Key Points
The DropArray technology is compatible with the retention of suspension cells in multistep procedures thus enabling novel assay methods.
This technology enabled visualization and quantification of specific killing events triggered by bispecific antibodies engaging T cells.
Abstract
Despite significant progresses, cell-based assays still have major limitations part to because of their plate format. Here, we present a wall-less plate technology based on unique liquid dynamics named DropArray that takes advantage of hydrophobic and hydrophilic surface properties. Liquid velocities within the DropArray plate were quantified through fluid dynamics simulation and complete retention of suspension cells experimentally demonstrated within the range of simulated shear stresses. Subsequently, we compared the DropArray technology with conventional microtiter plates in a cell-based protein-binding assay. Although the wall-less plate produced similar results with adherent cells, the advantage of the DropArray technology was absolutely clear when semiadherent or suspension cells were used in this multistep experimental procedure. The technology also was evaluated for the cell viability assay and generated similar results to conventional plate format while enabling significant reduction in toxic reagent use. Finally, we developed a DropArray cell-based assay to evaluate a bispecific antibody designed to engage cytotoxic T cells and trigger tumor cell killing. This assay enables for the first time the visualization and quantification of the specific killing events and represents a very powerful tool to further investigate functional aspects of the cancer immunotherapy.
Introduction
The demonstration that single cells could be grown in vitro1 combined with the development of specific growth media2 and later the establishment of the first cell lines3-6 definitively marks the birth of cell culture as a critical research tool. Since then, investigators have developed a myriad of in vitro cellular models to further the understanding of various normal and pathologic cellular processes as well as to screen and characterize potential therapeutic modalities. All this progress occurred in concert with the development of many technologies that have impacted the investigator's ability to establish specific culture conditions and measure or visualize different cellular signals. As a result, cell-based assays are today almost universally used in biology research laboratories.
During the past decade, cell-based assays have become fundamental and irreplaceable tools in the drug discovery and development industry. This trend was driven mainly by 2 rationales. First, the completion of the human genome,7 combined with advances in functional genomics and proteomics,8 has led to the need to evaluate thousand of potential new targets. Second, the high attrition rate of therapeutic candidates at the preclinical and clinical stages generated the need for more biologically relevant high-throughput screening approaches, bringing cell-based assay technologies to the forefront of the drug-discovery strategy. As a result, investigators' ability to develop rapid, flexible, robust, and cost-effective high-throughput cell-based assays became of paramount importance.
Despite significant technological progress in enabling technologies such as molecular labeling and the advent of high content screening approaches, cell-based assays, in part because of their well-plate format, continue to have major limitations. Although wells are an efficient and simple strategy to segregate experimental conditions in formats from 6- to 1536-well plates, they have major restrictions when suspension, loosely adherent, and in some cases fully adherent cells are used, especially with high-throughput formats such as 96-, 384-, and 1536-well plates. Microwell plates not only limit the use of certain cells but also significantly reduce the spectrum of experimental procedures that can be implemented in high-throughput cell-based screenings. Because the addition and removal of reagents to the wells could definitively compromise the cells, various technologies have been developed to circumvent these limitations. The homogeneous assay, developed in recent years, provides a convenient “add, mix, and read” approach to explore a broad spectrum of biologic events from cell viability/death and proliferation9-13 to the regulation of specific genes through reporter gene activation based on luciferase,14 β-galactosidase,15 and β-glucuronidase16 activities. Although these in vitro homogeneous assays are intensively used in drug target screening, increasing skepticism has arisen about the actual benefits of current cell-based reporter or homogeneous assays that are vulnerable to false-positive/negatives hits as well as misleading interpretations.17 Therefore, new methods or derivatives of exciting methods should be developed to better select, evaluate, and characterize potential therapeutic candidates in vitro.
Here, we report the development of multiple cell-based assays using a novel, wall-less plate technology named DropArray. Our data clearly demonstrate the possibility of using suspension cells in existing multistep experimental procedures and establish the DropArray technology as a new plate format enabling investigators to circumvent many current cell-based assay limitations. In addition, the DropArray technology provides unprecedented opportunities to develop new cell-based assay strategies by allowing the visualization and quantification of complex biologic events involving multiple cell types, including suspension cells. As proof of principle, we developed a method to characterize bispecific antibodies designed to engage cytotoxic T cells and trigger tumor cell killing. This new approach enables for the first time the visualization and quantification of the specific tumor B-cell killing events in real time and represents a very powerful tool to further investigate functional aspects of cancer immunotherapy in conditions that better mimic in vivo situations than the conventional protocol.
Methods
Cell lines
COS7, 293T, and S, U937, K562, and BJAB cell lines were purchased from ATCC and maintained in culture according to ATCC recommendations. CD8+ T cells were purified from human PBMCs isolated from peripheral blood of random healthy donors via the use of a CD8+ T-cell isolation kit (130-094-156; Miltenyi Biotec).
Reagents
Antibodies and stains used were as follows: mouse anti-CD8–APC (555369; BD Biosciences), Hoechst 33342 (H-3570; Molecular Probes), phalloidin-TRITC (P1951; Sigma-Aldrich), rabbit anti–ZO-1 (61-7300; Invitrogen) and anti–human IgG (H+L) AlexaFluor 488 (A-11013; Invitrogen). Anti-CD3/CD19 bispecific antibody (Genentech) was produced by Chinese hamster ovary cells and purified from cell culture supernatant as described elsewhere.18 For live imaging applications, the endocytic inhibitor Dynasore hydrate (D7693; Sigma-Aldrich) was used. Cell viability tests were done using the following reagents: CellTiter-Glo (G7570; Promega), CellTracker Green 5-Chloromethylfluorescein Diacetate (C7025; CMFDA; Molecular Probes), ethidium bromide (EB; E1510; Sigma-Aldrich), acridine orange (AO; 318337; Sigma-Aldrich), and propidium iodide (PI; P3566; Molecular Probes).
Flow chamber experiments
The flow chamber was fabricated from polydimethylsiloxane (Sylgard 184; Dow Corning) with the use of soft lithography and replica molding. K562 cell suspension (0.5 × 106 cells/mL) was seeded into the DropArray (2 μL droplet/spot) and incubated in a humidified incubator at 37°C, 5% CO2 overnight (∼ 16 hours). The flow chamber was placed over the cell droplets and adhered to the DropArray via the application of a vacuum suction to the attachment channels. The cell medium was loaded into a syringe (Hamilton) and a 22-gauge needle threaded into PE 20 Polyethylene Tubing (BD Intramedic) and attached to the inlet port of the flow chamber after all air was expelled. A syringe pump (Harvard Instruments) controlled the rate of fluid flow through the flow chamber, and images were captured every 5 minutes with the use of an Olympus IX71 inverted Microscope (Nikon). Images were analyzed with ImageJ Version 1.46 software.
Fluid velocity simulations
Comsol Multiphysics Software Package Version 4 was used to construct a 2-dimensional model of the washing chamber of the DropArray System. The chambers dimensions were 128 mm (l) × 10 mm (h). We used a 2-phase flow, phase field simulation that used incompressible flow to simulate the fluid velocities within the chamber. The chamber was simulated to move horizontally at an oscillation of 0.667 Hz (44 rpm) or 1.4667Hz (88 rpm). Fluid velocities 10 μm above the bottom of the chamber were extrapolated during the 10-second washing period.
Cell transfections
COS7 cells were transfected with cDNA by Fugene 6 (E2691; Promega) according to the manufacturer's protocol at a ratio of 6 μL of Fugene 6 per 1 μg of DNA diluted in OPTI-MEM I (31985-070; Invitrogen). Cells were grown for 48 hours before we began the staining procedures.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 20 minutes, washed with 1× PBS, then blocked with 1% BSA in PBS for 30 minutes. Human Fc-tagged bait proteins or specific antibodies were incubated with the cells for 1 hour in 1% BSA in PBS, washed with 1× PBS, and incubated with appropriate secondary detection antibodies for 45 minutes at room temperature. After secondary antibody incubation, the cells were washed with 1× PBS and imaged.
Cell viability assays
For all cell viability assays, cells were treated with various concentrations of monomethyl auristatin E (MMAE) for 72 hours before the assay. For the CellTiter-Glo assays, 2-μL drops of CellTiter-Glo were added on top of the 2 μL drops of cells on the DropArray plate. After a 5-minute incubation at room temperature, the plate was read on an Envision 2102 Multilabel Reader (Perkin Elmer). For fluorescence-based assays, cells were labeled with CMFDA when we added MMAE. After it was washed to remove dead cells and cellular debris, the plate was imaged on an IN CELL Analyzer 2000 (GE Healthcare), and the number of green cells left on the plate was quantified with the IN CELL Developer Version 1.9 image analysis software. For the EB/AO assay, a mix of both dyes at 100 μg/mL in PBS was prepared. The DropArray plate was washed once, and a 1:10 dilution of the dye mix in PBS was added to the drops. After a 15-minute incubation, the plate was imaged on an IN CELL Analyzer 2000, and the number of orange cells and the number of green cells was quantified.
Bispecific T-cell killing assay
For assays without the addition of the bispecific antibody, BJAB cells were seeded onto the DropArray plates and labeled with 5μM CMFDA. The next day, purified CD8+ T cells were labeled with an anti-CD8 antibody for 1 hour on ice. The labeled T cells were washed with complete media and added 1:1 on top of the BJAB cells, a 2-μL drop added to a 2-μL drop. For assays with the addition of a bispecific antibody, the same procedure varied as follows. The BJAB cells seeded on a DropArray plate were washed with complete media, then the bispecific or control antibody was added along with PI at 1:1000 and incubated at 37°C for 1 hour. Labeled CD8+ cells were then added 1:1 on top of the BJAB cells and observed on the IN CELL Analyzer 2000 at the indicated time points.
Microscopy
Fluorescence images were acquired using either IsoCyte laser imager (ImageXpress Velos, Molecular Devices) or IN CELL Analyzer 2000 (GE Healthcare). Image analysis was performed using IN CELL Developer Version 1.9 or BBIsoCyteDL Version 5.0.0.106 software.
Results
DropArray wall-less plate technology
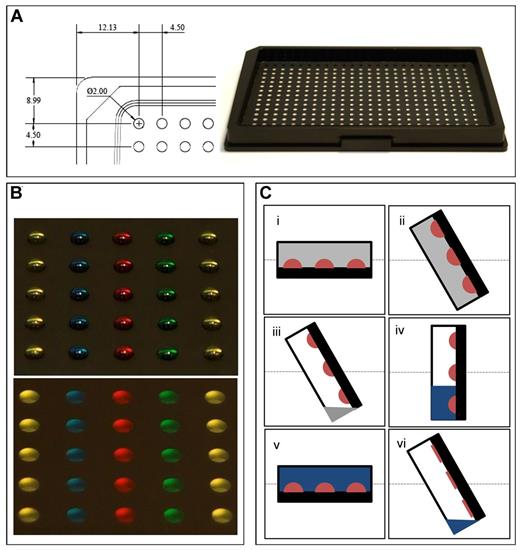
The DropArray wall-less plate technology consists of an array of 384 circular areas on a polytetrafluoroethylene resin–coated glass slide that is attached to a Society for Biomolecular Sciences (SBS) format aluminum frame (Figure 1A). Each circular area or experimental spot has a diameter equal to 2 mm, and the spots are 4.5-mm apart. Each spot functions similar to a well on a microtiter plate and houses a 2-μL drop of cell culture media containing the cells, allowing the assay within the drop to be performed in an individual controllable manner. Because each spot is surrounded by polytetrafluoroethylene resin, the 2-μL drop is very stable on the spot (Figure 1B top). To eliminate evaporation and enable long-term cell culture experiments, the array of droplets is immersed in perfluorocarbon-based immiscible liquid oil (Figure 1B bottom). Again, the retention forces maintaining the drop of media onto the hydrophilic area are strong enough to absolutely insure the integrity of the drop while the DropArray plate is filled with the incubation oil. Washing steps are performed using a fully automated rinsing station that uses gravity and a vacuum pump for complete drainage of the oil and wash buffers (supplemental Figure 1 and supplemental Video 1; see the Supplemental Materials link at the top of the article). To summarize, during the washing procedure the DropArray plate is initially sealed to create a rinsing chamber (Figure 1Ci) and progressively inclined to drain the oil (Figure 1Cii-iii). Next, wash buffer is introduced through an inlet at the bottom of the rinsing chamber (Figure 1Civ) and is subsequently returned to the horizontal position to undergo linear shaking (Figure 1Cv). The rinsing chamber is subsequently inclined vertically again to remove the wash buffer (Figure 1Cvi).
DropArray plate characteristics and processing. (A) DropArray plates consist of an array of circular areas on a polytetrafluoroethylene resin–coated glass slide attached to an aluminum support frame. (B) Each 2-mm diameter well is loaded with 2 μL of food coloring for better visualization. During an assay, the array of droplets is immersed in a perfluorocarbon-based liquid oil (gray), which virtually eliminates evaporation. (C) Steps involved during the plate washing procedure include the following: (i) the plate is sealed to the washer to create a large rinsing chamber; (ii) the rinsing chamber tilts to allow drainage of incubation oil; (iii) drainage of the incubation oil through gravity assist and a liquid pump; (iv) the rinsing chamber is filled with wash buffer (1× PBS, blue); (v) the rinsing chamber returns to the horizontal position for linear shaking; and (vi) after linear shaking, the rinsing chamber tilts to allow drainage of wash buffer.
DropArray plate characteristics and processing. (A) DropArray plates consist of an array of circular areas on a polytetrafluoroethylene resin–coated glass slide attached to an aluminum support frame. (B) Each 2-mm diameter well is loaded with 2 μL of food coloring for better visualization. During an assay, the array of droplets is immersed in a perfluorocarbon-based liquid oil (gray), which virtually eliminates evaporation. (C) Steps involved during the plate washing procedure include the following: (i) the plate is sealed to the washer to create a large rinsing chamber; (ii) the rinsing chamber tilts to allow drainage of incubation oil; (iii) drainage of the incubation oil through gravity assist and a liquid pump; (iv) the rinsing chamber is filled with wash buffer (1× PBS, blue); (v) the rinsing chamber returns to the horizontal position for linear shaking; and (vi) after linear shaking, the rinsing chamber tilts to allow drainage of wash buffer.
Optimal washing conditions for suspension cells with the DropArray technology
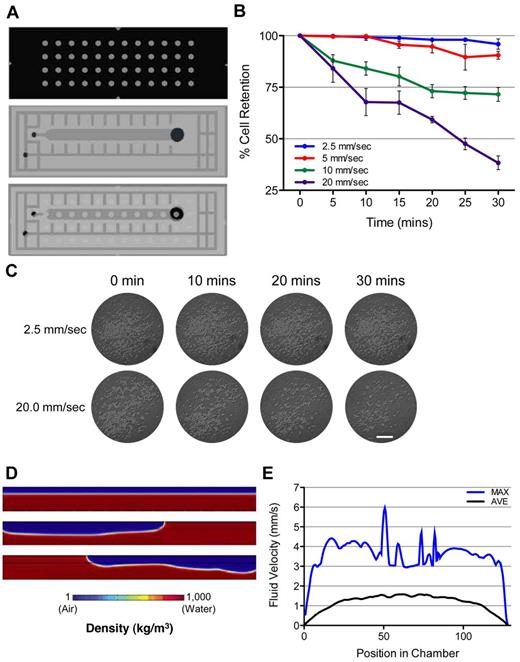
Cell retention within the DropArray plates is dependent on a fine balance of cell on the glass and shear forces created by mechanical washing movement of the system. Although the fluid movement that creates these shear forces is detrimental to cell retention, it is paramount to maximize the washing of small molecules, proteins, and staining reagents away from the cells. To reconcile these 2 opposing objectives, we used a microfluidic flow chamber to expose K562 human myelogenous leukemia cells to 30 minutes of constant shear force (2.5, 5.0, 10.0, and 20.0 mm/s) after being seeded onto the DropArray system (Figure 2A). Cell media was introduced via syringe pump (PHD 2200; Harvard Instruments) at flow rates of 1.0, 2.0, 4.0, and 8.0 μL/s, respectively, to induce the corresponding shear force on the cells.
Flow chamber and fluid velocity simulation. (A) Schematic of the flow chamber platform used for analysis. Cells were seeded on a DropArray plate, and flow chamber was overlaid on top of the cells and DropArray plate. (B) Quantification of the percentage of cells remaining in the well after 30 minutes of constant exposure to fluid velocities of 2.5, 5.0, 10, and 20 mm/s and the standard error. As the fluid velocity increases, the number of cells retained decreases. (C) Time series micrographs of K562 cells seeded within a 2-mm droplet during exposure of constant fluid sheer force (2.5 and 20 mm/s) over 30 minutes. Scale bar = 0.5 mm. (D) Visualization of simulated fluid movement within the chamber. Before the application of horizontal washing action of 44 RPM, the air (blue) and liquid media (red) span the length of the chamber. During washing, the fluid and air components oscillate between the ends of the chamber. (E) Quantification of simulated fluid velocity with a washing speed of 44 rpm at 10 μm above the chamber floor. The average fluid velocity (mm/s) at each location along the chamber floor is plotted as well as the standard error over the duration of washing (10 seconds). In addition, the maximum velocity at each location is plotted, representing the fastest fluid movement a cell on the chamber floor could potentially be exposed.
Flow chamber and fluid velocity simulation. (A) Schematic of the flow chamber platform used for analysis. Cells were seeded on a DropArray plate, and flow chamber was overlaid on top of the cells and DropArray plate. (B) Quantification of the percentage of cells remaining in the well after 30 minutes of constant exposure to fluid velocities of 2.5, 5.0, 10, and 20 mm/s and the standard error. As the fluid velocity increases, the number of cells retained decreases. (C) Time series micrographs of K562 cells seeded within a 2-mm droplet during exposure of constant fluid sheer force (2.5 and 20 mm/s) over 30 minutes. Scale bar = 0.5 mm. (D) Visualization of simulated fluid movement within the chamber. Before the application of horizontal washing action of 44 RPM, the air (blue) and liquid media (red) span the length of the chamber. During washing, the fluid and air components oscillate between the ends of the chamber. (E) Quantification of simulated fluid velocity with a washing speed of 44 rpm at 10 μm above the chamber floor. The average fluid velocity (mm/s) at each location along the chamber floor is plotted as well as the standard error over the duration of washing (10 seconds). In addition, the maximum velocity at each location is plotted, representing the fastest fluid movement a cell on the chamber floor could potentially be exposed.
To ensure that all cells adhered to the glass, each trial was first expose to 2 minutes of fluid flow of 0.4 μL/s (1.0 mm/s) to clear nonadhered cells. Images of each well were taken at 5-minute intervals and cells counted using Image J (Figure 2C). High fluid velocities resulted in immediate loss of cells with 72% ± 3% of cells being retained after 30 minutes of 10 mm/s shear force and 38% ± 3% cell retention when exposed to 20 mm/s (Figure 2B). Lower fluid velocities resulted in minimal cell loss with 96% ± 3% and 90% ± 2% of cells being retained after 30 minutes of shear forces of 2.5 mm/s (Figure 2B) and 5.0 mm/s, respectively. To maximize the washing ability of the DropArray system, a small amount of cell loss is acceptable to ensure complete removal of small molecules. From the flow chamber analysis, we determined that a flow rate of 5 mm/s maximum would be optimal to ensure a high percentage of cell retention and greatest washing ability.
Considering the fact that fluid movements are generated by the DropArray washing system, which moves the DropArray plate laterally, we subsequently needed to optimize the washing motion to be within the acceptable shear force limits determined through the flow chamber analysis. To achieve this step, a computation model of the fluid dynamics was developed with the use of Comsol Multiphysics Software Version 4.0. A deterministic 2-dimensional model of 2-phase flow was used to simulate the fluid dynamics of the cell culture media and air within the washing chamber. The cross section of the simulated 128-mm × 10-mm washing chamber was filled with 70% water and 30% air as it is in the actual system. The plate was simulated to move in an oscillatory motion with a period of 0.7 Hz (44 rpm; Figure 2D) and 1.5 Hz (88 rpm) during the standard 10-second washing period (supplemental Videos 2 and 3, respectively). The simulated fluid velocity was determined 10 μm above the plate bottom and the average and maximum shear force calculated every 0.5 μm across the bottom of the plate. Moving the plate at a rate of 44 rpm resulted in a maximum shear force of 5.96 mm/s and on average all positions 10 μm above the plate bottom were exposed to a shear force of 1.17 mm/s (Figure 2E). In these conditions, fluid speed is consistent along the length of the chamber and is optimal for uniform washing. In contrast, increasing the speed to 88 rpm results in a fluid velocity profile that is not uniform. Because of the quick oscillation, the fluid is not permitted to transverse the entire plate before the velocity vector is reversed (supplemental Video 3). In addition, as expected, the increased plate speed results in greater shear forces with a maximum of 12 mm/s and an average of 3.43 mm/s. To ensure maximum cell retention and washing, a speed of 44 rpm is optimal.
Cell-surface protein-protein interaction investigations via the DropArray versus microtiter plate
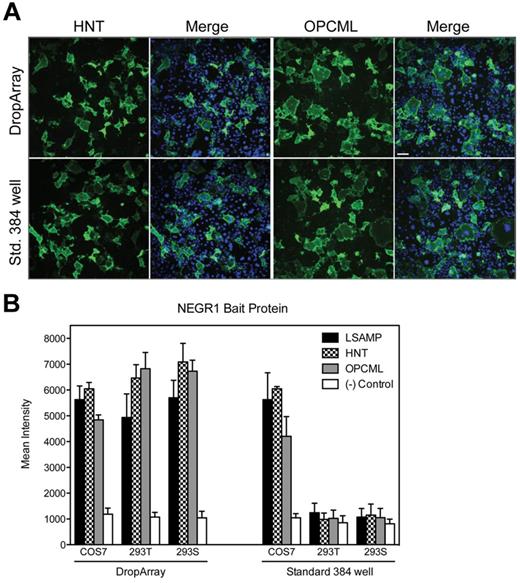
To assess the performance of the DropArray plate in a multistep cell-based assay procedure, we performed an expression cloning experiment to investigate protein-protein interactions among IgLON family members. IgLONs are glycorylphosphatidylinositol-anchored neural cell adhesion molecules comprising LSAMP19 (limbic system-associated membrane protein), HNT (neurotrimin),20 OPCML (opioid binding protein/cell adhesion molecule-like),21 and NEGR1 (neuronal growth regulator 1/neurotractin).22 These proteins have been shown to be involved in cell-to-cell adhesion and to interact homophilically and heterophilically within the family with various binding affinities.23-26 Using the optimized washing conditions described previously, we compared the DropArray technology with conventional 384-well plates using a regular automated washing procedure. Here, COS7 (adherent), HEK293T (semiadherent), and HEK293S (suspension-adapted) were transiently transfected with LSAMP, HNT, or OPCML constructs. No change in cell number or density was observed 48 hours after transfection with any of these constructs compared with nontransfected or mock-transfected controls (data not shown). Cell expressing LSAMP, HNT, or OPCML were then incubated with NEGR1-hFc bait protein. Subsequently, NEGR1-hFc binding was detected with the use of an anti–human AlexaFluor 488 antibody. As expected, COS7 cells expressing NEGR1 displayed specific binding to LSAMP (data not shown), HNT, and OPCML, and binding was similarly detected using either plate format (Figure 3A). Quantitation of the fluorescent signal results in at least a 5-fold increase in mean intensity in response to the specific binding of NEGR1-hFc to LSAMP, HNT or OPCML compared with the nonspecific binding observed in nontransfected cells on either plate format (Figure 3B).
Cell-based extracellular protein-protein interactions. (A) COS7 cells transfected with either HNT or OPCML expression constructs were fixed and then incubated with NEGR1-hFc bait protein. The cells were then washed and stained with anti–human AF488 (green) and scanned on the IN Cell Analyzer 2000. The same reagents for transfection and staining were used on a standard 384-well microtiter plate (Aurora, Brooks Automation) and a DropArray plate for a side-by-side comparison of the 2 formats. Hoechst staining (blue) of the nuclei shows the same cell density on both plate formats. Scale bar = 70 μm. (B) Quantification of the mean intensity of the green channel from the images acquired. With the use of 3 different cell types, equivalent intensity values are seen when NEGR1-hFc binds to cells expressing 3 different expression constructs of known NEGR1 binding partners. The same values are generated from the standard 384-well format, but only for the adherent cell line (COS7), whereas the weakly adherent (293T) and suspension adapted (293S) cell lines were not retained during the washing procedure on the standard 384-well plate format.
Cell-based extracellular protein-protein interactions. (A) COS7 cells transfected with either HNT or OPCML expression constructs were fixed and then incubated with NEGR1-hFc bait protein. The cells were then washed and stained with anti–human AF488 (green) and scanned on the IN Cell Analyzer 2000. The same reagents for transfection and staining were used on a standard 384-well microtiter plate (Aurora, Brooks Automation) and a DropArray plate for a side-by-side comparison of the 2 formats. Hoechst staining (blue) of the nuclei shows the same cell density on both plate formats. Scale bar = 70 μm. (B) Quantification of the mean intensity of the green channel from the images acquired. With the use of 3 different cell types, equivalent intensity values are seen when NEGR1-hFc binds to cells expressing 3 different expression constructs of known NEGR1 binding partners. The same values are generated from the standard 384-well format, but only for the adherent cell line (COS7), whereas the weakly adherent (293T) and suspension adapted (293S) cell lines were not retained during the washing procedure on the standard 384-well plate format.
As expected, NEGR1-Fc binding to LSAMP-, HNT-, or OPCML-transfected HEK293T or S cells was not detected in conventional 384 well-plate because these semiadherent and suspension-adapted cells, respectively, are washed out of the well during the washing procedure (Figure 3B). However, when we used the DropArray plates and the optimized washing conditions, NEGR1-Fc binding to LSAMP-, HNT-, or OPCML-transfected HEK293T or S cells was perfectly detectable and mean fluorescence intensity equivalent or greater than COS7 cells. This experiment involving several cycles of washes was possible exclusively because the DropArray technology enables 15 or more wash cycles of suspension cells with minimum cell loss (live or fixed; supplemental Figure 2). These results clearly demonstrate the ability to use semiadherent and suspension-adapted cells in multistep experimental procedures with the DropArray technology. In addition to detecting and quantifying protein binding events using an image-based readout, we also demonstrated the compatibility of the DropArray technology with flow cytometry (supplemental Figure 3). In this case, COS7 cells were detached from the DropArray plates after the protein interaction staining procedure and subsequently harvested with the use of a HyperCyt sampling system (Intellicyt) and analyzed using a bench-top flow cytometer (Accuri). Identical protein-protein interaction data were generated using either readout.
Evaluation of multiple cell viability assays using the DropArray technology with suspension cells
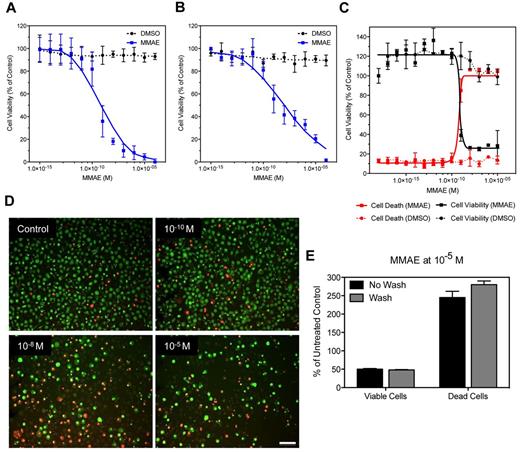
Next, we evaluated the DropArray technology in various cell viability assays using the human leukemic monocyte lymphoma cell line U937. Cells were treated with increasing concentrations of the MMAE, a synthetic analog of the natural product dolastatin 10 and very potent antimitotic agent, which inhibits cell division by blocking the polymerization of tubulin.27 We first measured the U937 cell viability after 72 hours' treatment with MMAE concentration ranging from 10−15 to 10−5M using CellTiter-Glo luminescent cell viability assay, a homogeneous method of determining the number of viable cells in culture determined by the quantitation of the ATP present, an indicator of metabolically active cells (Figure 4A). Using this method, we applied a plastic 384-well plate grid on the DropArray plate to individualize each spot to avoid cross-talk between spots during the acquisition of the luminescent signal. In these conditions, the calculated half maximal inhibitory concentration (IC50) was equal to 0.5nM with a standard deviation equal to 0.27 × 10−9 (Figure 4A). Similar data were observed with U937 cells treated with MMAE concentration ranging from 10−15 to 10−5M in standard 384-well plate when we used the CellTiter-Glo luminescent cell viability readout (supplemental Figure 4). We compared this homogeneous cell viability assay readout to a passive immunofluorescence-based viability readout using CMFDA. The CMFDA fluorescent probe freely passes through cell membranes and is converted to a cell-impermeant reaction product. To summarize, U937 cells were treated with MMAE (same dose response curve described previously) in the presence of CMFDA. After 72 hours incubation cells treated on the DropArray plates were washed 3 times and counted by an image analysis method (see “Methods”). In these conditions, the MMAE IC50 was equal to 0.81nM with an SD equal to 0.43 × 10−9 (Figure 4B).
Fluorescence and luminescence cell viability assays. (A-C) Quantitation of cell viability of U937 cells after a 72-hour treatment with increasing concentrations of the antimitotic agent MMAE. (A) The cells were then lysed and incubated with CellTiter-Glo to measure the amount of ATP in the cells. This is a correlative readout for cell viability and proliferation. The plate was read on a microplate luminometer after we placed a custom-made plastic grid over the plate to prevent well-to-well light contamination. (B) U937 cells were labeled with CMFDA 24 hours before the addition of MMAE. After the 72-hour MMAE treatment, the plate was washed to remove dead cells and cellular debris and then scanned on an Image Xpress Velos laser scanner and the number of cells remaining was calculated. (C) U937 cells were dual stained with EB and AO to measure live, apoptotic, and necrotic cells simultaneously. This dual stain causes live cells to fluoresce green while apoptotic cells retain a red-orange fluorescence. (D) Visualization of the images used to acquire data for (C). Scale bar = 70 μm. (E) Quantitation of the effect of washing the plates after MMAE treatment and EB/AO staining on the number of dead versus live cells remaining on the plate surface reveals that the washing steps do not affect the overall counts for live or dead cells.
Fluorescence and luminescence cell viability assays. (A-C) Quantitation of cell viability of U937 cells after a 72-hour treatment with increasing concentrations of the antimitotic agent MMAE. (A) The cells were then lysed and incubated with CellTiter-Glo to measure the amount of ATP in the cells. This is a correlative readout for cell viability and proliferation. The plate was read on a microplate luminometer after we placed a custom-made plastic grid over the plate to prevent well-to-well light contamination. (B) U937 cells were labeled with CMFDA 24 hours before the addition of MMAE. After the 72-hour MMAE treatment, the plate was washed to remove dead cells and cellular debris and then scanned on an Image Xpress Velos laser scanner and the number of cells remaining was calculated. (C) U937 cells were dual stained with EB and AO to measure live, apoptotic, and necrotic cells simultaneously. This dual stain causes live cells to fluoresce green while apoptotic cells retain a red-orange fluorescence. (D) Visualization of the images used to acquire data for (C). Scale bar = 70 μm. (E) Quantitation of the effect of washing the plates after MMAE treatment and EB/AO staining on the number of dead versus live cells remaining on the plate surface reveals that the washing steps do not affect the overall counts for live or dead cells.
Finally, we evaluated the EB and AO staining assay method.28,29 In this assay AO permeates all cells and makes the nuclei appear green (Figure 4C-D). EB is only taken up by cells that are dying resulting in loss of cytoplasmic membrane integrity and stains the nucleus red. EB also dominates over AO. Thus, live cells have a green nucleus whereas apoptotic/necrotic cells retain the distinctive red-orange fluorescence (Figure 4D). Using the DropArray technology, we performed this assay with or without washing after treating U937 cells with MMAE for 72 hours has described previously. Using a simple image-based analysis to count either the decrease in viable cells (green) or the increase in apoptotic/necrotic cells (red-orange), we found that the MMAE IC50 was equal to 0.67nM with an SD equal to 0.27 × 10−7 (Figure 4C-D). Performing the image analysis either in presence of the AO/EB stains (homogeneous assay format) or after a 3-wash cycle of the DropArray plate did not modify the results of the assay (Figure 4E), demonstrating the ability of the DropArray technology to preserve the integrity of the apoptotic/necrotic as much as the viable cells on the DropArray plate even with a rinsing step. Finally, the 3 different cell viability readouts provided comparable IC50 values for MMAE on U937 suspension cell lines (average 0.66 ± 0.15nM). These values are within the IC50 range described for this antimitotic agent on various human lymphoma cell line when standard plate format is used.30
In addition, we also successfully demonstrated the possibility of using the DropArray plates for high-resolution immunofluorescence (supplemental Figure 5) and real-time imaging (supplemental Figure 6) applications. The technology also was successfully implemented to grow and maintain (up to 10 days) various cell types from various species, including mouse and rat (supplemental Figure 7). Overall, our results demonstrate the broad compatibility of the DropArray technology with a wide range of cell-based experimental procedures and assay readouts, including suspension cells without the need for any centrifugation before or after the wash/rinsing steps.
Application of the DropArray technology for the evaluation of bispecific antibodies
Stimulation of the immune response is a promising strategy to treat oncologic diseases. Among the different cancer immunotherapy modalities, the use of bispecific antibodies designed to engage cytotoxic T cells and trigger tumor cell killing represent a very active field of investigations.31 To date in vitro characterization of bispecific antibody potency is primarily performed with flow cytometry analysis.31 Although this approach provides valuable data, it has significant limitations: it is performed in conventional U-bottom microtiter plates that force the physical interactions between T and B lymphoma cells; it does not distinguish between cell killing events mediated through the bispecific antibody versus random killing; and, finally, the method relies on an end-point readout and therefore does not provide any information regarding killing event kinetic. Considering the great performances of the DropArray technology when suspension cells in different cell-based applications are used, it became apparent that the platform could have an exquisite application toward the in vitro evaluation of bispecific antibodies. This technology provides the unique ability to generate qualitative and quantitative data in a single experimental set up to better characterize the potency and mode of action of bispecific antibodies.
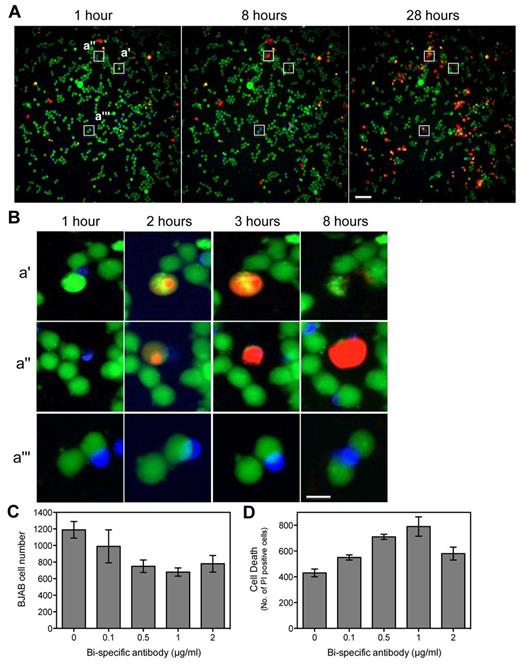
To summarize, the B lymphoma cell line BJAB was seeded on the DropArray plate and stained overnight with CMFDA. The next day BJAB cells were treated for 1 hour with increasing doses of anti-CD3/CD19 Bispecific T-cell engager antibody18 from 0.1 to 2 μg/mL. Meanwhile primary human CD8+ cells were purified and subsequently stained with the use of an APC-conjugated anti-CD8 antibody. Subsequently, the stained T cells were added on top of the anti-CD3/CD19 bispecific antibody treated BAJB at effector to target ratio of 1:1 and incubated for 1 hour before the DropArray plate was washed and PI added. The DropArray plate was then incubated for 28 hours. Image acquisition was performed at various time points to visualize the interaction between CMFDA-stained B cells (Figure 5 green cells) and anti-CD8–APC-stained T cells (Figure 5 blue cells) and monitor the appearance of PI-positive B cells (Figure 5 red cells) over time. Over time, the number of CMFDA+ BJAB cells significantly decreased in response to 1 μg/mL of anti-CD3/CD19 bispecific antibody whereas the number of PI+ BJAB cells increased (Figure 5A).
Visualizing T cell–mediated B-cell killing using bispecific antibodies. (A) CMFDA-labeled BJAB cells (green) were incubated with the bispecific antibody and anti-CD8–APC-labeled T cells (blue) on the Drop-Array plate. T cell–mediated killing of the BJAB cells is seen as positive PI staining (red) in a green cell adjacent to a blue cell. Over time, the number of PI-positive cells increases. The inset regions indicate the areas magnified in panel B. Scale bar = 70 μm. (B) Three different interaction events are indicated. Panel i shows an event where a T cell is adjacent to a BJAC cell and causes the death of that cell as indicated by positive PI staining. Panel ii shows an event in which the T cells cause cell killing of the BJAB cell but the PI positive cell does not burst until much later than the 8-hour time point. Panel iii shows an event in which a T cell is adjacent to 2 BJAB cells and fails to kill either of them. Scale bar = 14 μm. (C) Quantitation of the number of BJAB cells (green) remaining at 28 hours with increasing concentrations of the bispecific antibody. (D) Quantitation of the number of PI-positive cells (red) after 28 hours with increasing concentrations of the bispecific antibody.
Visualizing T cell–mediated B-cell killing using bispecific antibodies. (A) CMFDA-labeled BJAB cells (green) were incubated with the bispecific antibody and anti-CD8–APC-labeled T cells (blue) on the Drop-Array plate. T cell–mediated killing of the BJAB cells is seen as positive PI staining (red) in a green cell adjacent to a blue cell. Over time, the number of PI-positive cells increases. The inset regions indicate the areas magnified in panel B. Scale bar = 70 μm. (B) Three different interaction events are indicated. Panel i shows an event where a T cell is adjacent to a BJAC cell and causes the death of that cell as indicated by positive PI staining. Panel ii shows an event in which the T cells cause cell killing of the BJAB cell but the PI positive cell does not burst until much later than the 8-hour time point. Panel iii shows an event in which a T cell is adjacent to 2 BJAB cells and fails to kill either of them. Scale bar = 14 μm. (C) Quantitation of the number of BJAB cells (green) remaining at 28 hours with increasing concentrations of the bispecific antibody. (D) Quantitation of the number of PI-positive cells (red) after 28 hours with increasing concentrations of the bispecific antibody.
Multiple fields in which T cells were in contact with B lymphoma cells were monitored over time. Three representative fields (i-iii) are presented in Figure 5B. In the first case (i) the interaction between the BJAB and T cells resulted in the death of the B lymphoma cell as early as 2 hours with a complete bursting of the lymphoma cell at the 8-hour time point. In the second case (ii), the interaction between the BJAB and T cell resulted in the death of the B lymphoma cell as early as 2 hours. The lymphoma cell is entirely PI positive by 8 hours but does not burst until the 24-hour time point (data not shown). In other cases, despite the clear interaction between the BJAB and T cells at the 1 hour time point, either no cellular death was observed at any subsequent time points up to 28 hours (Figure 5iii) or very late at 24 hours despite continuous cell contact (data not shown). Using image analysis software (IN Cell Developer v1.9, GE Healthcare), we determined the number of CMFDA+ BJAB cells, PI+ cells, and anti-CD8–APC+ T cells in every condition at every time point. The maximum BJAB cell killing either defined as the lowest number of CMFDA+ cells (Figure 5C) or the highest number of PI+ BJAB cells (Figure 5D) was observed in response to 1 μg/mL of bispecific antibody at any time point. No change in the number of anti-CD8–APC+ T cells was observed at any bispecific antibody concentrations tested regardless of time point (data not shown). Significant B lymphoma cell killing compared with untreated condition was observed as early as 4 hours and as late as 12 hours, depending on the primary T-cell donor (data not shown).
Discussion
Here, we report for the first time the development and implementation of cell-based assay conditions via the use of a novel wall-less plate technology termed DropArray. This technology uses a unique liquid dynamic, taking advantage of hydrophobic and hydrophilic surface areas, and provides a great opportunity to potentially overcome the major limitations encountered with a conventional microtiter plate format. Using a flow chamber, we determined experimentally that a flow rate of 5 mm/s maximum would be optimal to ensure minimum loss of suspension cells on the DropArray spots while enabling optimum washing. Subsequently, we demonstrated that oscillation of the plate during washing at a rate of 44 rpm resulted in a shear force average compatible with the retention of suspension cells across the entire plate. Knowing that suspension cells would be retained on the DropArray plate during the wash steps using our optimized conditions, we then investigated the use of this novel wall-less plate technology in various cell-based assay formats. Implementation of the DropArray technology was successfully carried on in the context of an expression cloning platform applied to the detection of specific protein-protein interactions using adherent, semiadherent and suspension cells.
This specific application benefited from several key features of this novel plate technology compared with conventional 384-well plates. First, the DropArray enabled the use of semiadherent and suspension cells in multistep experimental procedures involving numerous washing steps. To date, using conventional plate formats this application was mostly suitable to adherent cells. This finding represents a critical advantage of the DropArray because protein expression could significantly vary depending on the cell line, providing the investigator with the ability to use the best mammalian expression system regardless of the cell adhesion properties. In addition, because the DropArray technology use minute amounts of media (2-4 μL per spot), its implementation results in a significant reduction of the reagents consumption and consequently could reduce by up to 10-fold the cost of the assay compared with the conventional microtiter plate approaches.
Investigating multiple cell viability assay readouts, we also demonstrated that the DropArray technology could be used in conjunction with a broad spectrum of experimental procedures and readouts. Furthermore, despite the use of suspension cells in a killing assay, we demonstrated that the use of wash steps on the DropArray does not impact the number of viable and dead cells, indicating that the optimized washing conditions not only warrant the optimum retention of the viable cells but also dying/dead cells. These results suggest that the DropArray technology could be used with both adherent and suspension cells in a variety of existing cell-based assay procedures using well-established readouts. Similarly to the standard plate format, the DropArray plate format could be implemented in various laboratory settings using specific washing stations designed for low- or high-throughput applications while manual washing implementation could also be considered for occasional use.
To further demonstrate how enabling the DropArray technology could be when using suspension cells, we turned our attention to the in vitro characterization of bispecific antibodies designed to engage cytotoxic T cells and trigger tumor cell killing. More specifically we evaluated the ability of a recombinant anti-CD3/CD19 bispecific T-cell engager antibody to induce highly efficacious lymphoma-directed cytotoxicity mediated by unstimulated peripheral T lymphocytes.32 To date, the characterization of the bispecific antibodies in the context of T cell–mediated B lymphoma cell killing was mostly limited to FACS based analysis using a conventional round-bottom 96-well plate format. Although this approach provides valuable data about the in vitro potency of the antibody, it is fairly limited in term of visualization of the killing events, preventing the investigator to differentiate the random death of individual B cells versus killing events associated with a direct interaction between B and T cells. In addition, the use of round-bottomed plates biases the experiment toward more B and T cells contact independently of the presence of the bispecific antibody. Here the DropArray provided the unique opportunity to develop a completely new assay method that overcomes these limitations. Using this technology, we have visualized and quantified B-cell death events specifically associated with a T-cell contact. Interestingly, the cytotoxicity activity of the anti-CD3/CD19 bispecific antibody observed with the DropArray technology was significantly lower than described with the conventional FACS-based analysis in a conventional round-bottom 96-well plate format.32 This result might represent a more physiologically relevant estimation of the cytotoxic activity of this bispecific antibody for several reasons: the excess of antibody was washed off after priming incubation, the cells were treated on a flat surface avoiding bias toward nonspecific B- and T-cell interactions and B-cell killing events associated with T-cell interaction at one point in time were specifically tracked. Furthermore the DropArray method enabled us to monitor T cell–mediated B lymphoma cell killing in response to bispecific antibody over time, allowing us to evidence significant difference in the kinetic of the cell killing between T lymphocyte donors. The ability of the method to track cell killing events in real time also provides us the opportunity to further characterize bispecific antibodies mechanism of action.
In conclusion, the DropArray technology described here enables the application of existing as well as the development of novel cell-based assay methods via the use of various cell types, including suspension cells. This technology provides the opportunity to overcome many of the current limitations in using suspension cells in complex multistep experimental procedures and will undoubtedly expand the use and relevance of high-throughput cell-based assays in multiple research areas including target discovery, therapeutic lead selection, and characterization.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Namyong Kim, Melvin Lye, and Dr John Zhong Wang from Curiox Biosystems for their expert assistance and support; Carmen Chan for providing purified cDNA; and Dr Wai Lee T. Wong for her support and enthusiasm.
Authorship
Contribution: G.A.Q. and T.I.M. designed and performed the research and analyzed data; K.N., H.L., and S.K. performed the research; L.S. contributed vital new reagents; N.L.J. designed the research; and J.-P.S. designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: G.A.Q., K.N., L.S., and J.-P.S. are employees of Genentech Inc. The remaining authors declare no competing financial interests.
Correspondence: Jean-Philippe Stephan, Protein Chemistry Department, Genentech Inc, 1 DNA Way, South San Francisco, CA 94080; e-mail: stephanj@gene.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal