In this issue of Blood, Hilcenko et al show that the USB1 gene (alias C16orf57 and MPN1), which is mutated in poikiloderma with neutropenia, is an exonuclease active in processing spliceosomal U6snRNA, a fascinating but enigmatic observation that opens up investigation of the pathogenesis of this rare condition.1
Poikiloderma with neutropenia (PN) is a rare autosomal recessive disease characterized by early onset poikiloderma, pachyonychia, palmo-plantar hyperkeratosis, skeletal defects, and noncyclic neutropenia.2 Since 2010 it has been known that the causative mutations are mainly biallelic truncating mutations in a gene of unknown function, C16orf57, located at 16q21.3,4 Three recent papers, including one in this issue of Blood, have reported that the gene encodes a protein that is essential for the processing and stability of U6 snRNA, a molecule with a crucial role in RNA splicing.1,5,6
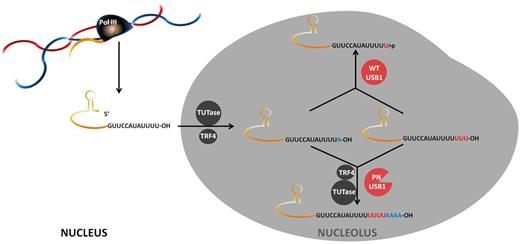
U6 snRNA along with 4 other snRNAs (U1, U2, U4 and U5) and their associated proteins make up the spliceosome complex that catalyses the removal of introns from mRNA. U6 is associated with the 5′ end of the intron by base pairing before lariat formation. U6 snRNA is transcribed by RNA polymerase III, unlike the other spliceosomal snRNAs, which use RNA pol II. In yeast newly transcribed U6 terminates at a uridine tract, a typical Pol III termination signal while human U6 is oligouridylated posttranscriptionally. Curiously, 90% of human U6 molecules have short 3′ terminal U tracts consisting of 5 Us blocked by a cyclic 2′3′ phosphate group at the 3′ end while the other 10% can have long 3′ U tracts, up to 20 Us in length and terminating with an -OH group.7 The functional significance of this variation is unknown, although dynamic interactions of the 3′ end of U6 with protein components of the spliceosome are involved in the function and recycling of the snRNA. The structure at the U6 3′ end is determined by a balance between a terminal uridyltransferase (TUTase) and a nuclease that trims the 3′ terminus. The identification of this nuclease in yeast and humans as the enzyme mutated in PN is the subject of these new papers.
The work that led to this finding took different routes in the 3 groups. Mroczek et al constructed Saccharomyces cerevisiae strains in which the homolog of the C16orf57 gene, which they call USB1 (for U 6 Biogenesis 1)was inducible; cells without Usp1p had a growth defect which was suppressed by overexpression of either wild-type Usb1p or U6-snRNA, suggesting Usb1p is involved in the regulation of U6.5 Shchepachev et al were screening for mutants in Schizosaccharomyces pombe that regulated transcription at telomeres and found a gene, which they call mpn1 (for mutated in poikiloderma with neutropenia), whose deletion leads to high levels of telomere transcripts (TERRA) and shorter telomeres.6 Further investigation revealed that Mpn1p lacking cells had a generalized pre-mRNA splicing defect including accumulation of unspliced U6-snRNA precursor (the gene contains an intron in S pombe) and this led to further investigation of U6 RNA biogenesis. Finally, Hilcenko et al deduced from the crystal structure of USB1 that the protein was a 3′-5′ exoribonuclease with structural homology to some cyclic nucleotide phophodiesterases.1 Elegant biochemical experiments led Hilcenko et al to the conclusion that the enzyme was a 3′-5′ exonuclease that leaves a uridine with a cyclic phosphate group at the 3′ end, the type of enzyme hypothesized to be involved in U6-snRNA 3′ end processing. They went on to demonstrate by complementation that it did indeed act on U6 snRNA.
Clues as to what goes wrong in PN were provided by all 3 groups, showing that U6 3′ end processing was aberrant in patient cells with some decrease in the stability of U6 snRNA. Mutants that were found in patients could not correct the defect caused by lack of Usb1p in yeast whereas WT human USB1 could. A model emerged whereby U residues are added to the 3′ end of U6 by TUTase and USB1 is the exonuclease that trims the oligo(U) tails and leaves 5Us at the end with a terminal 3′ cyclic phosphate. Hilcenko et al further found that USB1 was capable of degrading oligo A tracts and showed that in PN patient cells A residues were present in the 3′ U6 tails. They speculate that failure to remove these A residues may target the U6 RNA for degradation by the exosome, because oligoA tracts are a hallmark of RNA surveillance targets (see figure). Interestingly, all the papers agreed that no general splicing defects could be detected in PN patient cells, unlike in Usb1p deficient yeast.
Model for the function of USB1 in U6 snRNA processing according to Hilcenko et al.1 The U6 is transcribed by RNA polIII and terminates with UUUU and a 3′OH. The transcript is then extended with U residues by TUTase. A residues can also be incorporated. USB1 chews back the 3′ tail, leaving the RNA with 5 terminal U residues and a 3′ cyclic phosphate, which cannot be a substrate for polyA polymerase TRF4. When USB1 is defective in PN, TRF4 can use the 3′OH terminus as a substrate and oligoadenylate the RNA, which can then be degraded by the exosome. Illustration by Nieves Perdigones.
Model for the function of USB1 in U6 snRNA processing according to Hilcenko et al.1 The U6 is transcribed by RNA polIII and terminates with UUUU and a 3′OH. The transcript is then extended with U residues by TUTase. A residues can also be incorporated. USB1 chews back the 3′ tail, leaving the RNA with 5 terminal U residues and a 3′ cyclic phosphate, which cannot be a substrate for polyA polymerase TRF4. When USB1 is defective in PN, TRF4 can use the 3′OH terminus as a substrate and oligoadenylate the RNA, which can then be degraded by the exosome. Illustration by Nieves Perdigones.
With apologies to Winston Churchill, these remarkable discoveries at the end of U6 mark the beginning of our understanding of PN pathogenesis. Although the culprit molecule has been found, there is still no indication how PN develops. Failure to find a splicing defect does not support aberrant splicing as a mechanism, although it is possible that the defect was not present in the lymphocyte cell lines used in these experiments but may be expressed in the important cells in vivo, neutrophil precursors in the bone marrow, or in the keratinocyte. It is also possible that USB1 acts on targets other than U6 snRNA, perhaps on RNAs not involved in splicing. Interestingly, patients with USB1 mutations have also been diagnosed with Rothmund-Thomson syndrome (RTS) and dyskeratosis congenita (DC) due to considerable phenotypic overlap in the presentation of these 3 conditions. All 3 show poikiloderma; DC and PN both have bone marrow manifestations; and DC and RTS share predisposition to malignancies.4 A subset of RTS patients have bone marrow failure. RTS is usually caused by mutations in RECQL4, encoding a helicase that functions in DNA damage repair and has recently been shown to be involved in telomere maintenance.8 DC is caused by mutations in a number of genes encoding products essential for telomere maintenance and usually presents with very short telomeres. Although PN patients have normal length telomeres, it is interesting that Shchepachev et al picked up the yeast Mpn1 gene in a screen for modulators of telomere transcription and Mpn1 deletion was associated with short telomeres in yeast.5 Although short telomeres are not found in peripheral blood cells of RTS or PN patients, telomere integrity may be compromised only in the affected cells, the myelocytes and keratinocytes, and possibly without measureable shortening. Telomeres, which cap and protect the ends of chromosomes, may be a common factor in these 3 conditions.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal