Key Points
Ldb1 complexes bind to and positively regulate the expression of a large number of erythroid genes including most known Gata1-regulated genes.
Ldb1 complexes and Klf1 frequently bind together and coregulate erythroid gene expression.
Abstract
Erythropoiesis is dependent on the lineage-specific transcription factors Gata1, Tal1, and Klf1. Several erythroid genes have been shown to require all 3 factors for their expression, suggesting that they function synergistically; however, there is little direct evidence for widespread cooperation. Gata1 and Tal1 can assemble within higher-order protein complexes (Ldb1 complexes) that include the adapter molecules Lmo2 and Ldb1. Ldb1 proteins are capable of coassociation, and long-range Ldb1-mediated oligomerization of enhancer- and promoter-bound Ldb1 complexes has been shown to be required for β-globin gene expression. In this study, we generated a genomewide map of Ldb1 complex binding sites that revealed widespread binding at erythroid genes and at known erythroid enhancer elements. Ldb1 complex binding sites frequently colocalized with Klf1 binding sites and with consensus binding motifs for other erythroid transcription factors. Transcriptomic analysis demonstrated a strong correlation between Ldb1 complex binding and Ldb1 dependency for gene expression and identified a large cohort of genes coregulated by Ldb1 complexes and Klf1. Together, these results provide a foundation for defining the mechanism and scope of Ldb1 complex activity during erythropoiesis.
Introduction
Critical functions have been demonstrated for 3 lineage-restricted transcription factors, Gata1, Tal1/Scl, and EKLF/Klf1, in erythroid development. Ablation of any one of the genes that encode these proteins causes profound defects in erythropoiesis.1-3 Furthermore, in the absence of Gata1, Tal1, or Klf1, transcription of β−globin and several other key erythroid genes is markedly down-regulated, suggesting a broad coregulatory function for Gata1, Tal1, and Klf1 for erythroid gene expression.4-6
In erythroid cells, Gata1 and Tal1 can assemble within a multimeric protein complex that includes the adapter molecules Lmo2 and Ldb1.7 These complexes (hereafter referred to as Ldb1 complexes) bind to paired E-box/GATA DNA motifs and are thought to function primarily to positively regulate erythroid gene expression.7-10 A distinguishing feature of Ldb1 complexes is their ability to oligomerize through Ldb1 coassociation. Ldb1-mediated oligomerization can facilitate long-range associations between promoters and enhancers, and in erythroid cells, this activity has been shown to be essential for β-globin, Myb, and Epb4.2 gene expression.10-12 The importance of Ldb1 complexes for erythropoiesis has also been established by documenting that each of its subunits is essential for erythroid development.1,2,13,14 However, whether Ldb1 complexes control the expression of a few critical genes necessary for erythropoiesis or instead have a more generalized role in regulating erythroid gene expression remains to be determined.
Chromatin immunoprecipitation (ChIP)-on-chip and ChIP coupled with massively parallel sequencing (ChIP-Seq) were recently used to map Ldb1 complex binding sites in erythroid lineage cells.9,15 In these studies, Ldb1 complex binding was detected at many erythroid genes that are almost exclusively induced during terminal erythropoiesis; however, a direct role for Ldb1 complexes in the global activation of target genes was not demonstrated. Also unclear is the extent to which Ldb1 complexes function in concert with other transcription factors to positively regulate erythroid gene transcription. Cobinding of Ldb1 complexes and Klf1 at a distal regulatory element for Myb has been recently demonstrated, and Ldb1 was shown to be required for recruitment of Klf1 to the Myb promoter and for Myb transcription.11 However, the results of genomewide ChIP-Seq studies have been inconsistent regarding the extent of overlap between Klf1 and known or presumed Ldb1 complex interactomes; thus, the degree of functional synergy between Ldb1 complexes and Klf1 remains unclear.6,16-18
Here, we used ChIP-Seq to generate a global map of Ldb1/Tal1/Gata1 complex binding in primary hematopoietic cells. We show that Ldb1 complexes stabilize DNA binding of Tal1 and Gata1 and target a vast number of erythroid genes, as well virtually all known erythroid cis-regulatory elements. By integrating ChIP-Seq data with data from microarrays that evaluated the effect of short hairpin RNA (shRNA)-mediated Ldb1 KD on gene expression in erythroid cells, we also provide the first demonstration of a direct role for Ldb1 in the widespread activation of erythroid genes and in the regulation of multiple pathways critical for terminal erythroid development. Finally, we document that Ldb1 complexes frequently bind near and function in concert with Klf1 to activate expression of many erythroid genes. Our results suggest that this activity occurs by direct promoter co-occupancy by Ldb1 complexes and Klf1 or by Ldb1-mediated recruitment of distal regulatory elements that bind Klf1. These data establish Ldb1 complexes as key instruments of Tal1/Gata1-mediated transcriptional activity and suggest a broad function for Ldb1 complex binding and Ldb1 oligomerization in the regulation of erythroid gene expression.
Methods
ChIP-Seq and microarray analysis
Motif searches
De novo motif search was performed using the Multiple EM for Motif Elicitation (MEME) tool20 with 0 or 1 motif occurrence per sequence and search window 6-20. Search for known motifs was performed using the Motif Alignment and Search Tool (MAST).21 To determine the distribution of spacer lengths between the E-box motifs and GATA motifs, we used text-based string matching. Relevant sequences around Ldb1 complex sites were retrieved from the reference genome using both the forward strand as well as the reverse strand. The distance between motifs was measured in number of nucleotides.
Quantitative reverse transcription-polymerase chain reaction
The murine erythroleukemia (MEL) cell line expressing Ldb1 shRNA has been described previously.10 For gene expression studies, quantitative reverse transcription-polymerase chain reaction was performed as described.13,19 Primer sequences will be provided on request.
Animal Care and Use Committee approval was obtained for all experiments in this study.
Data access
Results
Genomewide mapping of Ldb1, Tal1, and Gata1 binding sites in total bone marrow cells
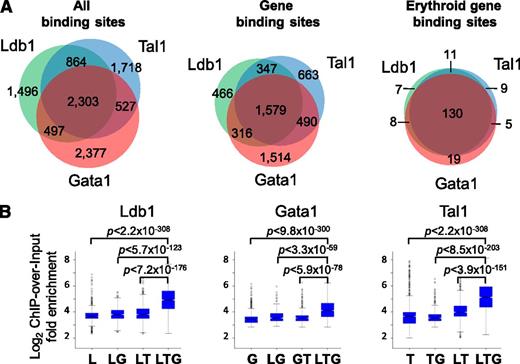
We performed whole-genome deep sequencing (ChIP-Seq) experiments on unfractionated primary bone marrow cells from adult C57BL/6 mice with antibodies against Ldb1 or against Tal1 or Gata1, the known DNA binding subunits of erythroid Ldb1 protein complexes.7 Site Identification from Short Sequence Reads analysis of ChIP-Seq reads22 identified 5160 Ldb1 binding sites, 5412 Tal1 binding sites, and 5704 Gata1 binding sites (Figure 1A, left). Forty-five percent of the Ldb1 binding sites colocalized with both Tal1 and Gata1 binding sites (Figure 1A). Similar percentages of Tal1 binding sites and Gata1 binding sites colocalized with Ldb1 and Gata1 or with Ldb1 and Tal1, respectively (Figure 1A). The Ldb1/Tal1/Gata1 binding site overlap is likely underestimated because we excluded binding sites where occupancy of all 3 proteins was detectable but where 1 or 2 peaks failed to meet our criteria for significance. An exception was that many “Gata1-only” sites lacked detectable Ldb1 or Tal1 binding, indicating that they may be sites where Gata1 either binds in isolation or in association with factors other than Ldb1 and Tal1 (supplemental Table 1; supplemental Figure 1).
ChIP-Seq analysis of Ldb1/Tal1/Gata1 complexes (Ldb1 complexes). (A) Frequency of colocalized Ldb1, Tal1, and Gata1 binding sites. Shown are (left) all sites , (center) gene binding sites, and (right) erythroid fingerprint gene binding sites. Gene binding sites are defined as those sites within 5 kb of a TSS or within a gene body. Erythroid fingerprint genes are as described.24 (B) ChIP-over-input fold enrichment at Ldb1, Tal1, and Gata1 binding sites. G, Gata1; L, Ldb1; T, Tal1.
ChIP-Seq analysis of Ldb1/Tal1/Gata1 complexes (Ldb1 complexes). (A) Frequency of colocalized Ldb1, Tal1, and Gata1 binding sites. Shown are (left) all sites , (center) gene binding sites, and (right) erythroid fingerprint gene binding sites. Gene binding sites are defined as those sites within 5 kb of a TSS or within a gene body. Erythroid fingerprint genes are as described.24 (B) ChIP-over-input fold enrichment at Ldb1, Tal1, and Gata1 binding sites. G, Gata1; L, Ldb1; T, Tal1.
ChIP-Seq tag densities for Ldb1, Tal1, and Gata1 at shared binding sites were significantly higher than at binding sites for any single or pairwise combinations of these proteins for each of the ChIP-Seq runs (Figure 1B). Notably, Tal1 and Gata1 sequence tag numbers were significantly higher when Tal1 and Gata1 were part of an Ldb1 complex in comparison with all other potential complexes, suggesting that the DNA binding affinity of Tal1 and Gata1 is increased when they function within the context of Ldb1/Tal1/Gata1 protein complexes.22
Analysis of Ldb1 complex binding sites reveals selective targeting of erythroid genes
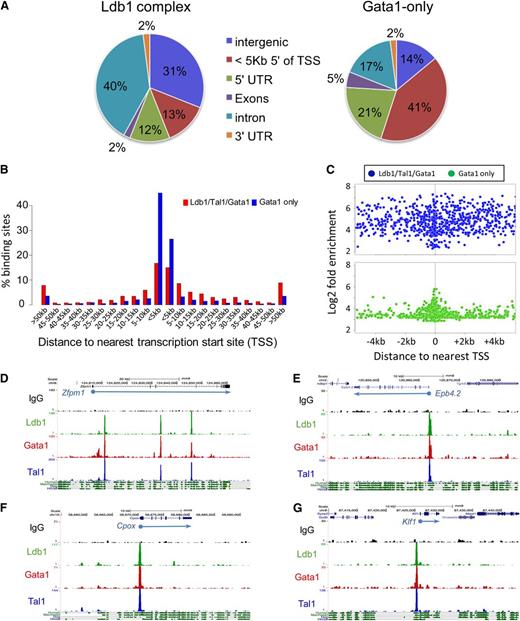
To determine which genes are targeted by Ldb1 complexes, we considered overlapping Ldb1/Tal1/Gata1 binding sites that were within 5 kb of a transcription start site (TSS) and/or within a gene body (supplemental Table 1). Using these criteria, 69% of all DNA bound Ldb1 complexes mapped to known genes (Figure 1A, center). The majority of Ldb1 complex gene binding sites were within introns, with only 25% of sites localized within putative promoter regions (in the 5-prime UTR or within 5 kb of the TSS; Figure 2A). Interestingly, similar to a previous study,9 we found that many intronic binding sites were within the first intron (supplemental Figure 2; data not shown) at locations where erythroid-specific promoters have been recently identified.18 Among the remaining third of Ldb1 complex binding sites, many were far removed (>50 kb) from any known gene (Figure 2B). In contrast to Ldb1 complex binding sites, non-Ldb1/Tal1-associated Gata1 binding sites were mostly clustered near promoters with 62% of sites in the 5-prime UTR or within 5 kb of the TSS (Figure 2A-C).
Location of Ldb1 complex binding sites. (A) Genomic distribution of (left) Ldb1 complex binding sites or (right) non-Ldb1/Tal1-associated (Gata1 only) binding sites. (B) Location of all Ldb1 complex binding sites and Gata1-only binding sites relative to the nearest gene. Bar heights indicate percentage of all Ldb1 complex binding sites (red) or non-Ldb1/Tal1 associated Gata1-only binding sites (blue) that map to the indicated distance from a known gene. (C) Distribution of all Ldb1 complex binding sites (blue) and all Gata1-only binding sites (green) located within 4 kb of a TSS. y-axis shows Log2 fold enrichment of ChIP-Seq tag number/input tag number. (D-G) UCSC Genome Browser shots showing Ldb1, Tal1, and Gata1 binding sites at (D) Zfpm1 (Fog1), (E) Epb4.2, (F) Cpox, and (G) Klf1. y-axis shows number of sequence reads. Mammalian conservation tracks are shown at the bottom.
Location of Ldb1 complex binding sites. (A) Genomic distribution of (left) Ldb1 complex binding sites or (right) non-Ldb1/Tal1-associated (Gata1 only) binding sites. (B) Location of all Ldb1 complex binding sites and Gata1-only binding sites relative to the nearest gene. Bar heights indicate percentage of all Ldb1 complex binding sites (red) or non-Ldb1/Tal1 associated Gata1-only binding sites (blue) that map to the indicated distance from a known gene. (C) Distribution of all Ldb1 complex binding sites (blue) and all Gata1-only binding sites (green) located within 4 kb of a TSS. y-axis shows Log2 fold enrichment of ChIP-Seq tag number/input tag number. (D-G) UCSC Genome Browser shots showing Ldb1, Tal1, and Gata1 binding sites at (D) Zfpm1 (Fog1), (E) Epb4.2, (F) Cpox, and (G) Klf1. y-axis shows number of sequence reads. Mammalian conservation tracks are shown at the bottom.
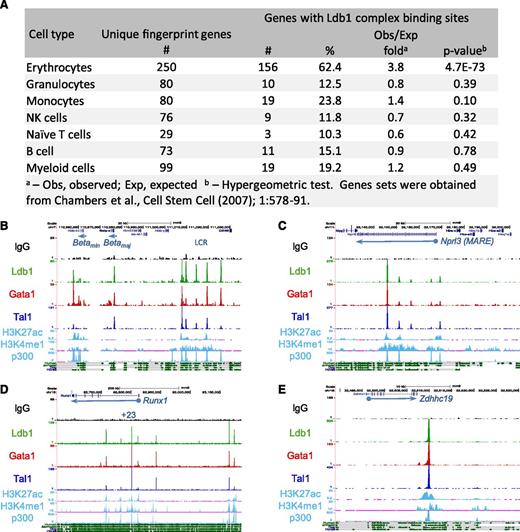
Ontology analysis of genes targeted by Ldb1 complexes23 revealed a strong enrichment in functional categories relevant to erythropoiesis including heme/porphryin biosynthesis, erythrocyte differentiation/hemostasis, gas transport, and iron homeostasis (supplemental Table 2). Using a keyword search algorithm (supplemental Methods), we generated a list of 533 erythroid genes (supplemental Table 3). Ldb1 complex binding was significantly overrepresented at these genes relative to randomly selected gene sets of the same size (P < 1.6 × 10−16; hypergeometric test). Erythroid specificity was also evident when we analyzed Ldb1 complex binding at lineage-specific hematopopoietic “fingerprint” genes24 (Figure 3A). The extent of coincident Ldb1, Tal1, and Gata1 binding sites was higher for gene binding sites (29%) (Figure 1A, center) than for all sites (24%) (Figure 1A, left) and was higher still for erythroid fingerprint genes (69%) (Figure 1A, right), likely reflecting the selective targeting of erythroid genes by Ldb1 complexes.
Enrichment of Ldb1 complex binding sites at erythroid genes and erythroid cis-regulatory elements. (A) Ldb1 complex binding sites at erythroid “fingerprint” genes.24 (B-E) UCSC Genome Browser shots of Ldb1 complex binding at known or presumed cis-regulatory elements: (B) β-globin LCR, (C) α-globin (MARE) cis-regulatory region, (D) Runx1 locus, and (E) presumed cis-regulatory element near Zdhhc19. Binding sites for p300 and H3K27ac, H3K4me1 profiles in MEL cells, and mammalian conservation tracks are shown at the bottom.
Enrichment of Ldb1 complex binding sites at erythroid genes and erythroid cis-regulatory elements. (A) Ldb1 complex binding sites at erythroid “fingerprint” genes.24 (B-E) UCSC Genome Browser shots of Ldb1 complex binding at known or presumed cis-regulatory elements: (B) β-globin LCR, (C) α-globin (MARE) cis-regulatory region, (D) Runx1 locus, and (E) presumed cis-regulatory element near Zdhhc19. Binding sites for p300 and H3K27ac, H3K4me1 profiles in MEL cells, and mammalian conservation tracks are shown at the bottom.
Ldb1 complexes bound to genes involved in a wide range of erythroid cellular processes including transcription (eg, Klf1, Zfpm1/Fog1, Nfe2), heme synthesis (eg, Alad, Alas2, Cpox, Fech, Ppox), and cytoskeletal organization (eg, Add1/2, Ank1, Epb4.1, Epb4.2, Spna1) (Table 1, Figure 2D-G; supplemental Table 1; supplemental Figure 3). We also detected Ldb1 complex binding at genes encoding erythrocyte cytosolic proteins (eg, β-globin, Hepb1, Prdx2), as well as genes encoding key erythrocyte cell membrane proteins, including Aqp1, Gypa, Gypc, solute carrier proteins, and all known red cell surface antigens (Table 1). Together, these results suggested that Ldb1 complexes have a global role in erythroid gene transcription.
Erythroid genes with Ldb1 complex binding sites
| Cellular process . | Gene . |
|---|---|
| Nuclear/transcription | Bach1, Btg2*, Cbfa2t3, Ccne1*, Cdc6*, Cdyl, Cldn13*, Ddit3*, Dpy30*, E2f2, E2f4, Egr1, Elf1, Foxo3, Gata1, Gfi1b, Hemgn*, Hipk2*, Hist1h1c, ikzf1, Klf1*, Klf11, Klf13, Lyl1, Mbd2*, Mybl2, Mxi1*, Nfe2, Nfe2l2, Nprl3 (MARE), Nrf1, Rngtt*, Runx1, Runx2, Sox6*, Tal1, Tom1l1, Trim10*, Xpo7, Zfpm1*, Ikzf1/Zfpn1a1 |
| Heme/Hb | Abcb6*, Abcg2*, Alad*, Alas2*, Cpox*, Fech*, Glrx5, Hebp1*, Hbb*, Isca1, Ppox*, Slc25a37*, Slc40a1, Tfrc*, Tmem14c*, Urod*, Hmbs/Uros1, Wwp2 |
| Cytoskeleton | Add1, Add2*, Add3, Ank1*, Asb1*, Asb17*, Ckap2l, Epb4.1*, Epb4.2*, Icam4, Micall2, Slc11a2*, Spire1*, Spna1*, Spnb1, Tmod1*, Tpm1, Tubb3* |
| Cytoplasmic | Arrb1*, Arhgef12*, Asb1*, Atpif1, Car2*, Cela1, Cpeb3, Ctsb, Dcun1d1, Dusp1*, Eif2ak1*, Fn3k, Grap2*, Gtpbp1*, Hebp1*, Mbp, Mgll*, Ppp1r15a/Myd116*, Naga, Pcx*, Pik3r1, Pklr*, Pkm2*, Prdx2*, Prkaa1, Pycr2*, Rab3il1*, Rgs10*, Sla*, Sphk1*, Stx11*, Tgm2*, Tnk1, Trim25, Txnrd2*, Uba7*, Ube2l6, Ube2o* |
| Membrane/blood group | Abca1, Abcb10, Abcc3*, Abcg4, Ache, Aqp1*, Aqp8, Aqp9*, Art4, Atp8a1, B4galt1, Bsg, Butr1, Cd59a*, Cldn13*, Darc, Epor, Ermap*, Gp9, Gypa*, Gypc*, Itga2b*, Itga4*, Kcnn4*, Kel*, Pigq, Prnp, Prokr1*, Ptdss2*, Rhag*, Rhd, Slc1a2, Slc4a1*, Slc6a9*, Slc11a2*, Slc14a1*, Slc2a4, Slc22a4*, Slc22a23, Slc30a9*, Slc38a5*, Slc43a3*, Spnb1*, Stom, Tm7sf3, Tmcc2*, Sgms1/Tmem23*, Tspan33*, Xk |
| Cellular process . | Gene . |
|---|---|
| Nuclear/transcription | Bach1, Btg2*, Cbfa2t3, Ccne1*, Cdc6*, Cdyl, Cldn13*, Ddit3*, Dpy30*, E2f2, E2f4, Egr1, Elf1, Foxo3, Gata1, Gfi1b, Hemgn*, Hipk2*, Hist1h1c, ikzf1, Klf1*, Klf11, Klf13, Lyl1, Mbd2*, Mybl2, Mxi1*, Nfe2, Nfe2l2, Nprl3 (MARE), Nrf1, Rngtt*, Runx1, Runx2, Sox6*, Tal1, Tom1l1, Trim10*, Xpo7, Zfpm1*, Ikzf1/Zfpn1a1 |
| Heme/Hb | Abcb6*, Abcg2*, Alad*, Alas2*, Cpox*, Fech*, Glrx5, Hebp1*, Hbb*, Isca1, Ppox*, Slc25a37*, Slc40a1, Tfrc*, Tmem14c*, Urod*, Hmbs/Uros1, Wwp2 |
| Cytoskeleton | Add1, Add2*, Add3, Ank1*, Asb1*, Asb17*, Ckap2l, Epb4.1*, Epb4.2*, Icam4, Micall2, Slc11a2*, Spire1*, Spna1*, Spnb1, Tmod1*, Tpm1, Tubb3* |
| Cytoplasmic | Arrb1*, Arhgef12*, Asb1*, Atpif1, Car2*, Cela1, Cpeb3, Ctsb, Dcun1d1, Dusp1*, Eif2ak1*, Fn3k, Grap2*, Gtpbp1*, Hebp1*, Mbp, Mgll*, Ppp1r15a/Myd116*, Naga, Pcx*, Pik3r1, Pklr*, Pkm2*, Prdx2*, Prkaa1, Pycr2*, Rab3il1*, Rgs10*, Sla*, Sphk1*, Stx11*, Tgm2*, Tnk1, Trim25, Txnrd2*, Uba7*, Ube2l6, Ube2o* |
| Membrane/blood group | Abca1, Abcb10, Abcc3*, Abcg4, Ache, Aqp1*, Aqp8, Aqp9*, Art4, Atp8a1, B4galt1, Bsg, Butr1, Cd59a*, Cldn13*, Darc, Epor, Ermap*, Gp9, Gypa*, Gypc*, Itga2b*, Itga4*, Kcnn4*, Kel*, Pigq, Prnp, Prokr1*, Ptdss2*, Rhag*, Rhd, Slc1a2, Slc4a1*, Slc6a9*, Slc11a2*, Slc14a1*, Slc2a4, Slc22a4*, Slc22a23, Slc30a9*, Slc38a5*, Slc43a3*, Spnb1*, Stom, Tm7sf3, Tmcc2*, Sgms1/Tmem23*, Tspan33*, Xk |
Significantly down-regulated (False Recovery Rate < 0.01) in MEL cells expressing Ldb1 shRNA (see text).
Ldb1 complexes bind at erythroid cis-regulatory elements
In erythroid progenitors, binding of Ldb1 complexes has been documented at the β-globin locus control region (LCR).10 Confirming those results, we detected Ldb1 complexes at DNase hypersensitive sites within the β-globin LCR, as well as at the β-maj and β-min globin promoters (Figure 3B; supplemental Table 4). To determine if Ldb1 complexes bind to other erythroid cis-regulatory elements, we looked for binding at enhancers that had been experimentally validated in prior studies. We detected prominent Ldb1 complex binding at each of the known erythroid enhancers that we examined including distal regulatory elements for Runx1, Ctfab3 (Eto2), α-globin, and Myb (Figure 3C-D; supplemental Table 4; supplemental Figure 4; data not shown). Most Ldb1 complex binding sites at uncharacterized intergenic locations were within conserved domains that displayed signature enhancer marks (H3K4me1 and H3K27ac) and were bound by the transcriptional coactivator p300, suggesting that they lie within erythroid cis-regulatory elements (Figure 3E; supplemental Figure 4).
Motif analysis of Ldb1 complex binding sites reveals sequence and spacer preferences for binding
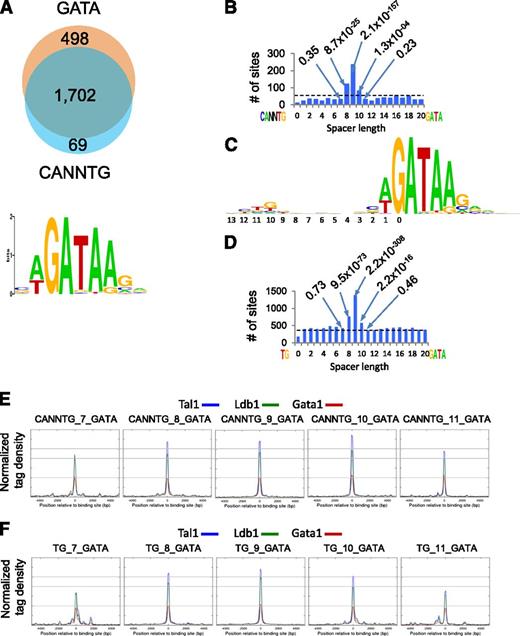
Ldb1 complexes bind preferentially to a conserved paired motif composed of a consensus E-box (CANNTG) and a GATA motif with restricted orientation and spacing: CANNTG-N8-10-GATA.7 Seventy-four percent of Ldb1 complex binding sites contained both CANNTG and GATA, and >90% of sites contained a WGATAR motif (Figure 4A), the known preferential Gata1 binding site.25 Notably, there was a significant enrichment for N8-N10 spacing, with N9 by far the most frequent within all DNA fragments that contained paired CANNTG-GATA motifs (Figure 4B).
Sequence analysis of Ldb1 complex–bound DNA fragments. (A) (Upper) Number of Ldb1 complex–bound fragments with GATA and/or E-box (CANNTG) motifs. (Lower) Consensus GATA motif within Ldb1 complex–bound DNA fragments discovered by MEME de novo motif search. (B) Number of Ldb1 complex binding sites that contain paired CANNTG/GATA motifs separated by 0-20 bp. Dotted line represents the expected number of peaks (binding sites) in randomly selected sites. Numbers in the plot represent P values of bars denoting fragments with the indicated number of base pairs separating the CANNTG and GATA motifs. (C) Consensus logo generated from the top 1000 Ldb1 complex binding sites. (D) Number of Ldb1 complex binding sites that contain paired TG/GATA motifs separated by 0-20 bp. Dotted line represents the expected number of peaks (binding sites) in randomly selected sites. Numbers in the plot represent P values of bars denoting fragments with the indicated number of base pairs separating the TG and GATA motifs. (E) Average Ldb1, Tal1, and Gata1 ChIP-Seq tag numbers for Ldb1 complexes bound to sites containing CANNTG from 7-11 bp 5-prime of a GATA. (F) Average Ldb1, Tal1, and Gata1 ChIP-Seq tag numbers for Ldb1 complexes bound to sites containing TG from 7-11 bp 5-prime of a GATA.
Sequence analysis of Ldb1 complex–bound DNA fragments. (A) (Upper) Number of Ldb1 complex–bound fragments with GATA and/or E-box (CANNTG) motifs. (Lower) Consensus GATA motif within Ldb1 complex–bound DNA fragments discovered by MEME de novo motif search. (B) Number of Ldb1 complex binding sites that contain paired CANNTG/GATA motifs separated by 0-20 bp. Dotted line represents the expected number of peaks (binding sites) in randomly selected sites. Numbers in the plot represent P values of bars denoting fragments with the indicated number of base pairs separating the CANNTG and GATA motifs. (C) Consensus logo generated from the top 1000 Ldb1 complex binding sites. (D) Number of Ldb1 complex binding sites that contain paired TG/GATA motifs separated by 0-20 bp. Dotted line represents the expected number of peaks (binding sites) in randomly selected sites. Numbers in the plot represent P values of bars denoting fragments with the indicated number of base pairs separating the TG and GATA motifs. (E) Average Ldb1, Tal1, and Gata1 ChIP-Seq tag numbers for Ldb1 complexes bound to sites containing CANNTG from 7-11 bp 5-prime of a GATA. (F) Average Ldb1, Tal1, and Gata1 ChIP-Seq tag numbers for Ldb1 complexes bound to sites containing TG from 7-11 bp 5-prime of a GATA.
The consensus logo generated from the top 1000 Ldb1 complex binding sites consisted of a paired motif that contained a weak partial E-box (TG) located 9 bp 5-prime of GATA: TG-N9-GATA (Figure 4C) that closely resembled the logos generated from 2 previous studies.4,9 We speculated that the relatively low apparent conservation of the partial E-box might be explained by flexibility in the spacing of the paired motif (ie, N8-N10). Indeed, when this was taken into consideration, 80% of the Ldb1 binding sites contained ≥1 TG-GATA paired motif separated by 8, 9, or 10 bp (Figure 4D).
Average Ldb1, Tal1, and Gata1 sequence tag numbers were similar at CANNTG/GATA paired motifs spaced either 8, 9, or 10 nucleotides apart, but dropped off outside of this range, suggesting that the N8-N10 spacing is important for high affinity binding of Ldb1 complexes to DNA (Figure 4E). Interestingly, a similar relative affinity trend was observed at binding sites containing either CANNTG/GATA or TG/GATA paired motifs provided that the N8-10 spacing was retained (Figure 4E-F).
Ldb1 complexes frequently bind near Klf1
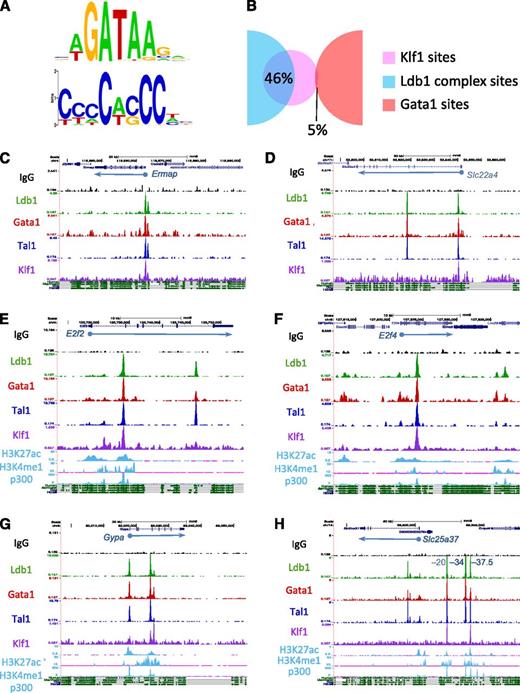
To ascertain if Ldb1 complexes bind near sites that recruit other transcription factors, we performed a nonbiased de novo (MEME)20 motif search on each of the ChIP-Seq fragments bound by Ldb1 complexes. The Gata1 binding consensus WGATAR was identified in all 7 possible categories of protein binding configurations supporting our speculation that many of these sites are weak Ldb1 complex binding sites rather than genuine single or 2-factor binding sites. (supplemental Figure 5A). Consensus binding sequences for Nrf1, Sp1, and Zfp143 were identified within the non–Ldb1-associated Gata1-bound DNA fragments (supplemental Figure 5A). Each of these transcription factors has been shown to interact functionally with Gata1 in erythroid cells and to bind primarily at the promoter regions of target genes.26-29 Within Ldb1 complex bound DNA fragments, we detected a conserved extended CACC motif (Figure 5A) that closely resembled the recently refined Klf1 (EKLF) binding consensus sequence.6
Ldb1 complexes bind at sites near those for Klf1. (A) GATA and CACC consensus motifs identified within Ldb1 complex–bound DNA fragments by MEME de novo motif search. (B) Overlap of the Klf1 binding sites from Tallack et al6 with Ldb1 complex (Ldb1/Tal1/Gata1) binding sites or with non-Ldb1/Tal1-associated Gata1-only binding sites. (C-H) UCSC Genome browser shots showing colocalized Ldb1, Tal1, Gata1, and Klf1 binding sites at (C) Ermap, (D) Slc22a4, (E) E2f2, (F) E2f4, (G) Gypa, and (H) Slc25a37 (positions of known cis-regulatory elements are indicated). Numbers on y-axis are number of sequence reads. Tracks for H3K27ac, H3K4me1, and p300 in MEL cells and mammalian conservation tracks are shown at the bottom.
Ldb1 complexes bind at sites near those for Klf1. (A) GATA and CACC consensus motifs identified within Ldb1 complex–bound DNA fragments by MEME de novo motif search. (B) Overlap of the Klf1 binding sites from Tallack et al6 with Ldb1 complex (Ldb1/Tal1/Gata1) binding sites or with non-Ldb1/Tal1-associated Gata1-only binding sites. (C-H) UCSC Genome browser shots showing colocalized Ldb1, Tal1, Gata1, and Klf1 binding sites at (C) Ermap, (D) Slc22a4, (E) E2f2, (F) E2f4, (G) Gypa, and (H) Slc25a37 (positions of known cis-regulatory elements are indicated). Numbers on y-axis are number of sequence reads. Tracks for H3K27ac, H3K4me1, and p300 in MEL cells and mammalian conservation tracks are shown at the bottom.
We next performed a motif search using a priori–defined binding motifs (MAST)21 to determine if Ldb1 complexes bind near less prevalent sites for other transcription factors. In agreement with the de novo search results, MAST analysis identified consensus Klf1 binding motifs within a high percentage (47%) of Ldb1 complex–bound DNA fragments (supplemental Figure 5B). Consensus binding sequences for several other transcription factors including Foxo3, Ets1, Egr1, Nrf1, Nfe2l2/Nrf2, Runx1, Sp1/Zbtb7a, and Nfe2 were found within 5% to 30% of the Ldb1 complex–bound fragments (supplemental Figure 5B). Interestingly, many of these genes are putative targets of Ldb1 complexes (Table 1).
We validated colocalization of Klf1 with Ldb1 complexes by integrating our ChIP-Seq data with Klf1 ChIP-Seq data from a recently published study.6 Analysis of the 2303 Ldb1 complex binding sites and the 370 Klf1 binding sites that met our criteria for significance revealed a striking overlap, with 46% of the Klf1 binding sites colocalizing with Ldb1 complex binding sites (Figure 5B). Strong overlap (30%) was also observed when we included all 840 Klf1 binding sites reported by Tallack et al,6 whereas very little overlap was observed between Ldb1-complex binding sites and binding sites for another transcription factor, Sfpi1(PU.1)30 (supplemental Figure 6A-B). Although different sample populations were used to identify Ldb1 complex binding sites (total bone marrow cells) and Klf1 binding sites (fetal liver cells), the strong overlap of Ldb1 complex binding with Klf1 binding suggested by our comparative analysis is consistent with the finding that 30% of Klf1 bound DNA fragments contained paired TG-N9-GATA Ldb1 complex binding motifs.6 In contrast to Ldb1 complexes, there was no enrichment for Klf1 motifs (supplemental Figure 5A) and very low (≤5%) overlap with Klf1 binding within non–Ldb1-associated Gata1 binding sites (Figure 5B).
Genome browser imaging of Ldb1, Tal1, Gata1, and Klf1 ChIP-Seq runs revealed colocalization of Klf1 and Ldb1 complexes at or near the TSS of many erythroid genes including βmaj-globin, Ermap, Slc22a4, Add2, and Pigq (Figure 5C-D; supplemental Figure 7; data not shown). As noted previously6 and similar to what we observed for Ldb1 complex binding sites, many Klf1 binding sites were intergenic or within introns rather than at known TSSs suggesting that Klf1 frequently binds at cis-regulatory elements (supplemental Figure 6C). Indeed, we detected colocalized Ldb1 complex and Klf1 binding at known intronic enhancers within the E2f2 and E2f4 genes (Figure 5E-F)6,31 and at the β-globin and α-globin cis-regulatory elements (supplemental Figure 7; data not shown), as well as at known or putative enhancers near several erythroid genes including Gypa, Slc25a37, and Slc4a1 (Figure 5G-H; supplemental Figure 7).
Ldb1 complexes mainly positively regulate gene expression in erythroid cells
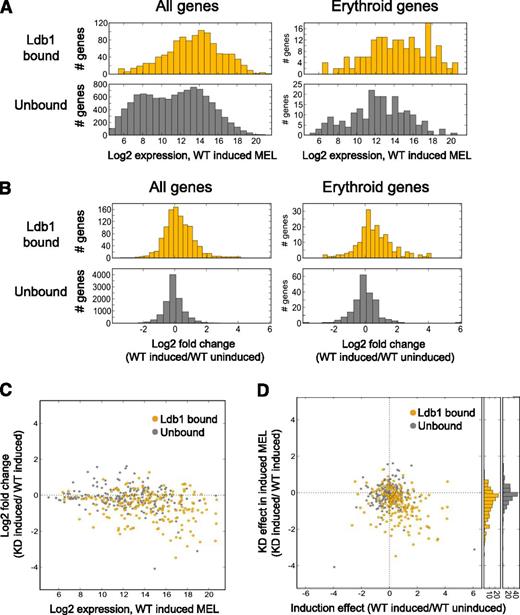
We next looked for a relationship between Ldb1 complex binding and gene expression in MEL cells which, on terminal differentiation, up-regulate erythroid genes.10 Prior work has shown a high degree of Ldb1 complex binding site identity in primary erythroblasts and differentiated MEL cells, demonstrating the suitability of these comparative analyses.9 Overall, gene binding by Ldb1 complexes was predominantly, although not exclusively, associated with transcriptional activation. Genes that were up-regulated in differentiated MEL cells were enriched for Ldb1 complex binding sites (P < 3.7 × 10−10; Fisher’s exact test [FET]), whereas down-regulated genes were not (P < .60; FET). In addition, all genes (P < 8 × 10−11, FET) as well as erythroid genes (P < 3.9 × 10−7, FET) that were up-regulated were enriched for Ldb1 complex binding sites. Genes with Ldb1 complex binding sites were also more highly expressed in differentiated MEL cells and were more strongly up-regulated on terminal erythroid differentiation compared with genes without Ldb1 complex binding sites (Figure 6A-B). Of the top 50 up-regulated genes in differentiated MEL cells, 40% were bound by Ldb1 complexes and 70% had binding sites within 10 kb of the gene locus in contrast to 16% and 26% of the top 50 down-regulated genes (supplemental Table 1).
Direct regulatory function for Ldb1 complexes in the induced expression of erythroid genes in MEL cells. (A) Ldb1 complex–bound genes are more highly expressed in differentiated MEL cells. (Left) All Ldb1-bound genes (yellow, upper panel) compared with all unbound genes (gray, lower panel), P < 7.7 × 10−74. (Right) Erythroid Ldb1 complex–bound genes (yellow, upper panel) compared with unbound erythroid genes (gray, lower panel), P < 1.4 × 10−9. (B) Ldb1 complex–bound genes are more strongly induced in differentiated MEL cells. (Left) All Ldb1 complex–bound genes (yellow, upper panel) compared with all unbound genes (gray, lower panel), P < 1.0 × 10−32. (Right) Ldb1 complex–bound erythroid genes (yellow, upper panel) compared with unbound erythroid genes (gray, lower panel), P < 7.9 × 10−11. Significance for all comparisons was calculated by Mann-Whitney U. (C-D) Ldb1-complexes directly regulate the expression of erythroid genes. Stable clones of MEL cells expressing Ldb1 shRNA or control shRNA were treated with 1.5% dimethylsulfoxide to induce erythroid differentiation. Total RNA was isolated and gene expression was assayed by microarray. (C) Highly expressed erythroid genes are the most strongly down-regulated in Ldb1 KD MEL cells. Genes bound by Ldb1 complexes are depicted in yellow; unbound genes are in gray. Zero line is indicated (dotted line). (D) Erythroid genes that are most strongly induced in differentiated control MEL cells are the most strongly down-regulated in Ldb1 KD MEL cells. Erythroid genes bound by Ldb1 complex are shown in yellow; unbound genes are shown in gray. Zero lines are indicated (dotted lines). y-axis histograms show stronger KD of Ldb1 complex–bound erythroid genes relative to all erythroid unbound genes, respectively (P < 6.7 × 10−10; Mann-Whitney U).
Direct regulatory function for Ldb1 complexes in the induced expression of erythroid genes in MEL cells. (A) Ldb1 complex–bound genes are more highly expressed in differentiated MEL cells. (Left) All Ldb1-bound genes (yellow, upper panel) compared with all unbound genes (gray, lower panel), P < 7.7 × 10−74. (Right) Erythroid Ldb1 complex–bound genes (yellow, upper panel) compared with unbound erythroid genes (gray, lower panel), P < 1.4 × 10−9. (B) Ldb1 complex–bound genes are more strongly induced in differentiated MEL cells. (Left) All Ldb1 complex–bound genes (yellow, upper panel) compared with all unbound genes (gray, lower panel), P < 1.0 × 10−32. (Right) Ldb1 complex–bound erythroid genes (yellow, upper panel) compared with unbound erythroid genes (gray, lower panel), P < 7.9 × 10−11. Significance for all comparisons was calculated by Mann-Whitney U. (C-D) Ldb1-complexes directly regulate the expression of erythroid genes. Stable clones of MEL cells expressing Ldb1 shRNA or control shRNA were treated with 1.5% dimethylsulfoxide to induce erythroid differentiation. Total RNA was isolated and gene expression was assayed by microarray. (C) Highly expressed erythroid genes are the most strongly down-regulated in Ldb1 KD MEL cells. Genes bound by Ldb1 complexes are depicted in yellow; unbound genes are in gray. Zero line is indicated (dotted line). (D) Erythroid genes that are most strongly induced in differentiated control MEL cells are the most strongly down-regulated in Ldb1 KD MEL cells. Erythroid genes bound by Ldb1 complex are shown in yellow; unbound genes are shown in gray. Zero lines are indicated (dotted lines). y-axis histograms show stronger KD of Ldb1 complex–bound erythroid genes relative to all erythroid unbound genes, respectively (P < 6.7 × 10−10; Mann-Whitney U).
To determine if Ldb1 complexes directly regulate transcription of target genes, we next analyzed gene expression in differentiated MEL cells that had been stably transduced with control or Ldb1 lentiviral shRNA.10 On Ldb1 knockdown (KD), both total (supplemental Figure 8A-B) and erythroid (Figure 6C-D) Ldb1 complex bound genes were more strongly down-regulated than non–Ldb1 complex–bound genes. Genomewide, Ldb1 KD inhibited gene induction during terminal erythroid differentiation (P < 2.0 × 10−4, analysis of covariance), and erythroid genes were enriched in the subset of genes that were down-regulated by Ldb1 KD (P < 8.9 × 10−12, FET).
Ldb1 KD inhibited the induction of genes encoding key erythroid transcription factors including Klf1, Zfpm1 (Fog1), Btg2, and Sox6 that were also bound by Ldb1 complexes (genes annotated with an asterisk in Table 1). The requirement for Ldb1 complexes for Klf1 activation is consistent with previous data demonstrating an essential role for a conserved E-box/GATA motif within the Klf1 promoter.32 The induction of genes critical for many other erythroid cellular processes were also significantly inhibited in Ldb1 KD MEL cells (Table 1; supplemental Table 1). Each of these genes contained ≥1 Ldb1 complex binding sites (Table 1) and, for selected genes, we confirmed down-regulation in differentiated Ldb1 KD MEL cells by quantitative reverse transcription-polymerase chain reaction (supplemental Figure 8C).
Many of the same genes that were dependent on Ldb1 for up-regulation in differentiated MEL cells were shown previously to be bound and induced by Gata1,5,29,33 indicating that, in these instances, Gata1 functions in the context of an Ldb1 complex to activate transcription of target genes. Gata1 binding has also been associated with gene repression during terminal erythroid differentiation.5,15,29,34,35 Interestingly, we noted that induction was more strongly associated with Ldb1 complex binding compared with non–Ldb1 complex Gata1 binding (P < 3 × 10−4, FET; supplemental Figure 8D).
Ldb1 complexes regulate critical events in terminal erythropoiesis
Terminal erythroid maturation requires the coordinated activation of multiple cellular responses including cytokenesis, enucleation, mitochondrial clearance (autophagy), oxidative stress tolerance, and the prevention of apoptosis.36,37 Many of the genes that regulate these pathways are bound and regulated by Ldb1 complexes in erythroid cells (supplemental Table 5). We reported previously that KD of Ldb1 in MEL cells blocks differentiation-associated induction of the prosurvival genes Bcl2l1(Bcl-xl) and Sox6 in MEL cells.13 Here, we also identified Ldb1 binding sites at or near genes known to be important for stress erythropoiesis (Cldn13 and Rb1) and for antioxidant responses (Mafk, Prdx2, Gpx1, and Ppp1r15) and document a requirement for Ldb1 for their induction in differentiated MEL cells (supplemental Tables 1 and 5). Key mediators of erythroblast enucleation, including Birc5 (Survivin), Mxi1, Hipk2, and Diap3 (mDia2), were also bound and regulated by Ldb1 complexes (supplemental Tables 1 and 5). It was recently shown that Gata1 is required for the induction of Map1lc3b/LC3B and Ctsb, 2 genes that regulate autophagy/mitophagy in erythroblasts.38 We found that both of these genes, as well as Bnip3l (Nix), which has been shown to be essential for targeting mitochondria to autophagosomes in developing erythroid cells,39 are bound and activated by Ldb1 complexes (supplemental Tables 1 and 5).
Ldb1 complexes and Klf1 coregulate erythroid gene expression
The striking overlap of Klf1 and Ldb1 complex binding sites suggested that these factors function cooperatively to regulate transcription of shared target genes during erythropoiesis. Klf1 has been shown to have an essential role in erythropoiesis and erythroid gene expression40,41 and to cooperate with Gata1 in the induction of several erythroid genes.28,42 We evaluated 106 Klf1-dependent genes (genes that require Klf1 for induction during terminal erythropoiesis) from lists compiled in 2 recently published studies6,43 (supplemental Table 6). Forty-seven (44%) of these genes were significantly down-regulated in Ldb1-KD MEL cells, revealing a strong overlap in regulatory function for Klf1 and Ldb1 complexes (supplemental Table 6). Furthermore, 36 of 47 (77%) of the Klf1/Ldb1 complex codependent genes were bound by Ldb1 complexes, supporting a direct regulatory role for Ldb1 complexes for their expression.
Of the 36 coregulated genes with Ldb1 complex binding sites, several, including Ermap, Ppox, and Slc22a4, had overlapping binding sites for Klf1, Tal1, Gata1, and Ldb1 near their TSS or within the first intron at putative erythroid lineage–specific promoters18 (Figure 5C-D; data not shown). Interestingly, many other coregulated genes had Ldb1 complexes but not Klf1 bound near their TSS. For several of these genes, including Gypa and Slc25a37, colocalized Klf, Tal1, Gata1, and Ldb1 binding was detected either within intron(s) of the same gene or at intergenic sites ranging from 10 to 100 kb away (Figure 5G-H). These intronic/intergenic Klf1/Ldb1 complex cobinding sites were within conserved regions bound by p300 and bearing epigenetic marks consistent with cis-regulatory elements. Indeed, coincident binding sites for Klf1 and Ldb1 complexes were detected at validated cis-regulatory elements for Slc25a37 (mitoferrin)44 (Figure 5H). Taken together, these findings indicate that Ldb1 complexes regulate erythroid gene activity through distinct mechanisms.
Discussion
Gata1 has been shown to be essential for the induction of most, if not all, erythroid genes.5,33 Here, we demonstrate that many genes that are induced by Gata1 are bound by Ldb1 complexes and require Ldb1 for their activation, indicating that Ldb1 complexes are important instruments through which Gata1 positively regulates erythroid gene transcription. Several genes that were down-regulated in response to Ldb1-KD lacked both Ldb1 complex and Gata1 binding sites. Many of these genes (eg, Dck, Cat, and Stx2) have been shown to be Klf1 dependent6,43 ; thus, their down-regulation could be explained by the regulation of Klf1 by Ldb1 complexes. Zfpm1 (Fog1) expression was positively regulated by Ldb1 complexes, raising the issue of whether the Ldb1 KD effect may, in some cases, be secondary to Fog1 down-regulation, as Fog1 is required for induction of many Gata1-dependent genes.45 However, it is notable that Fog1, Eif2ak1, and Klf1, which are Fog1-independent genes,45 were targets of Ldb1 complexes and required Ldb1 for their activation, demonstrating that the Ldb1 KD effect cannot be attributed only to Fog1 down-modulation.
Our findings confirm those from previous reports demonstrating that known or inferred Ldb1 complex binding is primarily associated with erythroid gene activation.9,15,46 However, it has also been shown that Gata1 can be either activating or repressive in different contexts.5,15,29,34,35 These results can be reconciled by a model in which Gata1, when within core Ldb1 complexes, functions as a transcriptional activator, whereas gene repression is mediated by distinct (non-Ldb1 nucleated) Gata1 repressive complexes.15,29,34,47 Nevertheless, for selected genes, such as Lyl1, Klf11, and Egr1, our results suggest a direct role for Ldb1 complexes in gene repression. In this regard, it is significant that Ldb1 complexes have been shown to acquire repressive activity through association with inhibitory molecules, including the chromatin remodeling protein BRG1,48 polycomb repressive complex 2,29,46 Eto2,9 and Pu.1.17
Evidence that Ldb1 complexes bind near and function in concert with other transcription factors to positively regulate erythroid gene expression has been limited, as are data addressing whether this activity is relatively selective or widespread during erythropoieisis. Analysis of merged ChIP-Seq data identified frequent colocalization of Klf1 and Ldb1 complex binding at erythroid gene promoters, at known erythroid enhancer elements, and at the β-globin and α-globin LCRs. We discovered a high frequency of Ldb1 complex binding and Ldb1 dependence for many known Klf1 activated genes. Thus, these results demonstrate widespread synergy between Ldb1 complexes and Klf1 in the regulation of erythroid gene expression.
A solid body of data supports the idea that Ldb1 complexes perform ≥2 important functions during erythropoiesis: the provision of critical transcription factors (Tal1/E2A and Gata1) through binding at conserved paired motifs at target gene promoters7-9,12,15 and the facilitation of long-range promoter/enhancer interactions through Ldb1 oligomerization.9-12,49 Examination of Ldb1 complex and Klf1 binding sites at or in the vicinity of genes that are dependent on both factors for their expression identified distinct binding configurations suggestive of different modes of Ldb1 complex–mediated gene regulation. Many Klf1/Ldb1 coregulated genes were bound by both Klf1 and Ldb1 complexes near their TSS. However, another group of genes lacked Klf1 promoter binding but had prominent Ldb1 binding sites near their TSS, as well as colocalized Ldb1 complex and Klf1 binding within intron(s) or at intergenic sites within known or suspected cis-regulatory elements. We propose that, in this context, Ldb1 complexes function to recruit Klf1 through Ldb1-mediated dimerization, similar to what has recently been shown to occur at the Myb locus.11 Thus, it seems probable that Ldb1 complexes function in distinct ways to orchestrate transcriptional activation on a global scale during terminal erythropoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jan Lee and Dalal El-Khoury for technical assistance.
This work was supported by the Intramural Research Programs of Eunice Kennedy Shriver National Institute of Child Health and Human Development project 1ZIAHD001803-19 (P.E.L.), National Institute of Environmental Health Sciences project 1ZIAES102625-04 (R.J.), National Heart, Lung and Blood Institute project 1ZHL006031-04 (K.Z.), and National Institute of Diabetes and Digestive and Kidney Diseases project DK075033-04 (A.D.).
Authorship
Contribution: L.L., J.F., K.C., R.D, S.-H.S., and R.J. conducted the experiments and/or performed data analysis; L.L. and P.E.L. wrote the paper; and L.L., A.D., K.Z., R.J., and P.E.L. designed the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul E. Love, Section on Cellular and Developmental Biology, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892; email: lovep@mail.nih.gov; and Raja Jothi, Laboratory of Molecular Carcinogenesis, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709; email: jothi@mail.nih.gov.
References
Author notes
L.L. and J.F. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal