copy Hdac1) display a more subtle reduction in positively selected cells.
Thymocytes deleted for Hdac1/2 fail to undergo positive selection. Experiments were performed on thymocytes isolated from mice at 3-4 weeks of age. (A) FACS analysis of TCRβ expression levels in mice of the indicated genotype. (B) Two-color FACS analysis on thymocytes isolated from mice of the indicated genotype (n > 6). (C) Percentage and cellularity (mean ± SEM n = 4) of wild-type and Hdac1Δ2/Δ2; Hdac2Δ2/WT thymocytes in the presence of an OTII-TCR transgene. (D) Cellularity of single positive (SP) thymocytes, total, CD4SP and CD8SP mean ± SEM are shown. (n = 4 for all genotypes).
Thymocytes deleted for Hdac1/2 fail to undergo positive selection. Experiments were performed on thymocytes isolated from mice at 3-4 weeks of age. (A) FACS analysis of TCRβ expression levels in mice of the indicated genotype. (B) Two-color FACS analysis on thymocytes isolated from mice of the indicated genotype (n > 6). (C) Percentage and cellularity (mean ± SEM n = 4) of wild-type and Hdac1Δ2/Δ2; Hdac2Δ2/WT thymocytes in the presence of an OTII-TCR transgene. (D) Cellularity of single positive (SP) thymocytes, total, CD4SP and CD8SP mean ± SEM are shown. (n = 4 for all genotypes).
To address whether this phenotype was TCR dependent we inter-crossed Hdac1Δ2/Δ2; Hdac2Δ2/WT and OT-II (TgOT-II-TCR) mice which express pre-rearranged transgenic forms of TCRα and TCRβ. The resulting Hdac1Δ2/Δ2; Hdac2Δ2/WT-TgOT-II mice help to test 2 hypotheses. Firstly, if defects in positive-selection are because of a nonfunctional TCR the presence of the TgOT-II-TCR should rescue the cellularity of mature T cells; and secondly, as the TCR produced by the TgOT-II-TCR transgene recognises only MHC class-II molecules (generating only CD4SP cells), this might be expected to counteract the bias toward development of CD8-positive cells in Hdac1/2 deleted T cells. Thus, CD4/CD8 co-receptor expression was analyzed on neonatal thymocytes from WT and Hdac1Δ2/Δ2; Hdac2Δ2/WT mice in the presence or absence of the TgOT-II-TCR transgene (Figure 2C). As expected, the majority of SP cells in WT-TgOT-II-TCR mice were CD4SP. However, the presence of the pre-arranged TCR failed to rescue the CD8-phenotype in Hdac1Δ2/Δ2; Hdac2Δ2/WT-TgOT-II-TCR thymocytes. Furthermore, the absolute number of mature SP thymocytes (CD4SP or CD8SP) was not significantly changed on the TgOT-II-TCR background (Figure 3D). These data indicate that the lack of positive selection and the accumulation of CD4low/CD8high cells cannot be rescued by the presence of an intact rearranged TCR complex.
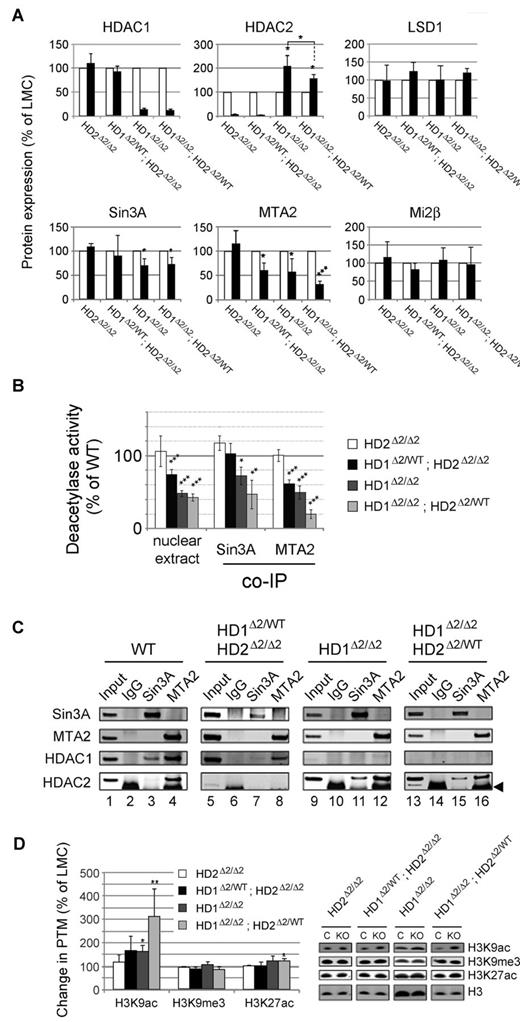
Loss of co-repressor complex integrity and increased histone acetylation in Hdac1/2 knock-out thymocytes. Experiments were performed on thymocytes isolated from 6- to 8-week-old mice. (A) Quantitative Western blot data for the indicated proteins. White and black bars denote protein levels measured in wild-type or knock-out cells, respectively from the indicated genotypes. Blots were quantified using a LiCOR scanner and normalized to the level of α-tubulin. Relative protein expression is presented as % of LMCs. Mean values (n = 3) ± SEM are plotted. (*P < .05, ***P < .001, paired t test). (B) Deacetylase activity was measured using a commercially available kit using 2μg of nuclear extract (left), or from individual HDAC1/2 containing complexes immunoprecipitated using anti-sera to Sin3A, or MTA2 as indicated. Nuclear extracts were prepared from thymocytes of the indicated genotype. (C) Western blot data for the indicated proteins co-immunoprecipitated in panel B. (D) Quantitative Western blotting was used to determine the levels of global histone acetylation. Acetylation levels were normalized to the total amount of H3 quantified using a LiCOR scanner. Mean values (n = 3) ± SEM are plotted. (*P < .05, **P < .01, paired t test).
Loss of co-repressor complex integrity and increased histone acetylation in Hdac1/2 knock-out thymocytes. Experiments were performed on thymocytes isolated from 6- to 8-week-old mice. (A) Quantitative Western blot data for the indicated proteins. White and black bars denote protein levels measured in wild-type or knock-out cells, respectively from the indicated genotypes. Blots were quantified using a LiCOR scanner and normalized to the level of α-tubulin. Relative protein expression is presented as % of LMCs. Mean values (n = 3) ± SEM are plotted. (*P < .05, ***P < .001, paired t test). (B) Deacetylase activity was measured using a commercially available kit using 2μg of nuclear extract (left), or from individual HDAC1/2 containing complexes immunoprecipitated using anti-sera to Sin3A, or MTA2 as indicated. Nuclear extracts were prepared from thymocytes of the indicated genotype. (C) Western blot data for the indicated proteins co-immunoprecipitated in panel B. (D) Quantitative Western blotting was used to determine the levels of global histone acetylation. Acetylation levels were normalized to the total amount of H3 quantified using a LiCOR scanner. Mean values (n = 3) ± SEM are plotted. (*P < .05, **P < .01, paired t test).
Loss of co-repressor complex integrity and increased histone acetylation in Hdac1/2 knock-out thymocytes
To address the biochemical mechanism underlying the block in T-cell development, we measured the enzymatic activity of HDAC1/2 containing co-repressor complexes, and levels of histone acetylation in knock-out cells. Consistent with previous reports,13,35 deletion of Hdac1 (Hdac1Δ2/Δ2) results in a 2-fold increase in the level of HDAC2 protein (Figure 3A and supplemental Figure 5). Significantly, a 50% reduction in Hdac2 gene dosage reduces this compensatory increase in HDAC2 protein (compare Hdac1Δ2/Δ2 and Hdac1Δ2/Δ2; Hdac2Δ2/WT, center panel), potentially explaining the difference in T-cell phenotype observed between these 2 genotypes (Figure 1C). In contrast, deletion of Hdac2 (Hdac2Δ2/Δ2) does not affect HDAC1 protein levels. Hdac1&2Δ2/Δ2 mice were not used in these analyses because of the severe reduction in neonatal cellularity and compensatory increase in HDAC2 levels at later time points.
We began by measuring the total deacetylase activity of nuclei isolated from single and compound Hdac1/2 knock-out T cells. Loss of Hdac2 alone had no overall effect (Figure 3B). In contrast, deletion of Hdac2 and a single copy of Hdac1 (Hdac1Δ2/WT; Hdac2Δ2/Δ2), Hdac1 alone, or Hdac1 and a single copy of Hdac2 (Hdac1Δ2/Δ2; Hdac2Δ2/WT), reduced total deacetylase activity significantly. Even with a single wild-type Hdac2 allele remaining in Hdac1Δ2/Δ2; Hdac2Δ2/WT mice, HDAC1/2 accounted for at least 60% of total deacetylase activity, making them the predominant HDAC enzymes in the T-cell nucleus. Within the nucleus they form the catalytic components of co-repressor complexes, principally, Sin3A, NuRD and CoREST.36 By immunoprecipitating Sin3A and MTA2 (a major component of NuRD) we were able to measure the residual deacetylase activity of the Sin3A and NuRD complexes in the absence of HDAC1/2 (Figure 3B-C). Deletion of Hdac1 alone, but not Hdac2, results in a reduction of deacetylase activity associated with both Sin3A and NuRD complexes. Interestingly, Sin3A deacetylase activity was unaltered in Hdac1Δ2/WT; Hdac2Δ2/ Δ2 compound mice, which correlates well with the almost complete absence of HDAC2 from the Sin3A complex in WT cells (Figure 3C compare lanes 3 and 11). In Hdac1Δ2/Δ2; Hdac2Δ2/WT mice, Sin3A activity was reduced 2-fold and NuRD 5-fold, prompting us test the integrity of complex components. Unexpectedly, we found that levels of both Sin3A and MTA2 protein, both of which interact with HDAC1/2 directly, were significantly reduced (Figure 3A). Whereas Mi2β and LSD1, which are thought to be recruited to the NuRD and CoREST complexes independently of HDAC1/2, were unaffected.
Because individual HDAC1/2 complexes were perturbed, we proceeded to measure levels of global histone acetylation (Figure 3D). Consistent with the loss of deacetylase activity, we observed a 3-fold increase in histone H3 K9 acetylation (H3K9Ac) in Hdac1Δ2/Δ2; Hdac2Δ2/WT cells. Global levels of histone H3 K14, K27 and H4 K12 acetylation were also increased significantly (supplemental Figure 5C), although to a lesser degree than H3K9Ac. In contrast, H3 K9 tri-methylation (H3K9me3) a marker of heterochromatic chromatin, which is largely devoid of HDAC1/2, was unaffected by deletion of either protein.
HDAC1/2 regulate the T-cell transcriptome
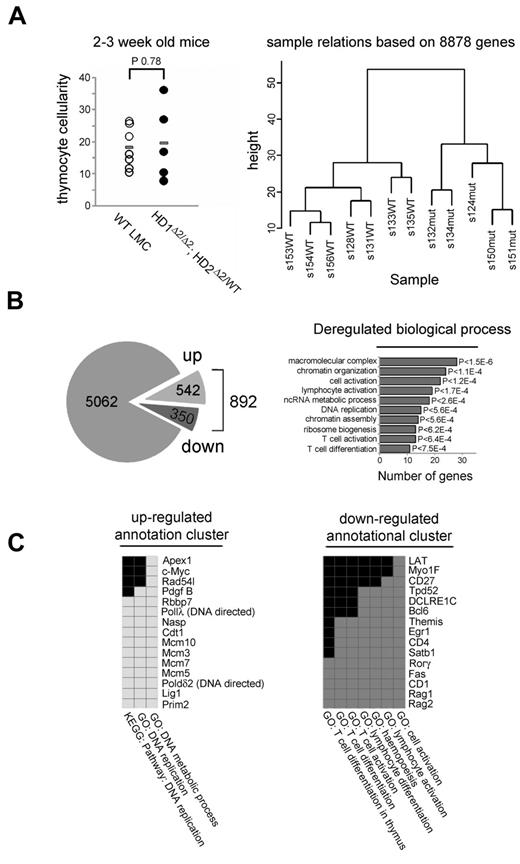
The alteration in histone acetylation in Hdac1/2 deleted cells (Figure 3D) is likely to affect a change in the gene expression program, potentially leading to the observed block in T-cell development. To analyze the transcriptome in Hdac1/2 deleted cells, thymocytes from 2-3 week old WT (n = 7) and Hdac1Δ2/Δ2; Hdac2Δ2/WT (n = 5) mice, in which cell number was unaffected (Figure 4A), were isolated, and RNA extracted from FACS sorted pre-selection populations (TCRβlow/CD5low). Comparative microarray analysis was performed and transcripts de-regulated ≥ 2-fold (Fc ≥ 2, adjusted P < .005) identified. Hierarchical clustering based on signal detection values of > 8000 genes revealed distinct transcriptional programs in WT vs. Hdac1Δ2/Δ2; Hdac2Δ2/WT thymocytes (Figure 4A right panel). A total of 5954 transcripts were detected in either genotype, with 892 de-regulated (Figure 4B and supplemental Table 1 for complete gene list). Consistent with a role for HDAC1/2 in transcriptional repression, more transcripts are up-regulated (542), than down-regulated (350). An analysis of functionally related gene groups among de-regulated genes using DAVID,32 reveals that genes involved in chromatin organization/assembly, DNA replication and T-cell activation/differentiation were among the top 10 significantly enriched gene clusters (Figure 4B). Performing the same analysis on separate up- and down-regulated gene lists identifies genes involved in DNA replication are up-regulated, conversely, genes assigned to T-cell activation/differentiation are down-regulated (Figure 4C). The down-regulated cluster includes a number of genes involved in V(D)J recombination (Rag1, Rag2 and DCLRE1C the gene that encodes Artemis). This suggests TCR recombination has been completed in (TCRβlow/CD5low) Hdac1Δ2/Δ2; Hdac2Δ2/WT thymocytes, but is still ongoing in wild-type DP cells which have yet to rearrange their TCRα chain. Consistent with the developmental block, and lack of selection thymocytes (TCRβint/CD69int), Cd69 is identified as down-regulated, as are key mediators of T-cell signaling such as LAT (linker of activated T cells), Themis (thymocyte expressed molecule involved in selection), Itk (IL-2 inducible T-cell kinase) and Tec (Tec protein-tyrosine kinase). Down-regulation of these genes suggests a model in which the absence of HDAC1/2 results in defective propagation of TCR signaling, thus blocking development.
Gene expression profiling of prelymphomic Hdac1Δ2/Δ2; Hdac2Δ2/WT thymocytes. (A) Total thymocyte cellularity is similar between Hdac1Δ2/Δ2; Hdac2Δ2/WT (n = 5) and WT LMCs (n = 7) of 2- to 3-week-old mice used for global transcription profiling. Bar indicates mean (P > .78, unpaired t test). Hierarchical clustering of samples based on signal detection values of 8878 genes indicates a distinct transcriptional program exists between Hdac1Δ2/Δ2; Hdac2Δ2/WT and WT LMCs. S indicates sample. Number indicates sample identifier, and WT or mut postfix indicates samples from WT LMCs or Hdac1Δ2/Δ2; Hdac2Δ2/WT genotypes, respectively. (B) Summary of transcriptome profiling. Number of transcripts detected in Hdac1Δ2/Δ2; Hdac2Δ2/WT or WT LMCs samples (left), presented as unregulated (gray), up-regulated (up), or down-regulated (down). Functional annotation clustering of all deregulated genes using DAVID (right). Represented are the top 10 statistically enriched biologic function gene ontology terms (BF-GO terms) and the number of deregulated genes of each annotational cluster. (C) DAVID analysis was also performed on up- and down-regulated genes, identifying enrichment of gene clusters as indicated. Gene names and associated BF-GO terms are listed; colored blocks indicate a corresponding GO term association positively correlated. Gene enrichment P values are provided by DAVID; calculated using EASE Score (modified Fisher exact P value).
Gene expression profiling of prelymphomic Hdac1Δ2/Δ2; Hdac2Δ2/WT thymocytes. (A) Total thymocyte cellularity is similar between Hdac1Δ2/Δ2; Hdac2Δ2/WT (n = 5) and WT LMCs (n = 7) of 2- to 3-week-old mice used for global transcription profiling. Bar indicates mean (P > .78, unpaired t test). Hierarchical clustering of samples based on signal detection values of 8878 genes indicates a distinct transcriptional program exists between Hdac1Δ2/Δ2; Hdac2Δ2/WT and WT LMCs. S indicates sample. Number indicates sample identifier, and WT or mut postfix indicates samples from WT LMCs or Hdac1Δ2/Δ2; Hdac2Δ2/WT genotypes, respectively. (B) Summary of transcriptome profiling. Number of transcripts detected in Hdac1Δ2/Δ2; Hdac2Δ2/WT or WT LMCs samples (left), presented as unregulated (gray), up-regulated (up), or down-regulated (down). Functional annotation clustering of all deregulated genes using DAVID (right). Represented are the top 10 statistically enriched biologic function gene ontology terms (BF-GO terms) and the number of deregulated genes of each annotational cluster. (C) DAVID analysis was also performed on up- and down-regulated genes, identifying enrichment of gene clusters as indicated. Gene names and associated BF-GO terms are listed; colored blocks indicate a corresponding GO term association positively correlated. Gene enrichment P values are provided by DAVID; calculated using EASE Score (modified Fisher exact P value).
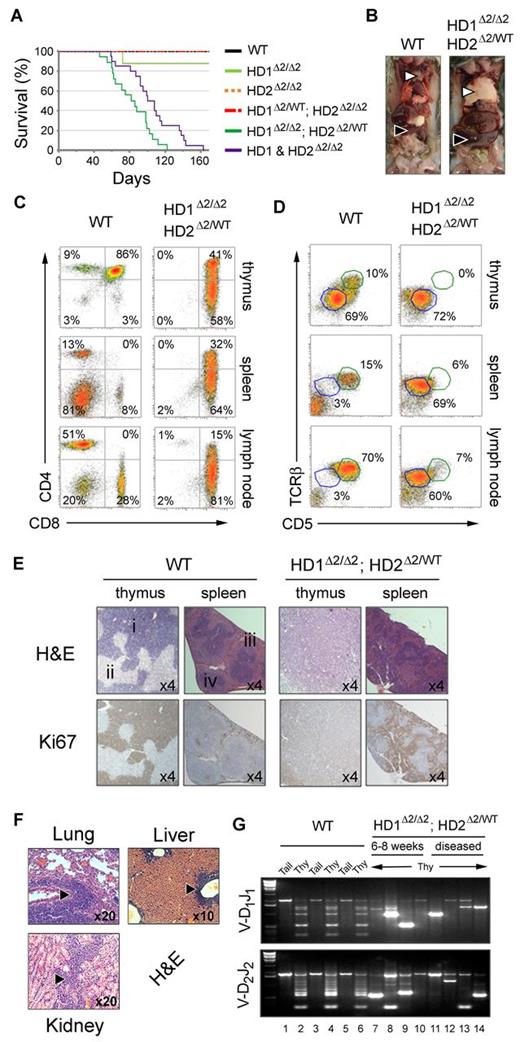
Loss of HDAC1/2 results in a lethal pathology as a result of intra-thymic T-cell neoplastic transformation
During the analysis it became apparent, that by 15 weeks of age a high proportion of Hdac1Δ2/Δ2; Hdac2Δ2/WT and Hdac1&2Δ2/Δ2 mice would become moribund, typified by an enlarged chest or abdomen with rapid respiration. We therefore monitored the survival of cohorts of all genotypes over a 5 month period (Figure 5A). Lethality of Hdac1Δ2/Δ2; Hdac2Δ2/WT and Hdac1&2Δ2/Δ2 mice occurred with 100% penetrance at an average of 12 and 15 weeks, respectively. The lag of 3 weeks in median survival of Hdac1&2Δ2/Δ2 mice likely reflects the time taken to overcome the 5-fold reduction in thymocyte cellularity at the neonatal stage (Figure 1C-D). By 6 weeks of age all Hdac1&2Δ2/Δ2 mice have undergone selection for cells which retain near wild-type levels of HDAC2 (supplemental Figure 3C), and thus tumor cells from double knock-out mice effectively phenocopy the Hdac1Δ2/Δ2; Hdac2Δ2/WT genotype. Tumor incidence correlates with cell types containing the lowest HDAC activity (Figures 3B and 5A). It is interesting to note therefore that 1 of 8 HDAC1Δ2/Δ2 mice also developed a tumor, suggesting that the level of HDAC activity in these cells is close to the threshold required for normal T-cell development. Necropsy of these mice revealed development of a pathologic diseased state characterized by massively enlarged thymus and splenomegaly (Figure 5B). FACS analysis of cells from these tissues reveals that they are overwhelmingly populated by immature (TCRβint/CD5Int) CD4low/CD8high and DP T cells (Figure 5C-D), which exhibited a side- and forward-scatter profile characteristic of a ‘blasting’ state, indicative of cellular proliferation (supplemental Figure 6A). Histologic analysis confirms that the thymus and spleen are inundated with proliferative (Ki67 positive) T cells, with complete effacement of medullary and cortical substructures in the diseased thymus (Figure 5E). Infiltration of T cells into nonlymphatic tissue (lung, liver and kidney) in 6 of 14 Hdac1Δ2/Δ2; Hdac2Δ2/WT mice examined was also detectable (Figure 5F). This aggressive and lethal increase in thymocytes could occur either as a consequence of a general hyperplasia (related to the developmental block) or expansion of cells with a monoclonal neoplastic transformation. To distinguish between these 2 possibilities we used PCR to determine the Dβ1-Jβ1 and Dβ2-Jβ2 rearrangements at the TCRβ locus. If the increase in cellularity was a consequence of general hyperplasia we would expect to detect multiple recombined events (similar to wild-type, Figure 5G lanes 2, 4, and 6), representative of a diverse and polyclonal population. In fact, thymocytes from 6-8 week Hdac1Δ2/Δ2; Hdac2Δ2/WT mice exhibited oligo-clonal TCRβ rearrangements which become increasingly monoclonal in diseased mice of the same genotype (Figure 5G 2/4 samples have a single rearranged PCR fragment, lanes 11 and 14); indicating monoclonal expansion rather than a general hyperproliferation of immature T cells. Thus, in addition to a block in T-cell development, Hdac1/2 haploinsufficiency in T cells results in neoplastic transformation and tumorigenesis.
Development of pathologic diseased state in Hdac1Δ2/Δ2; Hdac2Δ2/WT and Hdac1&2Δ2/Δ2 mice. (A) Kaplan-Meier survival curve. WT (n = 13), Hdac1Δ2/Δ2 (n = 8), Hdac2Δ2/Δ2 (n = 11), Hdac1Δ2/WT; Hdac2Δ2/Δ2 (n = 5), Hdac1Δ2/Δ2; Hdac2Δ2/WT (n = 18, median length of survival = 12 weeks) and Hdac1&2Δ2/Δ2 (n = 20, median length of survival = 15 weeks). (B) Enlarged thymus (white triangles) and spleen (black triangles) representative of Hdac1Δ2/Δ2; Hdac2Δ2/WT and Hdac1&2Δ2/Δ2 diseased mice. A WT LMC is also shown (left panel) for comparison. (C-D) CD4/CD8 and TCRβ/CD5 expression profiles of thymocytes, splenocytes and lymphocytes from diseased and aged matched WT LMCs. (E) Serial thymic sections from WT LMCs and diseased Hdac1Δ2/Δ2; Hdac2Δ2/WT mice stained with hematoxylin and eosin (H&E), or Ki67 counterstained with eosin, (i) cortex, (ii) medulla. (F) H&E-stained sections show infiltration of lung, liver and kidney in diseased Hdac1Δ2/Δ2; Hdac2Δ2/WT mice (6/14 cases analyzed). (G) Thymocyte clonality analysis: PCR was used to determine Dβ1-Jβ1 and Dβ2-Jβ2 rearrangement at the TCRβ locus of tail and thy (thymus) gDNA from the genotypes indicated.
Development of pathologic diseased state in Hdac1Δ2/Δ2; Hdac2Δ2/WT and Hdac1&2Δ2/Δ2 mice. (A) Kaplan-Meier survival curve. WT (n = 13), Hdac1Δ2/Δ2 (n = 8), Hdac2Δ2/Δ2 (n = 11), Hdac1Δ2/WT; Hdac2Δ2/Δ2 (n = 5), Hdac1Δ2/Δ2; Hdac2Δ2/WT (n = 18, median length of survival = 12 weeks) and Hdac1&2Δ2/Δ2 (n = 20, median length of survival = 15 weeks). (B) Enlarged thymus (white triangles) and spleen (black triangles) representative of Hdac1Δ2/Δ2; Hdac2Δ2/WT and Hdac1&2Δ2/Δ2 diseased mice. A WT LMC is also shown (left panel) for comparison. (C-D) CD4/CD8 and TCRβ/CD5 expression profiles of thymocytes, splenocytes and lymphocytes from diseased and aged matched WT LMCs. (E) Serial thymic sections from WT LMCs and diseased Hdac1Δ2/Δ2; Hdac2Δ2/WT mice stained with hematoxylin and eosin (H&E), or Ki67 counterstained with eosin, (i) cortex, (ii) medulla. (F) H&E-stained sections show infiltration of lung, liver and kidney in diseased Hdac1Δ2/Δ2; Hdac2Δ2/WT mice (6/14 cases analyzed). (G) Thymocyte clonality analysis: PCR was used to determine Dβ1-Jβ1 and Dβ2-Jβ2 rearrangement at the TCRβ locus of tail and thy (thymus) gDNA from the genotypes indicated.
Tumorigenic Hdac1Δ2/Δ2; Hdac2Δ2/WT T cells are associated with chromosomal instability
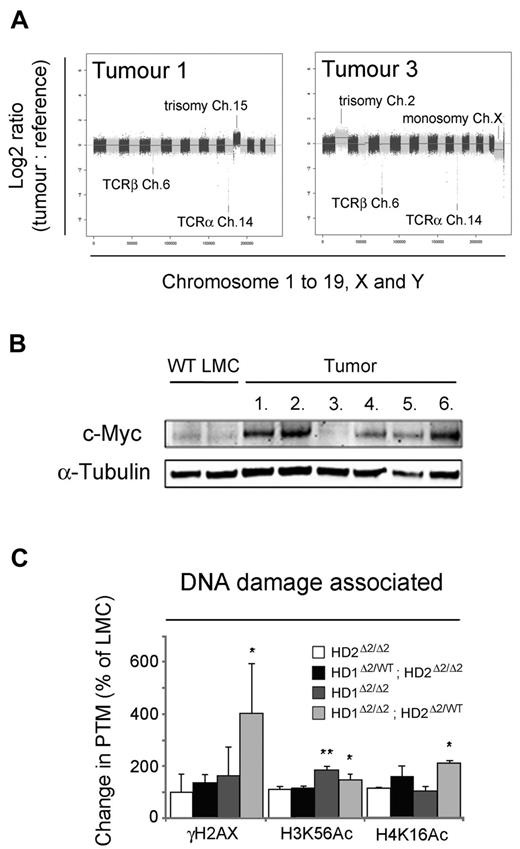
Chromosomal instability is a hallmark of human tumors37 and has been shown to play a critical role in the initiation and progression of hematologic malignancies.38 We therefore used array comparative genomic hybridization (aCGH) to detect gain or loss of genetic material in lymphomas derived from Hdac1Δ2/Δ2; Hdac2Δ2/WT mice (n = 6). All of the tumors displayed aneuploidy, with 4/6 containing trisomy 15 and the remainder (2/6) trisomy 2 (Figure 6A and supplemental Figure 6 for complete data series). There were 16 additional unique focal gains and deletions, including 1 sample with a homozygous deletion of the tumor suppressor Pten. Consistent with results assessing clonality (Figure 5G), all tumor samples revealed deletions at the Tcra (chromosome 14) and Tcrb loci (chromosome 6). Trisomy of chromosome 15 is characteristic of mouse T-cell tumors39,40 and is associated with the up-regulation of the proto-oncogene, c-Myc (located on chromosome 15 in mice). In agreement with microarray data (which show a 2.5-fold increase in c-Myc mRNA), Western blotting reveals elevation of c-Myc expression in 5/6 tumors compared with wild-type controls (Figure 6B) including all samples with trisomy 15. Although gene dosage may contribute to the increase in c-Myc protein, we also observed elevated c-Myc levels in a tumor sample wild-type for chromosome 15 (tumor 6, supplemental Figure 6), which suggests that additional factors may also play a role.
Hdac1Δ2/Δ2; Hdac2Δ2/WT T-cell tumors are associated with chromosomal instability. (A) Representative aCGH plots of gDNA isolated from Hdac1Δ2/Δ2; Hdac2Δ2/WT T-cell lymphomas (tumor) and tail (reference) from the same mouse. Log2 ratios of tumor versus reference samples are shown. Log2 ratios > ± 0.5 indicate single copy gains or losses, log2 ratios of > ± 1 indicate multiple copy gains or losses. Alternating dark and light shades represent individual chromosomes in sequence from 1 to 19 (autosomes), plus X and Y. (B) Western blot data of c-Myc using thymocyte protein extracts of tumor samples. α-tubulin is used as a loading control. (C) The global acetylation status of core histones from 6- to 8-week-old mice was detected using quantitative Western blotting. Acetylation levels were normalized to the total amount of H3 quantified using a LiCOR scanner. Mean values (n = 3) ± SEM are plotted. (*P < .05, **P < .01, paired t test).
Hdac1Δ2/Δ2; Hdac2Δ2/WT T-cell tumors are associated with chromosomal instability. (A) Representative aCGH plots of gDNA isolated from Hdac1Δ2/Δ2; Hdac2Δ2/WT T-cell lymphomas (tumor) and tail (reference) from the same mouse. Log2 ratios of tumor versus reference samples are shown. Log2 ratios > ± 0.5 indicate single copy gains or losses, log2 ratios of > ± 1 indicate multiple copy gains or losses. Alternating dark and light shades represent individual chromosomes in sequence from 1 to 19 (autosomes), plus X and Y. (B) Western blot data of c-Myc using thymocyte protein extracts of tumor samples. α-tubulin is used as a loading control. (C) The global acetylation status of core histones from 6- to 8-week-old mice was detected using quantitative Western blotting. Acetylation levels were normalized to the total amount of H3 quantified using a LiCOR scanner. Mean values (n = 3) ± SEM are plotted. (*P < .05, **P < .01, paired t test).
Chromosomal instability associated with tumor progression is generally coincident with elevated levels of DNA damage and a genome-wide loss of DNA methylation. HDAC1/2 have been suggested to stabilize Dnmt1 protein levels41 ; however, we found no difference in Dnmt1, or Dnmt3b, and little or no change in the DNA-methylation status of genomic repeats (supplemental Figure 7). HDAC1/2 also play a role in the DNA damage response (DDR) pathway, deacetylating chromatin in the vicinity of double strand breaks.42 Strikingly, we observed a 4-fold increase in γH2AX levels in Hdac1Δ2/Δ2; Hdac2Δ2/WT thymocytes compared with controls, indicative of substantial DNA damage in HDAC1/2 deleted cells (Figure 6C). Acetyl-lysines in both histone H3 (H3K56ac) and H4 (H4K16ac) are deacetylated to facilitate DNA repair.42,43 Hdac1Δ2/Δ2; Hdac2Δ2/WT thymocytes display a 1.5 and 2.1-fold increase in global levels of H3K56Ac and H4K16Ac, respectively, suggesting that these sites are regulated directly by HDAC1/2 containing complexes. These data suggest that HDAC1/2 play a crucial role in maintaining genomic stability independently of global DNA methylation status.
Discussion
HDAC1/2 and co-repressor complex integrity are required for normal T-cell development
We have demonstrated an essential role for HDAC1 and 2 (HDAC1/2) in the establishment of mature T-cell populations. Double homozygous deletion of Hdac1/2 (Hdac1&2Δ2/Δ2) in the T cells of neonatal mice, results in a 5-fold reduction in total thymocyte cellularity (Figure 1D) and a block in DN to DP transition (Figure 1E). Reduced cell numbers correlate with a significant increase in the percentage of DN cells (although absolute numbers of DN cells remains unchanged), indicating a block in DN to DP transition. The DN-DP transition is marked not only by expression of the CD4/CD8 co-receptors, but also by proliferative expansion; thus defects in cell cycle at this stage of development would explain the subsequent decrease in DP, CD4SP and CD8SP populations. Loss of both HDAC1/2 causes a severe cell cycle block in a number of systems, because of defects in gene regulation (eg, Cdk inhibitors p21 and p57)34,35,44 and mitosis.1 Conditional ablation of mSin3a in T cells results in a similar increase in DN cells, and a failure to proliferate in response to TCR signaling, suggesting that the DN to DP transition is mediated via the intact Sin3A:HDAC1/2 complex.26
Our analysis of mice with compound Hdac1/2 genotypes reveals a partial biochemical redundancy between the 2 enzymes. Retention of a single Hdac1 allele in the absence of Hdac2 (Hdac1Δ2/WT; Hdac2Δ2/Δ2) is less deleterious, in terms of total deacetylase activity and phenotype, than deletion of Hdac1 alone, or Hdac1 and a single copy of Hdac2 (Hdac1Δ2/Δ2; Hdac2Δ2/WT, Figure 3B). In agreement with our findings, a single Hdac1 allele, but not Hdac2, is sufficient to ensure normal B cell differentiation.36 This, along with data from embryonic stem cells,29 suggests that HDAC1 is the more dominant of the 2 enzymes. Removal of HDAC1/2 from the Sin3A and NuRD complexes causes increased protein turnover (mRNA levels are unaffected) of the direct HDAC binding subunits Sin3A and MTA2, respectively (Figure 3A), a phenomenon also observed in compound HDAC1/2 knock-out ES cells (O.M.D. and S.M.C., unpublished observations, 2012). This is of particular importance when considering the lymphoma that occurs in Hdac1/2 deleted mice (Figure 5) and the outcome of patients taking HDAC-inhibitors (HDACi) for the treatment of hematologic malignancies. Our data indicate that HDAC1/2 provide a structural role, in addition to their enzymatic function, within the Sin3A and NuRD co-repressor complexes; and thus the absence of the protein generates a much more pronounced phenotype in mice than inhibition of the same enzymes in situ with HDACi.
HDAC1/2 are required for the development of CD4SP cells
The reduction of deacetylase activity associated with Hdac1/2 deletion in T cells (Hdac1Δ2/Δ2; Hdac2Δ2/WT > Hdac1Δ2/Δ2 > Hdac1Δ2/WT; Hdac2Δ2/Δ2) correlates with a developmental block, in which immature CD4Low/CD8High and DP cells accumulate and fail to undergo positive selection (Figures 1C, 2B). Conditional inactivation of Mi2β, a central component of the NuRD complex and an ATP-dependent chromatin remodeler, results in a developmental phenotype with similar characteristics to Hdac1Δ2/Δ2; Hdac2Δ2/WT mice, that is, accumulation of DP-like thymocytes which have impaired CD4 expression.27 Mi2β is thought to activate CD4 expression in DP cells independently of HDAC1/2 (and therefore NuRD), by recruiting histone acetyltransferase enzymes, p300 and MOZ, to the Cd4 silencer, thus antagonizing the repressive function of Ikaros.45 However, given the similarity of the Mi2β and Hdac1/2 knock-out phenotypes and the down-regulation of MTA2 in cells lacking HDAC1/2, it is possible that the canonical NuRD complex may play a role in Cd4 activation. The classic notion of HDAC1/2 as exclusive mediators of transcriptional repression has been recently challenged by a ChIP-seq study (using peripheral CD4SP lymphocytes) which demonstrates that HDAC1/2 are predominantly located in the vicinity of active promoters, coincident with histone hyper-acetylation.11 Furthermore, HDAC1/2 activity can be negatively regulated via acetylation by p30046 (and potentially MOZ), which in the context of the NuRD complex would leave the chromatin remodeling activity of Mi2β as the dominant enzymatic function, and thus potentially able to function as a co-activator at the Cd4 silencer.
HDAC1/2 maintain global histone acetylation and chromosomal stability in developing thymocytes
All lymphomas isolated from Hdac1/2 deleted mice exhibited chromosomal instability (Figure 6A and supplemental Figure 6B). This is consistent with previous reports which have shown that HDAC activity is required for faithful chromosome segregation and maintenance of constitutive heterochromatin through subsequent cell divisions.47-49 The significance of this result in our system is that it occurs on a haploinsufficient (Hdac1Δ2/Δ2; Hdac2Δ2/WT) background. Indeed, Hdac1&2Δ2/Δ2 mice by 6 weeks have begun to for select cells which fail to inactivate Hdac2, and therefore phenocopy Hdac1Δ2/Δ2; Hdac2Δ2/WT mice (supplemental Figure 3). Complete loss of HDAC1/2 causes cell cycle arrest in numerous tissue systems,1,33,44,50 therefore retention of some HDAC1/2 activity, albeit at a significantly reduced level (Figure 3B), allows cells to divide while perturbing accurate chromosome segregation. This may be significant for the use of HDAC1/2 as therapeutic targets in the treatment of cancer. Complete inhibition of HDAC1/2 activity will cause tumor cells to arrest or undergo apoptosis,50 while partial inhibition might result in the opposite effect.
We have demonstrated an essential role for HDAC1/2 in the development of mature T-cell populations. Deletion of Hdac1/2 causes a marked reduction in thymocyte cellularity and a block in the DN-DP transition, most likely because of the down-regulation of key TCR signaling components. We have also shown, for the first time in a physiologic system, that HDAC1/2 activity is critical for maintaining genome stability. The occurrence of tumors (and the developmental block) is dose dependent, occurring in cells with the least amount of deacetylase activity, indicating that regulation of the acetyl-proteome, a balance between acetyltransferase and deacetylase enzymes, is crucial for normal cell development and viability.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Martin Dyer, Gerry Cohen, and Salvador Macip for useful discussions and critical reading of the manuscript. They also thank Christopher McGee and Ruben Bautista of the Wellcome Trust Sanger Institute Microarray Facility for performing Illumina microarray and aCGH analyses. They also acknowledge the Sanger Institute Core Facility for help in flow sorting.
This work was supported by a Medical Research Council studentship (to C.T.F.) and a Career Development Award (G0600135; to S.M.C.).
Authorship
Contribution: O.M.D., C.T.F., G.V., A.B., and S.M.C. designed transgenic mice and planned experimental approach; O.M.D., C.T.F., N.C., S.A.E., J.M.E., and R.S. performed experiments and analyzed data; and O.M.D., G.V., and S.M.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shaun M. Cowley, University of Leicester, Henry Wellcome Building, Lancaster Road, Leicester LE1 9HN, United Kingdom; e-mail: smc57@le.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal