Key Points
Immune monitoring models integrating multiple functions of HIV-1–specific CD8 T cells distinguish controllers from subjects with progressive HIV-1 infection.
This strategy may have important applications in predictive model development and immune monitoring of HIV-1 vaccine trials.
Abstract
The development of immunomonitoring models to determine HIV-1 vaccine efficacy is a major challenge. Studies suggest that HIV-1–specific CD8 T cells play a critical role in subjects achieving spontaneous viral control (HIV-1 controllers) and that they will be important in immune interventions. However, no single CD8 T-cell function is uniquely associated with controller status and the heterogeneity of responses targeting different epitopes further complicates the discovery of determinants of protective immunity. In the present study, we describe immunomonitoring models integrating multiple functions of epitope-specific CD8 T cells that distinguish controllers from subjects with treated or untreated progressive infection. Models integrating higher numbers of variables and trained with the least absolute shrinkage and selection operator (LASSO) variant of logistic regression and 10-fold cross-validation produce “diagnostic tests” that display an excellent capacity to delineate subject categories. The test accuracy reaches 75% area under the receiving operating characteristic curve in cohorts matched for prevalence of protective alleles. Linear mixed-effects model analyses show that the proliferative capacity, cytokine production, and kinetics of cytokine secretion are associated with HIV-1 control. Although proliferative capacity is the strongest single discriminant, integrated modeling of different dimensions of data leverages individual associations. This strategy may have important applications in predictive model development and immune monitoring of HIV-1 vaccine trials.
Introduction
Rare HIV-1–infected subjects (HIV-1 controllers) capable of restricting virus replication, maintaining high CD4 counts, and remaining disease free for decades without therapy provide evidence that HIV-1 can be controlled by the immune system.1-3 Multiple studies suggest that CD8 T cells contribute to this control.4-8 The high prevalence of specific HLA class I alleles (HLA-I) in controllers strongly supports a role of CD8 T cells in viral suppression.1,9,10 The clearest genotypic data have been established for HLA-B*5701, HLA-B*5703, HLA-B*2705,5,11-13 and, in African ethnicities, HLA-B*5801.14,15 Further studies suggest a beneficial impact of HLA-B*14, HLA-B*51, HLA-B*81, and HLA-Cw*1402. Viral escape mutations in CD8 T-cell epitopes that coincide with viremia increase demonstrate the relevance of these responses in vivo.16-18
Studies also suggest qualitative differences between HIV-1–specific CD8 T-cell responses of controllers and those of subjects with progressive disease. These characteristics include maintained proliferative capacity,4,19 higher production of IL-220,21 and IL-21,22 stronger “polyfunctionality” in terms of cytokine production,23,24 up-regulation of perforin after in vitro stimulation,4,25,26 higher expression of the transcription factor T-bet,26 decreased sensitivity of HLA-B*2705– and HLA-B*5701/03–restricted responses to inhibition by regulatory T cells,9 and stronger antiviral activity in vitro.27,28 In addition, products of the HIV-1 Gag gene are preferentially targeted by CD8 T cells in people with low viremia29,30 and HIV-1–specific CD8 T cells restricted by HLA-B are more polyfunctional than those restricted by HLA-A alleles.29,31 However, for any single variable studied, there is significant overlap among cohorts with distinct HIV-1 infection outcomes, in particular between subjects with spontaneous versus therapy-induced control of viremia.32,33 The tremendous heterogeneity of responses targeting different epitopes within subjects4,19,31,34 and the ensuing challenge of differentiating effective from irrelevant responses further complicates the discovery of determinants of protective immunity. Therefore, simultaneous consideration of different functions of HIV-1 epitope–specific CD8 T-cell responses may be more powerful in predicting protective immunity in HIV-1–infected subjects and vaccine recipients.
In the present study, we report the building of immunomonitoring models based on high-dimensional analysis of a set of CD8 T-cell functions easily measurable in vitro that accurately discriminates between HIV-1–infected subjects with different disease outcomes. Analyses identify links between HIV-1–specific CD8 T-cell functions, HLA-I alleles, and disease stage. Models integrating a higher number of variables and trained with the least absolute shrinkage and selection operator (LASSO) variant of logistic regression35 and 10-fold cross-validation produce “diagnostic tests” that display an excellent capacity to delineate subject categories. The models remain highly effective at discriminating classes of subjects in cohorts adjusted for prevalence of protective alleles. The results reveal a remarkable role of HIV-1–specific CD8 T-cell proliferation as a correlate of spontaneous viral control and suggest that whereas the quest for a critical determinant of protective cellular immunity against HIV-1 remains elusive, combining variables reflecting different aspects of the HIV-1–specific response may be an important alternative to inform on its overall efficacy. This strategy also substantiates the rationale for developing and testing such approaches in longitudinal models to predict disease outcome and to identify functional signatures of protective immunity elicited by preventive and therapeutic HIV-1 vaccines.
Methods
Study subjects
HIV-1–infected subjects (n = 84) were recruited from outpatient clinics at local Boston hospitals or were referred from providers throughout the United States. Written informed consent was obtained from every participant per the Declaration of Helsinki under institutional review board–approved protocols. HIV-1 controllers (n = 36) included elite controllers (n = 23) with HIV-1 RNA below the level of detection by ultrasensitive assay (< 50 copies/mL by PCR) and viremic controllers (n = 13) with HIV-1 RNA levels < 2000 copies/mL. In this study, elite controllers and viremic controllers were analyzed as 1 group commonly referred to as HIV-1 controllers. Chronic untreated progressors (n = 18) were defined as having HIV-1 RNA levels above 2000 copies. Chronically infected subjects on antiretroviral treatment (ART; n = 30) had undetectable HIV-1 plasma viral RNA on combination therapy (Table 1).
Baseline characteristics of the participants
| Characteristic . | Discovery cohort (n = 64) . | Validation cohort (n = 20) . | |||
|---|---|---|---|---|---|
| HC (n = 26) . | CP (n = 15) . | ARTC (n = 23) . | HC (n = 10) . | ARTC/CP (n = 10) . | |
| Sex, n (%) | |||||
| Male | 25 (96%) | 12 (80%) | 21 (91%) | 6 (60%) | 8 (80%) |
| Female | 1 (4%) | 3 (20%) | 2 (9%) | 4 (40%) | 2 (20%) |
| Age, y | |||||
| Median | 46 | 44 | 47 | 48 | 47 |
| Q1-Q3 | 39-54 | 37-48 | 43-52 | 45-64 | 45-59 |
| CD4+ T cells/mm3 | |||||
| Median | 761 | 477 | 463 | 745 | 371 |
| Q1-Q3 | 588-904 | 247-591 | 322-684 | 521-994 | 305-589 |
| HIV-1 log10 copies/mm3 | |||||
| Median | 1.8 | 4.2 | 1.7 | 1.9 | 1.9 |
| Q1-Q3 | 1.7-2.4 | 3.6-4.6 | 1.7-2.1 | 1.7-3.0 | 1.7-4.7 |
| Protective HLA alleles, n (%)* | |||||
| Any | 16 (62%) | 7 (47%) | 14 (61%) | 7 (70%) | 4 (40%) |
| B*5701/03 | 10 (38%) | 4 (27%) | 7 (30%) | 7 (70%) | 2 (20%) |
| B*2705 | 5 (19%) | 2 (13%) | 6 (26%) | 0 (0%) | 1 (10%) |
| B*5801 | 2 (8%) | 1 (7%) | 1 (4%) | 0 (0%) | 1 (10%) |
| Characteristic . | Discovery cohort (n = 64) . | Validation cohort (n = 20) . | |||
|---|---|---|---|---|---|
| HC (n = 26) . | CP (n = 15) . | ARTC (n = 23) . | HC (n = 10) . | ARTC/CP (n = 10) . | |
| Sex, n (%) | |||||
| Male | 25 (96%) | 12 (80%) | 21 (91%) | 6 (60%) | 8 (80%) |
| Female | 1 (4%) | 3 (20%) | 2 (9%) | 4 (40%) | 2 (20%) |
| Age, y | |||||
| Median | 46 | 44 | 47 | 48 | 47 |
| Q1-Q3 | 39-54 | 37-48 | 43-52 | 45-64 | 45-59 |
| CD4+ T cells/mm3 | |||||
| Median | 761 | 477 | 463 | 745 | 371 |
| Q1-Q3 | 588-904 | 247-591 | 322-684 | 521-994 | 305-589 |
| HIV-1 log10 copies/mm3 | |||||
| Median | 1.8 | 4.2 | 1.7 | 1.9 | 1.9 |
| Q1-Q3 | 1.7-2.4 | 3.6-4.6 | 1.7-2.1 | 1.7-3.0 | 1.7-4.7 |
| Protective HLA alleles, n (%)* | |||||
| Any | 16 (62%) | 7 (47%) | 14 (61%) | 7 (70%) | 4 (40%) |
| B*5701/03 | 10 (38%) | 4 (27%) | 7 (30%) | 7 (70%) | 2 (20%) |
| B*2705 | 5 (19%) | 2 (13%) | 6 (26%) | 0 (0%) | 1 (10%) |
| B*5801 | 2 (8%) | 1 (7%) | 1 (4%) | 0 (0%) | 1 (10%) |
HC indicates HIV-1 controllers; CP, chronic progressors; and ARTC, ART-treated subjects.
Protective alleles are defined here as B*5701, B*5703, B*2705, and B*5801.
Luminex assays and CFSE proliferation and assays
Freshly isolated PBMCs were either labeled with 0.5μM CFSE (Molecular Probes, Invitrogen) according to the manufacturer's protocols or left unstained before being stimulated in parallel with the same panel of donor HLA-I–matched optimal HIV-1 epitopes. To determine peptide concentrations that would induce optimal stimulation of controller and progressor PBMCs, we first tested serial peptide dilutions ranging from 100-0.0001 μg/mL. At least 3 different epitopes were tested in 3 controllers and 3 progressors and the peptide concentration that induced half maximum (EC50) responses were determined. (supplemental Figure 1a-b, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We picked 10 μg/mL because it induced maximum responses with the lowest background for all our stimulation conditions in all subject groups. Cells were plated at a concentration of 2.0 × 105 cells per well in a round-bottom 96-well plate. Supernatant of the unlabeled cells were harvested at 6, 24, and 72 hours after stimulation, inactivated with Triton X-100, and stored at −80 degrees until use. Secretion of the cytokines IFN-γ, IL-2, and TNF-α was measured at each time point by Luminex technology using the Milliplex Map Kit: High Sensitivity Human Cytokine (Millipore) according to the manufacturer's instructions. Cytokine levels were measured using the Bio-Rad Bioplex-200 System. CFSE-labeled PBMCs were harvested after 7 days of stimulation, stained with Abs against CD3, CD4, and CD8 (BD/Pharmingen), and assessed for functional proliferation by flow cytometry (LSRII flow cytometer; BD Biosciences). Proliferation was calculated by determining the fraction of CFSElow CD8 T cells using FlowJo Version 9.4 analysis software (TreeStar).
Combined ICS and CFSE assay
One million PBMCs were stained with CFSE and stimulated with HIV-1 optimal peptides. Cells were collected at 6, 24, 72, 96, and 168 hours after stimulation. After 1 hour, 10 ng/mL of brefeldin A (Sigma-Aldrich) was added and the cells were incubated for another 5 hours. Intracellular cytokine staining (ICS) was performed as described previously.36 Briefly, cells were first stained with dead cell dye for 10 minutes then washed and surface stained with anti-CD3, CD4, CD8, CD14, CD19, and CD56. The cells were then fixed and permeabilized with Cytofix/Cytoperm solution and stained with cytokine-specific Abs against IFN-γ and IL-2 and TNF-α (all from BD Biosciences). After staining, the cells were resuspended in PBS containing 2% paraformaldehyde and acquired on the BD LSRFortessa flow cytometer (BD Biosciences). Flow cytometry data were analyzed with the FlowJo Version 9.4 software package (TreeStar).
Statistical analysis
General linear mixed models controlling for clustering within subjects was used to compare the adjusted mean proliferation or cytokine secretion between groups. The analyses by general linear mixed models were extended to the kinetics of cytokine secretion integrating time as a continuous variable. We used a compound symmetry correlation structure to control for the correlation within each subject over time. We then compared characteristics of responses between groups stratified by protective versus nonprotective HLA alleles.
Based on the initial analysis, we developed multivariate models to predict whether a subject would be a controller or progressor. These models used LASSO logistic regression, which inherently determines the best factors to use for prediction. Models were assessed on their ability to correctly classify subjects as controller or progressor status, using the area under the receiving operating characteristic (ROC) curve (AUC) on out-of-sample data using 10-fold cross validation. Within each fold, nested 10-fold cross-validation was used to find the optimal value of the regularization parameter. To determine the probability that a given variable was predictive of control, we performed a bootstrap analysis also using LASSO logistic regression. For each bootstrap sample, we used 10-fold cross-validation to identify the optimal value of the regularization parameter and then used that value to train a single model on the entire bootstrap sample. The probability was then given by the fraction of bootstrap samples in which the variable weight was nonzero.
To determine whether 2 models predicted control to a better or worse degree, we determined P values using a permutation test. Each model assigned to each subject a probability that that subject was a controller. For each of the 2 models, we sorted subjects according to this probability forming a corresponding list of observed controller/noncontroller status. We then permuted these status labels in every row, and computed the absolute value of the AUC difference. We performed this permutation 100 000 times, noting how many times the AUC difference exceeded or was equal to the difference on the real data.
Results
Study design and subject characteristics
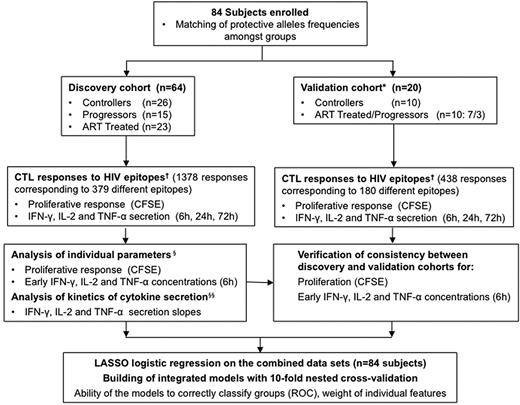
We used the study design presented in Figure 1 to investigate 84 subjects in whom high-resolution HLA-I genotype was determined at the A, B, and C loci.37 For the initial analysis, participants were categorized as a discovery cohort (n = 64) and a validation cohort (n = 20). Subjects were further classified as HIV-1 controllers (“controllers”), untreated subjects with progressive infection (“progressors”), and subjects with controlled viremia on antiviral therapy (“ART-treated”). There were no significant differences in clinical characteristics between the discovery and validation cohorts for the parameters considered, although there was a trend toward higher CD4 counts in the controller group (Table 1). The discovery and validation cohorts were merged for the cross-validation analysis.
Study design and flow chart of immunomonitoring model development. We investigated 84 subjects in whom HLA class I genotype was determined at the A, B, and C loci by high-resolution typing.37 For all alleles present at medium or high frequency in our cohorts, we defined panels of optimal HIV-1 epitopes referenced in the Los Alamos National Laboratory databases (http://www.hiv.lanl.gov) and published data.51,52 The 6 alleles of the HLA genotype of each subject then determined a panel of 16-26 (median 22) HIV-1 epitopes tested in functional assays. The participants were randomly subdivided into a discovery cohort and a validation cohort. Subjects were further classified as controllers, progressors, and ART-treated subjects. The progressor and ART-treated groups were enriched in subjects carrying protective HLA alleles to approximately match their prevalence in the controller groups. We used freshly isolated PBMCs to measure proliferation and cytokine secretion by HIV-1–specific CD8 T cells in response to the single optimal HIV-1 epitopes. We first analyzed individually the immunologic variables generated in the discovery cohort and subsequently combined them to build high-dimensional integrated models. The validation cohort was investigated to verify data consistency with the discovery cohort and to assess the ability of the models trained on the discovery dataset to appropriately discriminate among classes of subjects in independent groups of HIV-1–infected subjects. *Treated and untreated progressors were grouped in the validation dataset to develop a binary predictive model (controller vs treated/progressor classification). Detailed analyses of the discovery cohort dataset showed that combination of these groups is appropriate. †For each subject investigated, the panel of epitopes tested in the functional assays was determined by the HLA class I genotype. §Data were analyzed by general linear mixed models controlling for clustering within subjects. §§Data were analyzed by general linear mixed models controlling for clustering within subjects, with time integrated as a continuous variable.
Study design and flow chart of immunomonitoring model development. We investigated 84 subjects in whom HLA class I genotype was determined at the A, B, and C loci by high-resolution typing.37 For all alleles present at medium or high frequency in our cohorts, we defined panels of optimal HIV-1 epitopes referenced in the Los Alamos National Laboratory databases (http://www.hiv.lanl.gov) and published data.51,52 The 6 alleles of the HLA genotype of each subject then determined a panel of 16-26 (median 22) HIV-1 epitopes tested in functional assays. The participants were randomly subdivided into a discovery cohort and a validation cohort. Subjects were further classified as controllers, progressors, and ART-treated subjects. The progressor and ART-treated groups were enriched in subjects carrying protective HLA alleles to approximately match their prevalence in the controller groups. We used freshly isolated PBMCs to measure proliferation and cytokine secretion by HIV-1–specific CD8 T cells in response to the single optimal HIV-1 epitopes. We first analyzed individually the immunologic variables generated in the discovery cohort and subsequently combined them to build high-dimensional integrated models. The validation cohort was investigated to verify data consistency with the discovery cohort and to assess the ability of the models trained on the discovery dataset to appropriately discriminate among classes of subjects in independent groups of HIV-1–infected subjects. *Treated and untreated progressors were grouped in the validation dataset to develop a binary predictive model (controller vs treated/progressor classification). Detailed analyses of the discovery cohort dataset showed that combination of these groups is appropriate. †For each subject investigated, the panel of epitopes tested in the functional assays was determined by the HLA class I genotype. §Data were analyzed by general linear mixed models controlling for clustering within subjects. §§Data were analyzed by general linear mixed models controlling for clustering within subjects, with time integrated as a continuous variable.
Protective HLA-I alleles drive the remarkable HIV-1–specific CD8 T cell–proliferative capacity of HIV-1 controllers
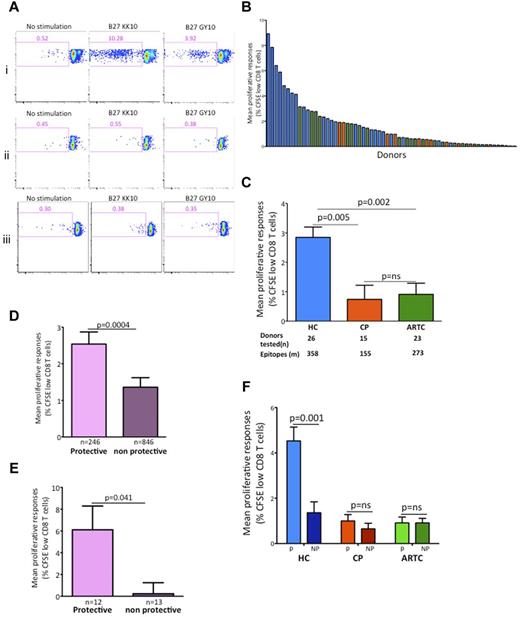
Maintained proliferative capacity of HIV-1–specific CD8 T cells in chronic infection has been described as a distinctive functional feature of controllers.4,19 To determine whether these findings could be generalized, we examined proliferation to optimal HIV-1–specific CD8 T-cell epitopes in the discovery cohort (Figure 2). As illustrated by representative subjects (Figure 2A), controllers typically exhibited stronger proliferative responses than progressors or ART-treated subjects. We calculated the mean of proliferative responses for each subject against all epitopes tested (Figure 2B). Comparison among groups (Figure 2C) demonstrated that proliferative responses were significantly stronger in controllers than in the other 2 groups, whereas progressors and ART-treated subjects did not significantly differ in these assays (Figure 2C), confirming previous findings on fewer epitopes and HLA-I alleles.4,38 Consistent with the data obtained with optimal epitopes, stimulation of CD4-depleted PBMCs with a pool of 66 overlapping peptides (14-18 mers) spanning HIV-1 Gag induced proliferative responses that were more robust in controllers than progressors (supplemental Figure 2). These results suggest that although the selected panels of HLA-matched peptides missed some epitopes, the responses they induced gave a representative picture of HIV-specific CD8 T-cell functionality.
Protective HLA class I alleles drive the differences in HIV-1–specific CD8 T-cell proliferative capacity between controllers and ART-treated or untreated progressors. (A) Flow cytometry of CFSE-labeled CD8 T cells from 3 representative HLA-B*2705 subjects incubated with no antigen, the B*2705-restricted HIV-1 optimal epitope B*2705-KK10, or the HIV-1 optimal epitope B*2705-GY10. Numbers in top left quadrants indicate percent CD3+CD8+CFSElow cells. Whereas robust proliferative responses were observed in the HIV-1 controller subject (i), no significant responses were present in the progressor (ii) or ART-treated (iii) subjects. Responses to both epitopes were detected by IFN-γ Luminex assay in all 3 subjects studied. (B) Hierarchical representation of HIV-1–infected subjects based on their mean proliferative response to panels of HLA-class I matched HIV-1 epitopes. Bars represent data for individual subjects. (C) Statistical analysis of mean proliferative responses among the 3 groups of HIV-1–infected subjects in the discovery cohort (n = 64). HC indicates HIV-1 controllers; CP, chronic progressors; and ARTC, ART-treated subject. (D) Statistical comparison of mean proliferative responses to epitopes restricted by protective versus nonprotective HLA-I alleles in all subjects. (E) Representative single-donor HIV-specific cytotoxic T-lymphocyte (CTL) proliferative responses stratified by protective and nonprotective restricting alleles. (F) Statistical analysis of intraindividual proliferative responses stratified by protective versus nonprotective HLA-I alleles in controllers, progressors, and ART-treated subjects. Throughout the figure, HC data are represented in blue; CP in red; and ARTC in green; protective HLA class I alleles (B*5701/03, B*2705 and B*5801) are shown in purple; and nonprotective HLA I alleles in orange. All comparisons of adjusted means were performed using generalized linear models controlling for clustering within subjects. Comparison of mean proliferative responses among the 3 groups were done by ANOVA, followed by posttest comparisons with Tukey. Comparisons of protective versus nonprotective alleles were performed with Student t test. Vertical interval bars correspond to the SEM.
Protective HLA class I alleles drive the differences in HIV-1–specific CD8 T-cell proliferative capacity between controllers and ART-treated or untreated progressors. (A) Flow cytometry of CFSE-labeled CD8 T cells from 3 representative HLA-B*2705 subjects incubated with no antigen, the B*2705-restricted HIV-1 optimal epitope B*2705-KK10, or the HIV-1 optimal epitope B*2705-GY10. Numbers in top left quadrants indicate percent CD3+CD8+CFSElow cells. Whereas robust proliferative responses were observed in the HIV-1 controller subject (i), no significant responses were present in the progressor (ii) or ART-treated (iii) subjects. Responses to both epitopes were detected by IFN-γ Luminex assay in all 3 subjects studied. (B) Hierarchical representation of HIV-1–infected subjects based on their mean proliferative response to panels of HLA-class I matched HIV-1 epitopes. Bars represent data for individual subjects. (C) Statistical analysis of mean proliferative responses among the 3 groups of HIV-1–infected subjects in the discovery cohort (n = 64). HC indicates HIV-1 controllers; CP, chronic progressors; and ARTC, ART-treated subject. (D) Statistical comparison of mean proliferative responses to epitopes restricted by protective versus nonprotective HLA-I alleles in all subjects. (E) Representative single-donor HIV-specific cytotoxic T-lymphocyte (CTL) proliferative responses stratified by protective and nonprotective restricting alleles. (F) Statistical analysis of intraindividual proliferative responses stratified by protective versus nonprotective HLA-I alleles in controllers, progressors, and ART-treated subjects. Throughout the figure, HC data are represented in blue; CP in red; and ARTC in green; protective HLA class I alleles (B*5701/03, B*2705 and B*5801) are shown in purple; and nonprotective HLA I alleles in orange. All comparisons of adjusted means were performed using generalized linear models controlling for clustering within subjects. Comparison of mean proliferative responses among the 3 groups were done by ANOVA, followed by posttest comparisons with Tukey. Comparisons of protective versus nonprotective alleles were performed with Student t test. Vertical interval bars correspond to the SEM.
To define the role played by restricting alleles, we next examined the responses classified as restricted by either protective (conservatively defined as HLA-B*5701, HLA-B*5703, HLA-B*2705, and HLA-B*5801) or nonprotective HLA-I molecules (all others). At the level of the entire discovery cohort, proliferative responses restricted by protective alleles were strongly immunodominant (Figure 2D). Differences between protective and nonprotective alleles were also found within controller subjects (Figure 2E). We next extended these intraindividual comparisons to the different groups. The proliferative responses were uniquely robust when restricted by protective alleles in controllers (Figure 2F). In contrast, responses restricted by nonprotective alleles in controllers and either protective or nonprotective alleles in progressors and ART-treated subjects were characterized by low proliferative capacity. These data obtained on a large number of epitopes indicate that the protective alleles HLA-B*2705, HLA-B*5701/03, and HLA-B*5801 restrict HIV-1–specific CD8 T cells with robust proliferative potential in controllers only: optimal control of antigen load by ART does not restore defects observed in progressive infection. Moreover, they show that even in controllers, proliferation is not a feature of all antigen-specific responses, only of those restricted by protective HLA-I alleles.
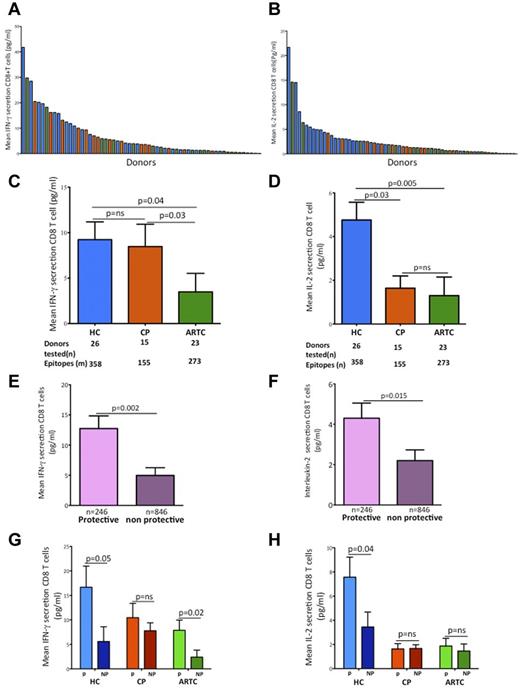
Secretion of IL-2, but not IFN-γ or TNF-α, early after HIV-1 antigen encounter distinguishes HIV-1–specific CD8 T cells of controllers
Having demonstrated differences in proliferative capacity among groups associated with protective HLA-I alleles, we next examined whether similar differences existed for effector functions. We used the HLA-I–matched panels of HIV-1 epitopes to stimulate PBMCs from the 3 cohorts and measure the concentrations of IFN-γ, IL-2, and TNF-α in culture supernatants by Luminex assays 6 hours after antigenic stimulation. For each subject, we calculated the mean responses for each cytokine (Figure 3A-B and supplemental Figure 3). Consistent with previous flow cytometry and ELISpot findings,39 controllers were indistinguishable from untreated progressors when ranked based on IFN-γ secretion (Figure 3C), whereas ART-treated subjects had the weakest IFN-γ responses. In contrast, subjects segregated differently on IL-2 secretion (Figure 3D), with controllers secreting higher amounts of IL-2 than ART-treated subjects and untreated progressors. Responses restricted by protective alleles dominated IFN-γ and IL-2 (Figure 3E-F) secretion when the entire cohort was evaluated. Intergroup comparison showed that this immunodominance of protective alleles was driven by the marked differences observed in controllers and was not present in untreated or treated progressors (Figure 3G-H), except for IFN-γ in ART-treated subjects. These data show that, like proliferation, cytokine secretion in controllers is dominated by epitopes restricted by protective HLA-I alleles. Moreover, they show that protective alleles are associated with qualitatively different responses in controllers compared with ART-treated and untreated progressors.
Analysis of early cytokine secretion by HIV-1–specific CD8 T cells shows that IL-2, but not IFN-γ, distinguishes controllers from treated or untreated progressors. (A-B) Summary data of the mean magnitude of IFN-γ (A) and IL-2 (B) secretion by HIV-1–specific CD8 T cells after a 6-hour stimulation with panels of individual HLA class I–matched HIV-1 epitopes. Each column corresponds to a study participant. (C-D) Statistical analysis of mean IFN-γ (C) and IL-2 (D) early (6 hours) secretion among the 3 groups of HIV-1–infected subjects of the discovery cohort (n = 64) Mean IFN-γ responses based on HLA-I restriction in all patients. HC indicates HIV-1 controllers; CP, chronic progressors; and ARTC, ART-treated subjects. (E-F) Statistical comparison of means early IFN-γ (E) and IL-2 (F) responses to epitopes restricted by protective and nonprotective HLA-I alleles in all subjects. (G-H) Statistical analysis of early IFN-γ (G) and early IL-2 (H) responses stratified by protective versus nonprotective HLA-I alleles in HC, CP, and ARTC subjects. Throughout the figure, HC data are represented in blue, CP in red, ARTC in green, protective HLA class I alleles (B*5701/03, B*2705 and B*5801) in purple, and nonprotective HLA I alleles in orange. All comparisons of adjusted means were done using generalized linear models controlling for clustering within subjects. Vertical interval bars correspond to the SEM.
Analysis of early cytokine secretion by HIV-1–specific CD8 T cells shows that IL-2, but not IFN-γ, distinguishes controllers from treated or untreated progressors. (A-B) Summary data of the mean magnitude of IFN-γ (A) and IL-2 (B) secretion by HIV-1–specific CD8 T cells after a 6-hour stimulation with panels of individual HLA class I–matched HIV-1 epitopes. Each column corresponds to a study participant. (C-D) Statistical analysis of mean IFN-γ (C) and IL-2 (D) early (6 hours) secretion among the 3 groups of HIV-1–infected subjects of the discovery cohort (n = 64) Mean IFN-γ responses based on HLA-I restriction in all patients. HC indicates HIV-1 controllers; CP, chronic progressors; and ARTC, ART-treated subjects. (E-F) Statistical comparison of means early IFN-γ (E) and IL-2 (F) responses to epitopes restricted by protective and nonprotective HLA-I alleles in all subjects. (G-H) Statistical analysis of early IFN-γ (G) and early IL-2 (H) responses stratified by protective versus nonprotective HLA-I alleles in HC, CP, and ARTC subjects. Throughout the figure, HC data are represented in blue, CP in red, ARTC in green, protective HLA class I alleles (B*5701/03, B*2705 and B*5801) in purple, and nonprotective HLA I alleles in orange. All comparisons of adjusted means were done using generalized linear models controlling for clustering within subjects. Vertical interval bars correspond to the SEM.
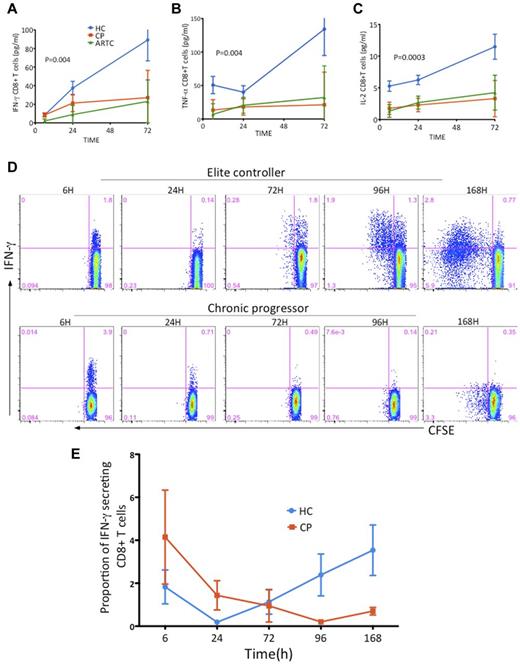
Kinetics of cytokine secretion by HIV-1–specific CD8 T cells present a unique pattern in controllers
Previous studies have suggested that controllers and progressors have a similar magnitude and breadth of HIV-1–specific CD8 T-cell responses.40,41 However, these data were based on measuring IFN-γ at a single and usually early time point after antigenic stimulation and would not reflect possible differences in the kinetics of these responses.42,43 Therefore, we examined the capacity of HIV-1–specific CD8 T cells to sustain cytokine production beyond the 6- or 12-hour incubation time used in standard ICS assays. We measured IFN-γ, IL-2, and TNF-α concentrations in PBMC supernatants at 6, 24, and 72 hours after epitope encounter and applied linear mixed models using time as a continuous variable to calculate the slope of secretion of each cytokine in the 3 subject groups. The slopes of IFN-γ, IL-2, and TNF-α secretion were markedly steeper in controllers than in ART-treated and untreated progressors (Figure 4A-C). As we did for proliferative capacity, we next confirmed that stimulation of CD4-depleted PBMCs with a pool of overlapping HIV-1 Gag peptides induced similar cytokine secretion kinetics (supplemental Figure 4). These data illustrate that HIV-1–specific CD8 T cells in controllers are characterized by prolonged and robust secretion of various cytokines, whereas the secretion slopes in ART-treated subjects and progressors were almost flat after the 6-hour time point. In contrast to what we observed for proliferative capacity, controller HIV-1–specific CD8 T cells restricted by both protective and nonprotective alleles are endowed with this extended capacity to exert effector functions. We next performed serial determinations of IFN-γ secretion and proliferation by epitope-specific CD8 T cells in combined ICS and CFSE assays over a 7-day period (Figure 4D-E). All subjects exhibited robust cytokine secretion in the 6-hour ICS and a large decrease in response at 24 hours. In contrast to progressors, controllers showed a biphasic pattern of IFN-γ production with a secondary and steady increase of IFN-γ production that was clear at 72 hours even though proliferation of both IFN-γ+ and IFN-γ− cells was still minimal. Further follow-up confirmed the remarkable capacity of HIV-1–specific CD8 T cells of controllers to sustain IFN-γ production after a single antigen encounter, contrasting with the attrition of the responses in progressors despite initial strong cytokine secretion. This prolonged functionality of controller CD8 T cells is highly suggestive of the behavior of central memory cells observed in adoptive transfer experiments in animal models.42,44
Kinetics of cytokine secretion by HIV-1–specific CD8 T cells present a unique pattern in controllers. (A-C) Secretion of cytokines was determined in supernatants of PBMCs 6, 24, and 72 hours after stimulation with HLA-I-–matched panels of optimal HIV-1 epitopes (discovery cohort, n = 64 subjects); comparative kinetics analysis of mean IFN-γ (A), IL-2 (B), and TNF-α (C) secretion in the 3 clinical cohorts after a single antigenic encounter. HC indicates HIV-1 controllers; CP, chronic progressors; and ARTC, ART-treated subjects. (D-E) CFSE-labeled PBMCs were incubated with optimal HIV-1 CD8 T-cell epitopes and ICS and CFSE intensity were measured in CD8 T cells at 6, 24, 72, 96, and 168 hours after stimulation. Control (no antigen) conditions show minimal background (data not shown. (D) Representative examples of responses of high magnitude in an elite controller (upper row) and a chronic progressor (lower row). (E) Summary data of the kinetics of IFN-γ secretion over a 7-day period after a single antigenic encounter for immunodominant responses in 3 HC subjects (blue line) and 3 CP subjects (red line). Mean cytokine responses for each subject was calculated at each time point (6, 24, and 72 hours). Vertical interval bars correspond to the SEM.
Kinetics of cytokine secretion by HIV-1–specific CD8 T cells present a unique pattern in controllers. (A-C) Secretion of cytokines was determined in supernatants of PBMCs 6, 24, and 72 hours after stimulation with HLA-I-–matched panels of optimal HIV-1 epitopes (discovery cohort, n = 64 subjects); comparative kinetics analysis of mean IFN-γ (A), IL-2 (B), and TNF-α (C) secretion in the 3 clinical cohorts after a single antigenic encounter. HC indicates HIV-1 controllers; CP, chronic progressors; and ARTC, ART-treated subjects. (D-E) CFSE-labeled PBMCs were incubated with optimal HIV-1 CD8 T-cell epitopes and ICS and CFSE intensity were measured in CD8 T cells at 6, 24, 72, 96, and 168 hours after stimulation. Control (no antigen) conditions show minimal background (data not shown. (D) Representative examples of responses of high magnitude in an elite controller (upper row) and a chronic progressor (lower row). (E) Summary data of the kinetics of IFN-γ secretion over a 7-day period after a single antigenic encounter for immunodominant responses in 3 HC subjects (blue line) and 3 CP subjects (red line). Mean cytokine responses for each subject was calculated at each time point (6, 24, and 72 hours). Vertical interval bars correspond to the SEM.
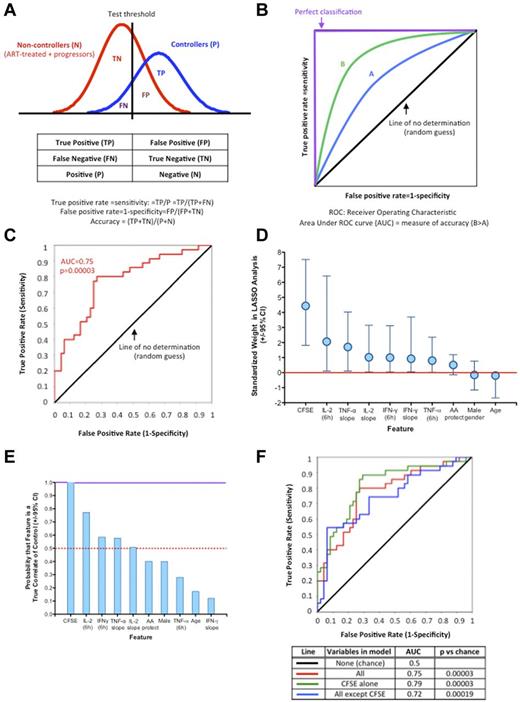
Diagnostic tests based on data integration into immunomonitoring models allow accurate discrimination of HIV-1 infection outcomes
In the last part of this study, we first verified the reproducibility of the data obtained on the discovery cohort by performing the same analyses described in Figures 1 and 2 on an independent validation cohort of 20 subjects (10 controllers and 10 noncontrollers). The results obtained on individual HIV-1–specific CD8 T-cell functions were very similar in both cohorts, as illustrated by the data obtained on proliferative capacity (Table 2). Profiles of cytokine responses were also highly reproducible (data not shown). We next assessed the additional information that could be derived from higher-level integration of the experimental results. To test an approach relevant for future clinical applications, we combined the discovery and validation datasets to build a palette of immunomonitoring models of HIV-1–specific CD8 T-cell functions (Figure 5). Applying statistical tools normally used to assess the performance of diagnostic tests in the clinical setting, we determined the capacity of each model to identify controllers in out-of-sample data. The strong similarities of findings in progressors and ART-treated subjects in our assays allowed applying statistical tests for a binary outcome (controllers vs noncontrollers; principles illustrated in Figure 5A-B). The schematic ROC curve (Figure 5B) illustrates the unavoidable compromise between sensitivity and specificity determined by the choice of the threshold of the test (Figure 5A). To obtain a robust estimation of how accurately the predictive models would perform, we use a nested cross-validation approach (10-fold cross-validation). Briefly, the total dataset was randomly partitioned into 10 subgroups. On a rotating basis, each subgroup was retained as the validation dataset and the 9 others used to train the models using LASSO logistical regression. The AUC obtained with the 10 models for their capacity to differentiate between controller and noncontrollers then led to a more robust measure of model accuracy. We used this approach to assess the ability of the “diagnostic test” based on combination of all 10 variables listed in Figure 5D to differentiate between controllers and noncontrollers (Figure 5C). This immunomonitoring model yielded an excellent AUC (0.75) with a highly significant ROC curve compared with chance (P = .00003). We next examined the contribution of each variable to the model and found a broad range in their respective weights for standardized variables in the LASSO analysis (Figure 5D): the proliferative capacity of HIV-1–specific CD8 T cells stood out as contributing remarkably to the model, although high weights were also observed for IL-2 at 6 hours and for the slope of TNF-α secretion. The low weight of HLA-I molecules here is attributable to the fact that the controller and noncontroller cohorts were both enriched for protective alleles by study design.
Proliferative capacity of HIV-1–specific CD8 T cells in the discovery and validation cohorts
| Paired comparison . | Discovery cohort . | Validation cohort . | ||||
|---|---|---|---|---|---|---|
| Data points* . | % CFSElow CTLs, mean (SEM) . | P . | Data points* . | % CFSElow CTLs, mean (SEM) . | P . | |
| HC | 358 | 2.85 (0.37) | 149 | 1.97 (0.3) | ||
| versus | .002 | .007 | ||||
| ARTC | 273 | 0.91 (0.42) | 74 | 0.35 (0.41) | ||
| HC | 358 | 2.85 (0.42) | 149 | 1.97 (0.32) | ||
| versus | .005 | .04 | ||||
| CP | 155 | 0.74 (0.57) | 35 | 0.32 (0.64) | ||
| ARTC | 273 | 0.91 (0.18) | 74 | 0.33 (0.18) | ||
| versus | .57 | .94 | ||||
| CP | 155 | 0.75 (0.22) | 35 | 0.31 (0.25) | ||
| Paired comparison . | Discovery cohort . | Validation cohort . | ||||
|---|---|---|---|---|---|---|
| Data points* . | % CFSElow CTLs, mean (SEM) . | P . | Data points* . | % CFSElow CTLs, mean (SEM) . | P . | |
| HC | 358 | 2.85 (0.37) | 149 | 1.97 (0.3) | ||
| versus | .002 | .007 | ||||
| ARTC | 273 | 0.91 (0.42) | 74 | 0.35 (0.41) | ||
| HC | 358 | 2.85 (0.42) | 149 | 1.97 (0.32) | ||
| versus | .005 | .04 | ||||
| CP | 155 | 0.74 (0.57) | 35 | 0.32 (0.64) | ||
| ARTC | 273 | 0.91 (0.18) | 74 | 0.33 (0.18) | ||
| versus | .57 | .94 | ||||
| CP | 155 | 0.75 (0.22) | 35 | 0.31 (0.25) | ||
CTLs indicates cytotoxic T lymphocytes; HC, HIV-1 controllers; CP, chronic progressors; and ARTC, ART-treated subjects.
Number of individual epitopes tested in the CFSE-based proliferation assays.
Diagnostic tests based on integration of diverse HIV-1–specific CD8 T-cell functions into immunomonitoring models allows accurate discrimination of disease outcomes. (A-B) Concepts of binary classifiers and ROC curves. (A) The goal of the modeling component of this study was to establish a diagnostic test that seeks to determine whether a person is a controller. The models based on HIV-1–specific CD8 T-cell functions will yield a continuous random variable that will be used as classifier (horizontal axis). As the outcome is binary (2 classes: controller or noncontroller), the boundary must be determined by a threshold value (vertical line). There are therefore 4 possible outcomes summarized in the “contingency table,” true positive (TP), false positive (FP), true negative (TN), and false negative (FN), which define the true-positive rate (sensitivity) and false-positive rate (1-specificity). (B) A ROC is defined by the false-positive rate and true-positive rate, which depicts relative trade-offs resulting from changing the test threshold. The best possible prediction method would yield a “square curve” reaching the upper left corner and representing 100% sensitivity and 100% specificity. A completely random guess (chance) would give a point along the diagonal line (line of nondiscrimination). In the present study, ROC curves can be used to compare tests derived from different immunomonitoring models: here model B is better than model A. ROC curves can be further characterized by the AUC, which is a measure of test accuracy (1.0 = the best possible test and 0.5 = no discrimination). (C) Graphical representation of the ROC curve of discrimination between controllers and noncontrollers corresponding to the averaged LASSO logistic regression model with 10-fold cross-validation obtained with all 10 variables (CFSE-based proliferation; IFN-γ, IL-2, and TNF-α secretion at 6 hours; slopes of IFN-γ, IL-2, and TNF-α secretion; sex; and age). (D) Weight of the individual standardized features in regard to their contribution to the 10-variable LASSO model. Vertical lines correspond to the 2.5 and 97.5 percentiles, respectively. (E) Bootstrap analysis of the probability that each feature of the 10-variable LASSO model corresponds to a true correlate of spontaneous viral control. (F) Comparative ROC curves and corresponding AUC characteristics corresponding to the averaged LASSO logistic regression models with 10 variables, CFSE-based proliferation only, and 9 variables (all but proliferation). The 3 ROC curves are not statistically different (CFSE alone vs all, P = .55; CFSE alone vs all others, P = .32). Details of the 10-fold cross-validation analyses are presented in Table 3.
Diagnostic tests based on integration of diverse HIV-1–specific CD8 T-cell functions into immunomonitoring models allows accurate discrimination of disease outcomes. (A-B) Concepts of binary classifiers and ROC curves. (A) The goal of the modeling component of this study was to establish a diagnostic test that seeks to determine whether a person is a controller. The models based on HIV-1–specific CD8 T-cell functions will yield a continuous random variable that will be used as classifier (horizontal axis). As the outcome is binary (2 classes: controller or noncontroller), the boundary must be determined by a threshold value (vertical line). There are therefore 4 possible outcomes summarized in the “contingency table,” true positive (TP), false positive (FP), true negative (TN), and false negative (FN), which define the true-positive rate (sensitivity) and false-positive rate (1-specificity). (B) A ROC is defined by the false-positive rate and true-positive rate, which depicts relative trade-offs resulting from changing the test threshold. The best possible prediction method would yield a “square curve” reaching the upper left corner and representing 100% sensitivity and 100% specificity. A completely random guess (chance) would give a point along the diagonal line (line of nondiscrimination). In the present study, ROC curves can be used to compare tests derived from different immunomonitoring models: here model B is better than model A. ROC curves can be further characterized by the AUC, which is a measure of test accuracy (1.0 = the best possible test and 0.5 = no discrimination). (C) Graphical representation of the ROC curve of discrimination between controllers and noncontrollers corresponding to the averaged LASSO logistic regression model with 10-fold cross-validation obtained with all 10 variables (CFSE-based proliferation; IFN-γ, IL-2, and TNF-α secretion at 6 hours; slopes of IFN-γ, IL-2, and TNF-α secretion; sex; and age). (D) Weight of the individual standardized features in regard to their contribution to the 10-variable LASSO model. Vertical lines correspond to the 2.5 and 97.5 percentiles, respectively. (E) Bootstrap analysis of the probability that each feature of the 10-variable LASSO model corresponds to a true correlate of spontaneous viral control. (F) Comparative ROC curves and corresponding AUC characteristics corresponding to the averaged LASSO logistic regression models with 10 variables, CFSE-based proliferation only, and 9 variables (all but proliferation). The 3 ROC curves are not statistically different (CFSE alone vs all, P = .55; CFSE alone vs all others, P = .32). Details of the 10-fold cross-validation analyses are presented in Table 3.
We next sought to determine by bootstrap analysis the probability that each variable was a true correlate of spontaneous viral control (Figure 5E). Proliferative capacity appeared to have a very high probability of being a true correlate of spontaneous viral control, whereas 4 other parameters (IL-2 at 6 hours, IFN-γ at 6 hours, TNF-α slope, and IL-2 slope) were more likely than not to be true correlates. However, in contrast to CFSE-based proliferation, the probabilities were for these variables below 95%, so investigation in a larger cohort would be necessary to confirm their individual contributions. These results suggest that the proliferative capacity of HIV-1–specific CD8 T cells is a uniquely strong discriminant for controller status. However, it is also a parameter that is more difficult to measure than cytokine secretion. This parameter is also notoriously sensitive to cell quality if performed with frozen samples, which is usually the situation in vaccine trials, whereas our studies were performed with fresh blood. Therefore, in the last part of the study, we used the LASSO and 10-fold cross-validation analytic approach to compare the accuracy of immune models based on: (1) all 10 variables, (2) CFSE-based proliferation alone, and (3) all variables except proliferation (Figure 5F). The data show that all 3 approaches give very good ROC curves that are not statistically different from one another (details of the cross-validation analyses are provided in Table 3). Therefore, in our dataset, proliferation alone was statistically as good at discriminating controllers from noncontrollers as integration of all parameters. However, if proliferation was dropped, high-dimensional analysis of the other variables leveraged independent associations among “weaker” factors and yielded a similarly robust immunomonitoring model.
Assessment of the different immunomonitoring models (AUC)
| Partition . | 10 variables (all) . | CFSE proliferation only . | All but CFSE . |
|---|---|---|---|
| 1 | 0.71 | 0.79 | 0.57 |
| 2 | 0.78 | 0.71 | 0.83 |
| 3 | 0.9 | 0.78 | 0.55 |
| 4 | 0.8 | 0.85 | 0.7 |
| 5 | 0.75 | 0.75 | 0.75 |
| 6 | 0.87 | 1 | 0.93 |
| 7 | 1 | 0.87 | 0.38 |
| 8 | 0.92 | 1 | 0.92 |
| 9 | 0.73 | 0.92 | 0.93 |
| 10 | 0.56 | 0.73 | 0.5 |
| Computed AUC | 0.75 | 0.79 | 0.72 |
| Partition . | 10 variables (all) . | CFSE proliferation only . | All but CFSE . |
|---|---|---|---|
| 1 | 0.71 | 0.79 | 0.57 |
| 2 | 0.78 | 0.71 | 0.83 |
| 3 | 0.9 | 0.78 | 0.55 |
| 4 | 0.8 | 0.85 | 0.7 |
| 5 | 0.75 | 0.75 | 0.75 |
| 6 | 0.87 | 1 | 0.93 |
| 7 | 1 | 0.87 | 0.38 |
| 8 | 0.92 | 1 | 0.92 |
| 9 | 0.73 | 0.92 | 0.93 |
| 10 | 0.56 | 0.73 | 0.5 |
| Computed AUC | 0.75 | 0.79 | 0.72 |
All data include a 10-fold cross-validation.
Discussion
Despite extensive efforts, unambiguous identification of correlates of protective immunity against HIV-1 remains elusive. In the present study, we describe a novel strategy to develop immunomonitoring models of HIV-1–specific immune responses that we believe present major advantages compared with traditional approaches. We demonstrate its capacity to generate diagnostic tests that accurately delineate HIV-1 controllers from subjects with progressive disease irrespective of treatment status. This approach builds on the fact that various coordinated functions are required for an effective immune response against a pathogen. It provides a rigorous methodology to proceed with the selection of immune parameters that should to be measured in clinical trials. Both current and new knowledge can be incrementally incorporated into evolving and flexible tools that are usable in translational research.
To adequately sample the diversity of immune responses in any given subject, we selected epitopes based solely on HLA-I genotype, thus avoiding the bias that would be introduced if a screening test (eg, IFN-γ ELISpot) was used to determine the epitopes tested in downstream functional assays. Building on previous findings,4,19 we clarify at a deeper level the relationships among epitope-specific CD8 T-cell proliferation, the nature of the restricting alleles, and disease stage. Our data show that the robust proliferative capacity of HIV-1–specific CD8 T-cell responses is unique to controllers, suggesting that protective alleles lose their ability to drive proliferation during progressive HIV-1 infection and that critical immune defects are not restored by ART.4,38 This is consistent with the observation that when therapy is withdrawn, viremia rapidly rebounds.45,46 It will be important to determine in future studies whether institution of ART at the time of acute infection is able to preserve some of the functional features identified herein as being associated with controller status.
The assessment of cytokine secretion by Luminex arrays at different time points after epitope stimulation allowed us to define functional differences at an unprecedented level of granularity. Data on cytokine secretion early after antigen encounter were consistent with results obtained by other approaches (eg, higher IL-2 secretion by HIV-1–specific CD8 T cells in controllers) and established that, in controllers, responses restricted by protective alleles dominated the early burst of cytokine release. However, kinetic analyses provided novel findings: the trajectories of IFN-γ, IL-2, and TNF-α secretion curves were strikingly different in controllers relative to ART-treated and untreated progressors. These differences were not a mere consequence of proliferation: both dividing and nondividing HIV-1–specific CD8 T cells of controllers were capable of prolonged secretion of various cytokines. Potent effector T cells at mucosal sites were associated with protective, nonsterilizing immunity in the SIV model of nonhuman primates vaccinated with CMV vectors.47,48 Future investigations should determine whether our results with PBMCs translate into other body compartments.
Limitations of our study include the size of the epitope panels tested, which was a compromise between obtaining an accurate picture of the heterogeneity of epitope-specific responses and practicality in the perspective of developing clinical useable immunomonitoring tools. There is a possibility of missing important responses in some subjects. However, the robustness of our results suggests that this sampling is sufficient to give a representative picture of the repertoire of HIV-1–specific CD8 T-cell responses. Because our study was performed with chronically infected subjects in whom the HIV-1 infection outcome was already established, our analyses yielded correlates of control but not of protection. Because the study was performed in already infected subjects, our results are more likely to be applicable to therapeutic, rather than prophylactic, vaccines. However, the overall modeling strategy would be equally applicable to preventive interventions. Longitudinal studies starting at the time of acute infection, when the viral set point is not yet established, will be critical to determine whether the variables we assessed are also predictors of subsequent viral control. We also concede that we did not have a large enough dataset to warrant generalization of our predictive model to the general population because we enriched the noncontroller groups in subjects carrying protective alleles. The model should therefore be further validated in larger, unbiased cohorts in which HLA-I alleles can be added to the model. Nonetheless, this does not detract from the model's strong illustration of the sensitivity and specificity of our assays that are independent of HLA-I alleles.
The models used in the present study can likely be further improved by including not only other parameters of HIV-1–specific CD8 T-cell responses showing association with controller status (eg, such as T-bet expression, perforin up-regulation, and MIP-1β secretion4,26,49,50 ), but also functions mediated by other cell types. Our approach thus allows for a generalization of the polyfunctionality concept initially explored by polychromatic flow cytometry.23,24 Each new variable should be assessed for its capacity to further improve accuracy and thus refine the model obtained with the previous dataset. In this study, we also measured perforin expression and the degranulation marker CD107a in a subset of donors. Both markers were significantly higher in controllers compared with chronic progressors (data not shown), which is consistent with previous findings.26,49 We believe that our experimental approach has broad applications for studies of immune responses in other infectious and inflammatory diseases, in particular in the immunocompromised host after transplantation or chemotherapy: there is currently no good surrogate to determine the “net state of immunosuppression” that critically stratifies the risk of infections in these patients. Our findings can also serve as a proof of principle for building longitudinal models based on immunologic data to predict disease outcome, especially during acute/early HIV-1 infection. Most critically, however, this strategy should be tested to evaluate the quality of HIV-1 vaccine–induced immune responses and to predict protective immunity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Rafi Ahmed, Amalio Telenti, and Sylvie Le Gall for input on the study and the clinical and laboratory staff at the Massachusetts General Hospital, the International HIV Controllers Study (www.hivcontrollers.org), and all study participants for their invaluable role in this project.
This study was supported by an Innovation Award of the Ragon Institute (to D.E.K. and P.L.D.J.), the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (RO1 AI30914 to B.D.W and PO1 AI-080192 to D.E.K.), the National Heart, Lung, and Blood Institute of the National Institutes of Health (RO1 HL-092565 to D.E.K.), the Mark and Lisa Schwartz Foundation (to B.D.W.), the Howard Hughes Medical Institute (to B.D.W.), and the Bill and Melinda Gates Foundation (to F. Pereyra and B.D.W.).
National Institutes of Health
Authorship
Contribution: Z.N.M., L.B.C., F. Pereyra, D.H., P.L.D.J., B.D.W., and D.E.K. provided intellectual input and contributed to the experimental design; Z.N.M., J.P., S.V., A.M., K.C., F. Porichis, R.B.J., D.M.A., M.G.H., and E.S. performed the experiments; A.P.-T. and F. Pereyra provided the clinical samples; L.B.C., C.K., D.H., and P.L.D.J. performed the statistical analyses and modeling; Z.N.M., L.B.C., P.L.D.J., B.D.W., and D.E.K. wrote the manuscript; B.D.W. and D.E.K. provided supervision; and D.E.K. was responsible for the overall design and conduct of the study.
Conflict-of-interest disclosure: C.K. and D.H. are employees of Microsoft. The remaining authors declare no competing financial interests.
Correspondence: Daniel E. Kaufmann, MD, Centre de Recherche du CHUM, Hôpital Saint-Luc, Pavillon Édouard Asselin, bureau 304, 264, Blvd René-Lévesque Est, H2X 1P1, Montréal, QC, Canada; e-mail: dkaufmann@partners.org.
References
Author notes
Z.M.N. and L.B.C. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal