Abstract
The integrin β3-mediated c-Src priming and activation, via the SH3 domain, is consistently associated with diseases, such as the formation of thrombosis and the migration of tumor cells. Conventionally, activation of c-Src is often induced by the binding of proline-rich sequences to its SH3 domain. Instead, integrin β3 uses R760GT762 for priming and activation. Because of the lack of structural information, it is not clear where RGT will bind to SH3, and under what mechanism this interaction can prime/activate c-Src. In this study, we present a 2.0-Å x-ray crystal structure in which SH3 is complexed with the RGT peptide. The binding site lies in the “N”-Src loop of the SH3 domain. Structure-based site-directed mutagenesis showed that perturbation on the “N”-Src loop disrupts the interaction between the SH3 domain and the RGT peptide. Furthermore, the simulated c-Src:β3 complex based on the crystal structure of SH3:RGT suggests that the binding of the RGT peptide might disrupt the intramolecular interaction between the SH3 and linker domains, leading to the disengagement of Trp260:“C”-helix and further activation of c-Src.
Key Points
Crystal structure of SH3:RGT complex reveals how integrin β3 primes/activates c-Src kinase.
Introduction
Src family tyrosine kinases, often found in diverse tissues including the brain, osteoclasts, and platelets, play essential roles in many signaling pathways that control cell proliferation, cell differentiation, apoptosis, and cell adhesion and migration.1-3 Moreover, it has been shown that the malfunction of c-Src can lead to various severe diseases, including cancers.3-5
The c-Src kinase contains several functional domains: the N-terminal segment with a myristoylation signal, an SH3 domain, an SH2 domain, an SH2-kinase linker, a kinase domain, and a C-terminal tail.2,6 The N-terminal myristoylation together with its adjacent residues are thought to help the protein to anchor into the inner leaflet of the cytoplasmic membrane.7 The rest of the structure can assemble into a conserved, closed architecture,8-10 in which the intramolecular interactions between domains are essential to keep the enzyme in its down-regulated state. Thus far, the activation of c-Src is thought be regulated by 3 different mechanisms: (1) The SH3-dominant activation. The SH3 domain is in direct contact with the linker and kinase domains. It has been shown that disruption of this intramolecular interaction by a proline-rich peptide can trigger activation.11-13 (2) The SH2-dominant activation. The SH2 domain binds to the kinase domain via the phosphorylated Tyr527 at the C-terminal tail of the enzyme. Dephosphorylation of Tyr527 can remove the conformational constraints that keep the enzyme in the inactive state.14 (3) The relative orientation and coupling between the SH3 and SH2 domains can provide another level of control. These 2 domains sit at the back of the kinase domain and lock the N- and C-terminal lobes in their inactive positions, preventing the final “switch-on” of the enzyme (ie, the phosphorylation of the Tyr416 in the active site).10,15 Therefore, all these activation pathways will eventually converge in the open-up of the enzyme that is often correlated with the conformational changes of Trp260 and the “C”-helix in the kinase domain. Hence, the disengagement of Trp260:“C”-helix is thought to be an important step in the activation pathway of c-Src.16-18
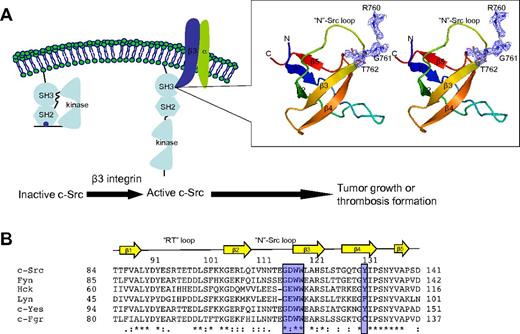
Very recently, it has been reported that integrin β3 can prime/activate c-Src (see Figure 1A).19,20 Integrin β3 is a key player of a bidirectional signaling pathway that regulates platelet aggregation and cell spreading.21,22 During the outside-in signaling, the integrin β3 is activated on binding to its cognate extracellular ligands, such as fibronectin, fibrinogen, and collagen. Its cytoplasmic domain can undergo conformational changes23 that, in turn, will enable the interaction of c-Src:β3.19 Furthermore, it was shown that the C-terminal end of integrin β3 with the sequence R760GT762 mediates a direct interaction with the SH3 domain of c-Src.20,24,25 Contrary to the canonical SH3 domain ligands with a typical poly-Pro type II helix,11,26,27 the integrin binding motif is much shorter and does not contain a proline residue. Recently, an in vitro study using synthetic peptides corresponding to the cytoplasmic tail of integrin showed that the sequence motif RGT can selectively inhibit outside-in signaling in human platelets.24 This together with the in vivo study using knock-in mouse suggested that peptidomimetics derived from the RGT moiety might be promising thrombosis drug candidates with little side effect toward the normal function of platelet.25 Furthermore, the complex of c-Src:β3 was also reported as an “oncogenic unit” promoting tumor growth and metastasis.28,29 Hence, these observations have led to the proposal and expectation that the interaction of c-Src:β3 could serve as an interesting therapeutic target, and better understanding of this interaction might lead to the discovery of new antithrombotic and anticancer strategies.30
However, until now, it is not yet clear where the RGT peptide binds to the SH3 domain and how integrin β3 can prime and activate c-Src. In this study, we report the crystal structure of the SH3 domain bound to a tripeptide derived from the integrin β3. The structure suggests that the RGT peptide binds to the “N”-Src loop of the SH3 domain. Site-directed mutagenesis was used to verify the binding. The in vitro strep-tactin pull-down assay, c-Src activity assay, and in vivo cell-spreading assay are all in good agreement with each other, showing that the perturbation of the binding site can disrupt the interaction of SH3:RGT and the signaling pathway controlled by integrin β3. These observations have led to the hypothesis of a novel RGT-mediated activation mechanism of c-Src.
Methods
Information concerning bacterial and eukaryotic cells, DNAs, and other materials used in this study is described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The experimental details of structure determination and functional characterizations using in vitro pull-down assays, in vitro c-Src enzymatic assays, in vivo cell spreading assays, and kinetic analysis are also shown in supplemental Methods. The statistics of the data collection and structure refinement are shown in Table 1. Coordinate of SH3:RGT has been deposited into the Protein Database Bank (entry code: 4HXJ).
Results
Crystal structure of SH3:RGT
The structure of SH3:RGT was determined by molecular replacement (Figure 1). The unit cell contains 2 SH3 polypeptide chains and a RGT peptide per asymmetric unit. The average B factor of RGT is 67 Å2, much bigger than that of overall protein (13.5 Å2; Table 1). To check whether the RGT modeling is correct, a simulated annealing omit map was used. In addition, structure refinement with the RGT peptide included can bring an extra 0.2% drop in Rfree factor, suggesting that the weak electron density and high B factor might be the results of the weak interaction of SH3:RGT. To quantify the binding, surface plasmon resonance-based technology was used (supplemental Figure 1). The KD between the SH3 domain and the cytoplasmic tail of integrin β3 is estimated to be ∼ 74.3 μM.
Crystal structure of SH3:RGT complex. (A) Schematic diagram of c-Src activation mediated by integrin β3. It has been reported that integrin β3 can activated c-Src via the interaction of SH3:RGT.19,36 The crystallographic characterization presented in this report is highlighted by an illustration box with a stereo diagram of SH3:RGT. The SH3 domain is shown as a diagram. The RGT peptide is shown in stick representation. The 2Fo-Fc electron density at the level of 0.5 σ is shown in blue. (B) Sequence alignment of the SH3 domains of Src family kinases. The secondary structural elements are annotated on top of the sequences. The residues that are in direct contact with the RGT peptide are highlighted with blue boxes. The highly and relatively conserved residues are indicated with “*” and “:,” respectively. The structural figure was prepared using Pymol.37
Crystal structure of SH3:RGT complex. (A) Schematic diagram of c-Src activation mediated by integrin β3. It has been reported that integrin β3 can activated c-Src via the interaction of SH3:RGT.19,36 The crystallographic characterization presented in this report is highlighted by an illustration box with a stereo diagram of SH3:RGT. The SH3 domain is shown as a diagram. The RGT peptide is shown in stick representation. The 2Fo-Fc electron density at the level of 0.5 σ is shown in blue. (B) Sequence alignment of the SH3 domains of Src family kinases. The secondary structural elements are annotated on top of the sequences. The residues that are in direct contact with the RGT peptide are highlighted with blue boxes. The highly and relatively conserved residues are indicated with “*” and “:,” respectively. The structural figure was prepared using Pymol.37
Data collection and structure refinement statistics of SH3:RGT
| Statistics . | Value . |
|---|---|
| Data collection | |
| Space group | P21 |
| Unit cell dimension, Å | |
| a | 27.9 |
| b | 40.8 |
| c | 43.3 |
| Molecule per ASU | 2SH3, 1RGT |
| Derivative | Native |
| Source/station* | BL17U |
| Wavelength, Å | 0.978 85 |
| Resolution range, Å | 29.2-2.0 |
| Observations, Ι/s(I) > 0 | 109110 |
| Unique reflections, Ι/s(I) > 0 | 6468 |
| High resolution shell, Å | 2.11-2.00 |
| Rsym, %†‡ | 17.0 (36.9) |
| <Ι/s(I)>‡ | 10.5 (7.0) |
| Completeness, %‡ | 99.9 (100.0) |
| Redundancy‡ | 8.0 (8.0) |
| Structure refinement | |
| Resolution range, Å | 29.2-2.0 |
| R-factor, % | 13.9 |
| R-factor (high resolution shell)§ | 13.5 |
| Rfree, %‖ | 19.3 |
| Rfree (high resolution shell) | 20.9 |
| Total no. of nonhydrogen atoms | |
| Protein atoms (SH3) | 922 |
| Water molecules | 95 |
| RGT peptide | 1 |
| R.m.s. deviations¶ | |
| Bond length, Å | 0.007 |
| Bond angle, ° | 1.025 |
| Main chain B-factors, Å2 | 2.600 |
| Side chain B-factors, Å2 | 3.933 |
| Wilson B-factor, Å2 | 9.5 |
| Average B-factor, Å2 | |
| Protein atoms | 13.5 |
| Solvent atoms | 27.3 |
| RGT atoms | 67.0 |
| Ramachandran statistics, % | |
| Most favored region | 95.7 |
| Allowed regions | 4.3 |
| Statistics . | Value . |
|---|---|
| Data collection | |
| Space group | P21 |
| Unit cell dimension, Å | |
| a | 27.9 |
| b | 40.8 |
| c | 43.3 |
| Molecule per ASU | 2SH3, 1RGT |
| Derivative | Native |
| Source/station* | BL17U |
| Wavelength, Å | 0.978 85 |
| Resolution range, Å | 29.2-2.0 |
| Observations, Ι/s(I) > 0 | 109110 |
| Unique reflections, Ι/s(I) > 0 | 6468 |
| High resolution shell, Å | 2.11-2.00 |
| Rsym, %†‡ | 17.0 (36.9) |
| <Ι/s(I)>‡ | 10.5 (7.0) |
| Completeness, %‡ | 99.9 (100.0) |
| Redundancy‡ | 8.0 (8.0) |
| Structure refinement | |
| Resolution range, Å | 29.2-2.0 |
| R-factor, % | 13.9 |
| R-factor (high resolution shell)§ | 13.5 |
| Rfree, %‖ | 19.3 |
| Rfree (high resolution shell) | 20.9 |
| Total no. of nonhydrogen atoms | |
| Protein atoms (SH3) | 922 |
| Water molecules | 95 |
| RGT peptide | 1 |
| R.m.s. deviations¶ | |
| Bond length, Å | 0.007 |
| Bond angle, ° | 1.025 |
| Main chain B-factors, Å2 | 2.600 |
| Side chain B-factors, Å2 | 3.933 |
| Wilson B-factor, Å2 | 9.5 |
| Average B-factor, Å2 | |
| Protein atoms | 13.5 |
| Solvent atoms | 27.3 |
| RGT atoms | 67.0 |
| Ramachandran statistics, % | |
| Most favored region | 95.7 |
| Allowed regions | 4.3 |
Beamline designations refer to the Shanghai Synchrotron Radiation Facility, Shanghai, People's Republic of China.
Rsym = Σ(I − <I>)2/ΣI2.
Overall, high resolution shell in parentheses.
High resolution shell: 2.080-2.000 Å.
Rfree calculated using 10% of total reflections omitted from refinement.
R.m.s. deviations report root mean square deviations from ideal bond lengths/angles and of B-factors between bonded atoms.38
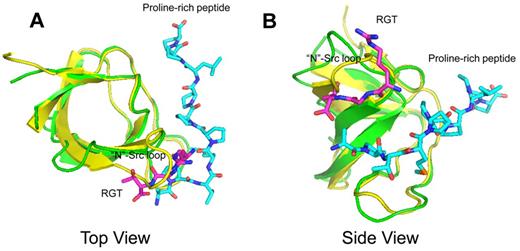
Conventionally, SH3-dominant activation of c-Src kinase is often associated with peptide-containing PXXP motif.11-13 In these structures, the proline-rich peptide binds to the SH3s domain with a poly-Pro type II helix, meandering in between the “N”-Src and “RT” loops (Figure 2). Similar mode of interaction with overlapping poly-Pro type II helix can also be observed in the structure complexed with non-PXXP peptides.27 In comparison, the crystal structure of SH3:RGT presented in this report reveals a novel binding mode (Figures 2 and 3). Because the core structure of SH3 domain is very similar with those characterized elsewhere with root mean square deviations of Cα positions between 1.2 and 1.5 Å, we will focus on the RGT peptide and its binding site. On binding, the RGT peptide adopts an L-shaped geometry with 2 arms formed by the side-chain of Arg760 and the backbone of the peptide (Figure 3A). The carboxyl groups flanking Gly761 make an uncommon cis configuration, contribute 2 hydrogen bonds to the overall binding, and enable a productive configuration of the RGT peptide conducive to binding (Figure 3B).
Structural superimposition between the complexes of SH3:RGT and SH3:PXXP (PDB code: 1ABO).26 The SH3:RGT are shown in green and magenta, respectively. The SH3:PXXP are shown in yellow and cyan, respectively.
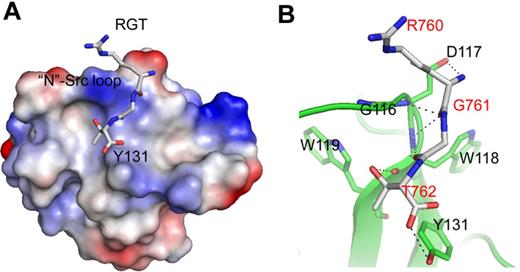
The binding site of the RGT peptide. (A) SH3 domain (rendered by electrostatic surface) complexed with the RGT peptide (shown in stick representation). (B) Enlarged view of the binding site. Hyphenated lines indicate the intermolecular hydrogen bonds. The SH3 domain is shown as a diagram, and the residues that are involved in direct contact are shown in stick representation. The label for the RGT peptide is colored in red.
The binding site of the RGT peptide. (A) SH3 domain (rendered by electrostatic surface) complexed with the RGT peptide (shown in stick representation). (B) Enlarged view of the binding site. Hyphenated lines indicate the intermolecular hydrogen bonds. The SH3 domain is shown as a diagram, and the residues that are involved in direct contact are shown in stick representation. The label for the RGT peptide is colored in red.
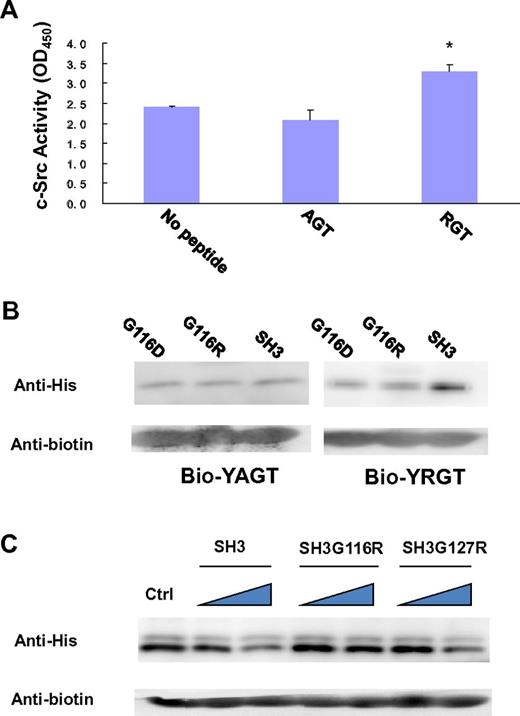
In the SH3 domain, the binding site is formed by the highly conserved residues G116DWW119 in the “N”-Src loop (Figures 1B and 3B). Gly116 and Asp117 are forming several backbone hydrogen bonds with Arg760 and Gly761 via their amino and carboxyl groups. Furthermore, the hydrophobic contact between the “N”-Src loop and Arg760 is important for the overall binding. The mutation of Arg760 to Ala in the RGT peptide can disrupt its activation activity toward c-Src (Figure 4A). When the RGT peptide was mixed with c-Src, the activity of the enzyme could be up-regulated by 40%. Similar results are observed in the pull-down assay, in which only YRGT, but not YAGT, shows binding preference toward the wild-type SH3 protein (Figure 4B). Another residue also important for binding is Thr762 (ie, the C-terminal charged residue of integrin β3). The current structure shows that this residue can make direct contact with Tyr131 via its charged carboxylate (Figure 3B). Sequence alignment shows that the aromatic residue in this position is conserved in all Src family kinases, except for c-Fgr (Figure 1B). Interestingly, all the SH3 domains, except for that of c-Fgr, can bind to integrin β3.19 To investigate this further, an SH3Y131C mutant was engineered (supplemental Figure 2C). The mutation has significantly disrupt the interaction of SH3:RGT, suggesting that the aromatic residue in position 131 is important for interaction, and might explain why c-Fgr does not bind to integrin β3.
Biochemical characterization of the interaction of SH3:RGT. (A) The side-chain of Arg760 in the RGT peptide is important for its activation activity. A standard c-Src activity assay was used to test whether mutation of Arg760 to Ala in the RGT peptide will affect its activation activity toward c-Src kinase. In the control experiment, no peptide was added into the reaction mixture containing c-Src kinase. The reading of OD450, an indicator for the phosphorylation of the c-Src substrate in the reaction mixture, was used to quantify the activity of the kinase. Values are mean ± SE; n = 3. *P < .05 (Student t test), statistically significant compared with control experiment. (B) The YAGT peptide shows no binding preference toward the SH3 domain. The wild-type SH3 protein or mutants were incubated with an excess amount of biotinylated YAGT or YRGT peptide. (C) Pull-down assays of SH3:YRGT. His-tagged SH3 domain was incubated with the biotinylated YRGT peptide in the presence of increasing concentrations of untagged wild-type SH3, SH3G116R, or SH3G127R, respectively. Each mixture was incubated with strep-tactin Sepharose solution. Strep-tactin resin was harvested by centrifugation and rinsed thoroughly. Proteins, which remain bound to the resin, were subjected to Western blot analysis. In the control experiment, no untagged SH3 protein was included in the pull-down assay.
Biochemical characterization of the interaction of SH3:RGT. (A) The side-chain of Arg760 in the RGT peptide is important for its activation activity. A standard c-Src activity assay was used to test whether mutation of Arg760 to Ala in the RGT peptide will affect its activation activity toward c-Src kinase. In the control experiment, no peptide was added into the reaction mixture containing c-Src kinase. The reading of OD450, an indicator for the phosphorylation of the c-Src substrate in the reaction mixture, was used to quantify the activity of the kinase. Values are mean ± SE; n = 3. *P < .05 (Student t test), statistically significant compared with control experiment. (B) The YAGT peptide shows no binding preference toward the SH3 domain. The wild-type SH3 protein or mutants were incubated with an excess amount of biotinylated YAGT or YRGT peptide. (C) Pull-down assays of SH3:YRGT. His-tagged SH3 domain was incubated with the biotinylated YRGT peptide in the presence of increasing concentrations of untagged wild-type SH3, SH3G116R, or SH3G127R, respectively. Each mixture was incubated with strep-tactin Sepharose solution. Strep-tactin resin was harvested by centrifugation and rinsed thoroughly. Proteins, which remain bound to the resin, were subjected to Western blot analysis. In the control experiment, no untagged SH3 protein was included in the pull-down assay.
To further verify the interaction of SH3:RGT, we designed 2 point mutants, G116R and G127R, located in and outside the binding site, respectively. The Gly116 to Arg mutation was designed to impose electrostatic and steric clashes that might prevent the binding of the RGT peptide to the c-Src SH3 domain. The substitution of Gly127, located ∼ 20 Å away from the peptide binding site was introduced as a negative control. The mutant proteins were stably expressed and were purified following the same protocol as for the wild-type protein, suggesting that the mutations have little effect on the overall conformation of the protein.
In our first set of experiments, we want to find out whether the perturbations in the “N”-Src loop will interfere with the interaction of SH3:RGT. Because the SH3:RGT interaction is predicted to be weak (supplemental Figure 1), a competitive pull-down assay was used. In this experiment, His-tagged SH3 protein was incubated with biotinylated YRGT peptide in the presence of untagged wild-type SH3 (which will act as a dominant negative probe to inhibit the interaction between the tagged SH3 and synthetic peptide). Western blot analysis was used to monitor the binding. We observed that increasing concentrations of untagged wild-type SH3 protein led to a concomitant decrease of tagged protein in the precipitate. In contrast, untagged SH3G116R (the Gly to Arg mutation on residue 116) is unable to compete off tagged wild-type SH3 domain from the biotinylated peptide, whereas untagged SH3G127R was indistinguishable from the wild-type (Figure 4C). This result prompted us to probe the significance of this interaction in the context of full-length c-Src.
Modeling the complex of c-Src bound to integrin β3
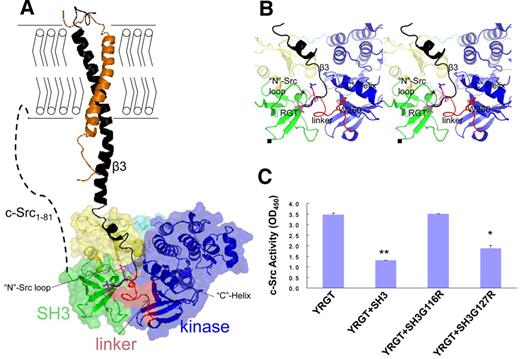
It has been shown that, on activation, the cytoplasmic membrane-proximal region of integrin β3 could undergo conformational changes23 to achieve an extended conformation.31 Such change in conformation can enable the binding of cytoplasmic proteins including c-Src.19 Based on the SH3:RGT complex, we constructed a model of c-Src:β3 complex, possibly at the early stage of the activation pathway (Figure 5A). Triggered by the binding of extracellular ligands, the cytoplasmic tail of integrin β3 is thought to adopt an extended conformation, whereby the cytoplasmic tail with the RGT motif is released from a membrane-proximal location and acts like a fishing hook (Figure 5A). Consistent with the published observations,20,24 the direct contact is mediated by the interaction of SH3:RGT (Figure 5B). The root mean square deviation (Cα) between the free and bound forms of the RGT peptides is 0.3 Å (supplemental Figure 2B). In the context of c-Src:β3 interaction, the RGT peptide is predicted to bind to a cleft decorated by residues including Asp117, Trp118, Tyr131, Arg318, and Lys257 from the SH3 and linker domains (Figure 5B; supplemental Figure 2A). Interestingly, these residues together with the nearby Trp260 and the “C”-helix in the kinase domain are thought to play important regulatory roles in keeping c-Src in its down-regulated state.8,18,32
Modeling the complex of c-Src bound to integrin β3. (A) The complex of c-Src:β3. The integrin β3 is modeled into the inactive c-Src based on the crystal structure of SH3:RGT. The cytoplasmic membrane is shown with bilayers. Activated integrin β3 (black) complexed with its nascent α subunit (orange) is shown as a diagram. The inactive c-Src kinase is shown as a diagram together with a transparent surface. Different functional domains, such as the SH3 domain, SH2 domain, SH2-kinase linker, kinase domain, and the C-terminal tail with phosphorylated Tyr527, are colored by green, yellow, red, blue, and cyan, respectively. The N-terminal membrane-anchoring sequence, designated as c-Src1-81, is represented by a dashed line. (B) The enlarged view of the putative binding site of c-Src:β3. The RGT peptide and its adjacent Trp260 are shown in stick representation. The choices/positions of mutations (ie, SH3G116R and SH3G127R) used in this study are shown by “*” and “■,” respectively. (C) Structure-based functional characterization of c-Src:β3 by in vitro c-Src activity assays. The YRGT peptide was used to activate c-Src in the presence of SH3, SH3G116R, or SH3G127R. In the controlled experiment, no recombinant SH3 protein was included in the reaction mixture. Values are mean ± SE; n = 3. *P < .05, statistically significant compared with control experiment. **P < .01, statistically significant compared with control experiment.
Modeling the complex of c-Src bound to integrin β3. (A) The complex of c-Src:β3. The integrin β3 is modeled into the inactive c-Src based on the crystal structure of SH3:RGT. The cytoplasmic membrane is shown with bilayers. Activated integrin β3 (black) complexed with its nascent α subunit (orange) is shown as a diagram. The inactive c-Src kinase is shown as a diagram together with a transparent surface. Different functional domains, such as the SH3 domain, SH2 domain, SH2-kinase linker, kinase domain, and the C-terminal tail with phosphorylated Tyr527, are colored by green, yellow, red, blue, and cyan, respectively. The N-terminal membrane-anchoring sequence, designated as c-Src1-81, is represented by a dashed line. (B) The enlarged view of the putative binding site of c-Src:β3. The RGT peptide and its adjacent Trp260 are shown in stick representation. The choices/positions of mutations (ie, SH3G116R and SH3G127R) used in this study are shown by “*” and “■,” respectively. (C) Structure-based functional characterization of c-Src:β3 by in vitro c-Src activity assays. The YRGT peptide was used to activate c-Src in the presence of SH3, SH3G116R, or SH3G127R. In the controlled experiment, no recombinant SH3 protein was included in the reaction mixture. Values are mean ± SE; n = 3. *P < .05, statistically significant compared with control experiment. **P < .01, statistically significant compared with control experiment.
Functional characterization of c-Src:β3
Next, we tested whether the interaction between the “N”-Src loop and the RGT peptide, suggested by the complex of c-Src:β3, is important for the activation of c-Src. Because the direct mutations of G116DWW119 in c-Src can trigger the activation of the enzyme8,18,32 and hence prevent further characterization of the interaction between the RGT peptide and c-Src, we used an indirect approach. In this in vitro assay, we investigated whether the isolated SH3 domain of c-Src, or mutants thereof, are able to interfere with integrin β3-mediated activation of c-Src kinase activity (Figure 5C). When synthetic peptide corresponding to the functional end of integrin β3 was incubated with c-Src in the presence of wild-type SH3 and SH3G127R mutant, the kinase activity dropped ∼ 60% and 50%, respectively. In comparison, SH3G116R showed no interference with the c-Src kinase activity (Figure 5C). Similar results could be observed in mutants, SH3D117A and SH3D117A&W118A (supplemental Figure 2C). The side-chain of Asp117 forms a hydrogen bond with the RGT peptide (Figure 3B). Hence, mutations involving Asp117 have disrupted the binding. In contrast, the side-chain of Trp118 dips downward away from the RGT peptide and the “N”-Src loop (Figure 3B). This is consistent with the observation that the mutation of Trp to Ala produces little impact on the interaction of SH3:RGT (supplemental Figure 2C).
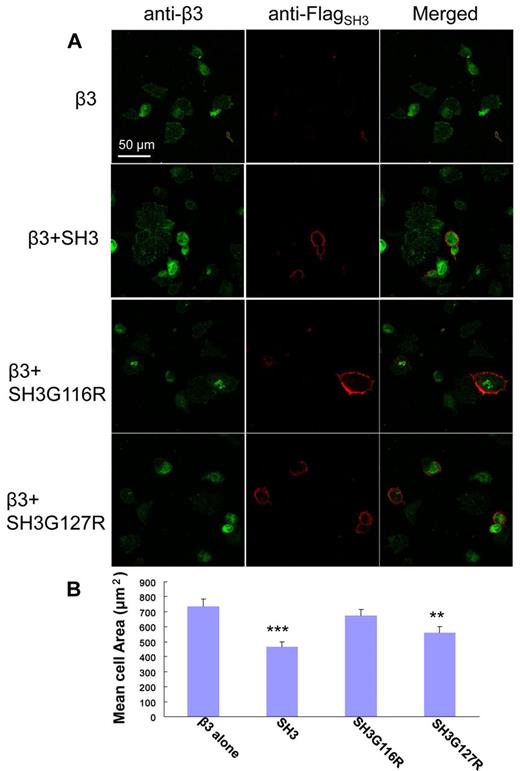
As demonstrated by Ylanne et al, the interaction of c-Src:β3 via the interaction of SH3:RGT can be monitored by a in vivo cell spreading assay.33 Similar to the working principles of in vitro pull-down and c-Src activity assays, a new set of SH3 including the membrane anchoring sequence (ie, SH31-141, SH31-141G116R, SH31-141G127R) were designed. As shown in Figure 6A, cells that constitutively express integrin β3, designated as CHOβ3, can spread on binding to fibrinogen. This behavior can be inhibited by the overexpression of SH31-141 and SH31-141G127R, but not by SH31-141G116R. Sixty randomly chosen cells for each coexpression were chosen for statistical analysis. The cells only expressing β3, designated as the control cells, had the biggest cell area of ∼ 750 μm2. The cells containing SH31-141G116R mimicked behavior of the control cells with averaged cell area of ∼ 700 μm2. However, the cells containing SH31-141 or SH31-141G127R, which are thought to interfere with the cell spreading via their interaction with the integrin β3, showed marked decrease in cell spreading with averaged cell areas of 450 μm2 and 550 μm2, respectively. In summary, the in vivo cell spreading assay is consistent with the in vitro pull-down and c-Src activity experiments, suggesting that the “N”-Src loop is the binding site of integrin β3, and the binding of R760GT762 in the cleft formed by the SH3 and linker domain is important for c-Src activation and signaling.
In vivo cell spreading assay. (A) Visualization of CHOβ3 cells with expression of SH31-141, SH31-141G116R, or SH31-141G127R. The expression of integrin β3 (green) and the SH3 protein (red) was detected using antibodies against integrin β3 and Flag-tag, respectively, and visualized under confocal microscopy. (B) A statistics analysis of cell spreading assay. The surface areas of adherent cells were measured using ImageJ software (National Institutes of Health). The averaged cell area for each coexpression was calculated over a dataset containing 60 randomly chosen cells. Values are mean ± SE; n = 60. **P < .01, statistically significant compared with control CHOβ3 cells. ***P < .001, statistically significant compared with control CHOβ3 cells.
In vivo cell spreading assay. (A) Visualization of CHOβ3 cells with expression of SH31-141, SH31-141G116R, or SH31-141G127R. The expression of integrin β3 (green) and the SH3 protein (red) was detected using antibodies against integrin β3 and Flag-tag, respectively, and visualized under confocal microscopy. (B) A statistics analysis of cell spreading assay. The surface areas of adherent cells were measured using ImageJ software (National Institutes of Health). The averaged cell area for each coexpression was calculated over a dataset containing 60 randomly chosen cells. Values are mean ± SE; n = 60. **P < .01, statistically significant compared with control CHOβ3 cells. ***P < .001, statistically significant compared with control CHOβ3 cells.
Discussion
It is well documented that the activation of c-Src via SH3 is often triggered by binding to peptides with PXXP motif. However, as first proposed by Arias-Salgado et al,19 the SH3 domain can make direct contact with the cytoplasmic tail of integrin β3 that contains no proline residues. This interaction can also prime/activate the enzyme. Further characterizations of integrin β3 have helped to identify that the last 3 residues, R760GT762, of integrin β3 makes the direct handshake. Compared with the PXXP peptide, the interaction between the RGT peptide and the SH3 domain is much weaker. A kinetic analysis using Biacore suggests that the KD of SH3:RGT is relatively weak with KD in the micromolar range (supplemental Figure 1). However, the strong binding ability is thought to be important for the activation by the peptides with PXXP motif. For example, it has been shown that the proline-rich peptide with a Kd value of 250nM is essential to compete against the SH2-kinase linker that are in association with the SH3 domain, and trigger the unfolding of the enzyme that leads to the final activation.34 Thus, how a RGT peptide with a KD value of 74.3μM can activate c-Src is very puzzling.
During the outside-in signaling, activated β3 can undergo oligomerization, a process known as microclustering.22 As a result, the local concentration of membrane-bound integrin β3 will increase dramatically. Furthermore, it has been shown that the N-terminal segment of c-Src containing the membrane-anchor signal is important for the interaction/signaling of c-Src:β3.19 It is reasonable to envisage that the majority of the interaction might happen while these 2 proteins still remain associated with the cytoplasmic membrane. Indeed, the membrane-bound forms will favor the interaction. Because of the presence of the membrane, the interacting event could be restricted in a 2-dimensional plane where the binding partners would enjoy higher probability of collision than in a 3-dimensional space. Interestingly, when the c-Src:β3 is modeled in the context of the membrane, the c-Src structure is positioned in such configuration that will give enough space for interaction (Figure 5A). Hence, we propose that the cytoplasmic membrane and the microclustering process during integrin signaling might be important to help the integrin β3 and c-Src to interact with each other at low-binding affinity(Figure 5A). Indeed, the interaction of c-Src:β3 is recognized as the “oncogenic unit” in cancer metastasis.29 The weak interaction regulated by the events of membrane anchoring, and microclustering might serve as another level of control to avoid leaky and harmful activation of c-Src by integrin β3.
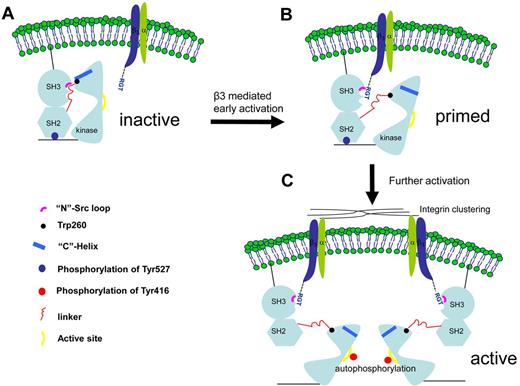
The structural and functional data presented in this report also suggest a novel activation mechanism involving the SH3 domain (Figure 7). In the canonical activation pathway, the PXXP peptide binds to the side of the SH3 in between the “N”-Src and “RT” loops. In the integrin β3-mediated activation, the RGT peptide adopts an “L”-shaped geometry, with Arg760 and Thr762 acting like 2 hands, clamping on the “N”-Src loop (Figure 2). Arg760 can contribute significant hydrophobic interaction toward the overall binding. Mutation of Arg to Ala disrupts the binding and activation activity of the RGT peptide (Figure 4A-B). Besides, on interaction, the side-chain of Arg760 can adopt different conformations that might interfere the formation of the hydrogen bond between Asp117 and Arg318 (supplemental Figure 2B-C). Interestingly, as shown in the structure of inactive c-Src,8 this intramolecular hydrogen bond is thought to favor the packing between the SH3, linker and kinase domains. At the C-terminal end of the RGT peptide, the charged carboxylate group of Thr762 is sandwiched by Tyr131 (from the SH3 domain) and Lys257 (from the linker domain). The presence of Thr762 clearly inhibits the formation of the hydrogen bond between Tyr131 and Lys257 observed in the inactive c-Src.8 In addition, Trp118, a highly conserved residue important for the kinase activity,32 is located close to the RGT peptide (Figure 3B). Although its side-chain does not interact directly with the RGT peptide (supplemental Figure 2C), it is reasonable to envisage that the binding of the RGT peptide in the cleft between the SH3 and linker domain might cause reshuffling of the region, leading to the disengagement between Trp118 and the linker domain. In turn, this might affect Trp260, which is located in the C-terminal end of the linker domain, 3 residues away from Lys257 that is predicted to be in direct contact with the RGT peptide (supplemental Figure 2A). Interestingly, the interaction between Trp260 and the “C”-helix of the kinase domain is thought to be a crucial structural element controlling the formation of the ATP binding site, leading to the final “switch-on” of the kinase.16-18
A novel c-Src activation mechanism by integrin β3. (A) The initial contact. The c-Src kinase is depicted in its inactive form. (B) A putative transition state in which the c-Src is primed by integrin β3. The intramolecular interactions between the SH3, SH2, SH2-linker, and kinase domain are thought to be important for the enzymatic activity. As suggested by the complexes of SH3:RGT and c-Src:β3, the binding of the RGT peptide could disrupt these intramolecular constraints, leading to the disengagement of Trp260 and “C”-helix. (C) As a result, the enzyme is primed for further activations, including the phosphorylation of Tyr416 in the active site in the kinase domain, a process that could be facilitated by integrin microclustering, autophosphorylation,19 and dephosphorylation of Tyr527 regulated by the tyrosine phosphatases and Csk kinase.19,35
A novel c-Src activation mechanism by integrin β3. (A) The initial contact. The c-Src kinase is depicted in its inactive form. (B) A putative transition state in which the c-Src is primed by integrin β3. The intramolecular interactions between the SH3, SH2, SH2-linker, and kinase domain are thought to be important for the enzymatic activity. As suggested by the complexes of SH3:RGT and c-Src:β3, the binding of the RGT peptide could disrupt these intramolecular constraints, leading to the disengagement of Trp260 and “C”-helix. (C) As a result, the enzyme is primed for further activations, including the phosphorylation of Tyr416 in the active site in the kinase domain, a process that could be facilitated by integrin microclustering, autophosphorylation,19 and dephosphorylation of Tyr527 regulated by the tyrosine phosphatases and Csk kinase.19,35
Therefore, the binding of the SH3 domain and the RGT peptide appears to be able to cause disruption of the intramolecular interactions between the SH3 and linker domains. This has led to the proposal of a novel c-Src activation mechanism by integrin β3 (Figure 7). On signaling, the integrin is activated and its cytoplasmic tail adopts an extended conformation with the RGT exposed for the binding of c-Src. The membrane anchoring and the microclustering process might help to raise the local protein concentration that enable integrin β3 and c-Src to make initial contact under a “fishing” mechanism, as suggested by the model of c-Src:β3 (Figure 5A). The binding of the RGT peptide to the SH3 domain of c-Src via the “N”-Src loop might trigger the conformational reshuffling in the nearby environment that might favor the disengagement of SH3:linker and Trp260:“C”-helix. As a result, the c-Src kinase is primed (Figure 7B). Further activations, including the phosphorylation of Tyr416 and the dephosphorylation of Tyr527, are thought to stabilize the activated state of c-Src in an open-up conformation19,35 (Figure 7C).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the personnel of beamline BL17U (Shanghai, China) for help during data collection.
This work was supported by Shanghai Municipal Science and Technology Commission (research grant 11JC1407200; G.M.), Shanghai Municipal Education (research grant 12ZZ109; G.M.), Program for New Century Excellent Talents in University (NCET-10-9571; G.M.), Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institute of Higher Learning (G.M.), and National Natural Science Foundation of China (research grant 31070645; G.M.).
Authorship
Contribution: R.X. and G.M. conceived, designed, and performed the experiments and prepared the manuscript; G.M. wrote the manuscript; and all authors analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guoyu Meng, Shanghai Institute of Hematology, Rui Jin Hospital, Shanghai JiaoTong University School of Medicine, 197 Rui Jin Er Road, Lu Wan District, Shanghai 200025 People's Republic of China; e-mail: guoyumeng@shsmu.edu.cn.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal