Key Points
We present an antidote for dabigatran that effectively reverses its anticoagulative effect in human plasma in vitro and in rats in vivo.
The antidote shares structural features with thrombin in the mode of binding but has no activity in coagulation tests.
Abstract
Dabigatran etexilate is a direct thrombin inhibitor and used widely as an anticoagulant for the prevention of stroke in patients with atrial fibrillation. However, anticoagulation therapy can be associated with an increased risk of bleeding. Here, we present data on the identification, humanization, and in vitro pharmacology of an antidote for dabigatran (aDabi-Fab). The X-ray crystal structure of dabigatran in complex with the antidote reveals many structural similarities of dabigatran recognition compared with thrombin. By a tighter network of interactions, the antidote achieves an affinity for dabigatran that is ∼350 times stronger than its affinity for thrombin. Despite the structural similarities in the mode of dabigatran binding, the antidote does not bind known thrombin substrates and has no activity in coagulation tests or platelet aggregation. In addition we demonstrate that the antidote rapidly reversed the anticoagulant activity of dabigatran in vivo in a rat model of anticoagulation. This is the first report of a specific antidote for a next-generation anticoagulant that may become a valuable tool in patients who require emergency procedures.
Introduction
Thromboembolic disorders such as myocardial infarction, stroke, and venous thromboembolism are the most common causes of mortality and morbidity in Western societies.1 These thromboembolic events can be triggered by excessive activation of coagulation, and thrombin plays a major role in these processes. The most widely used agents for antithrombotic therapy are indirect thrombin inhibitors (such as unfractionated and low-molecular-weight heparin) and vitamin K antagonists (VKAs) such as warfarin. Recently with the introduction of new oral anticoagulants on the market, new options for patients who require antithrombotic therapy are available. The first of these, dabigatran etexilate, is a novel, potent, nonpeptidic direct thrombin inhibitor.2 The orally administered double prodrug is hydrolized in vivo by esterases into the active form, dabigatran.3 It binds reversibly to the active site of both free and clot-bound thrombin, thus effectively preventing the formation of fibrin clots.4 Currently, dabigatran etexilate has been approved in more than 70 countries worldwide for the prophylaxis of thromboembolism in patients undergoing orthopedic surgery and for the prevention of stroke in patients with atrial fibrillation. In one study, treatment with dabigatran in patients with atrial fibrillation reduced the risk of stroke with a better safety profile compared with warfarin.5 In all indications, a fixed-dose regimen of dabigatran has provided effective anticoagulation with a favorable bleeding profile in patients without regular monitoring required.6
Despite these improvements in treatment with dabigatran, anticoagulation therapy is associated with an increased risk of bleeding. When VKAs are used, the anticoagulant effect in emergency situations is usually reversed with a combination of vitamin K, fresh-frozen plasma, prothrombin complex concentrate, or recombinant factor VIIa.7 Compared with VKAs, the new oral anticoagulants such as dabigatran and the factor Xa inhibitors rivaroxaban and apixaban have a shorter half life and shorter duration of anticoagulant effect.8 Although hemodialysis is recommended for the elimination of dabigatran9 and preclinical evidence suggests coagulation factor concentrates may also reverse bleeding,10,11 there is currently no specific antidote for dabigatran or for any of the other novel oral anticoagulants.
Antibodies and antibody fragments such as fragment antigen binding (Fab) can be used as antidotes for toxins and drugs. With Fabs, intoxications with digoxin12 and colchicine13 can be treated and the effect of desipramine can be reversed.14 The preclinical development of a single-chain fragment variable for the neutralization of methamphetamine15 and a monoclonal antibody to treat cocaine overdoses16 have been reported recently. Here we present data on the identification, humanization, and X-ray structure of the antibody fragment aDabi-Fab, which binds dabigatran and reverses its anticoagulant effects in vitro and in vivo.
Materials and methods
Antibody generation
A hapten (Figure 1A, compound 3), a derivative of dabigatran (Figure 1A, compound 2) was conjugated via a short linker to human immunoglobulin G (IgG) and hemocyanin as carrier proteins. These immunogens were used for immunization of 8 mice of different strains (Biozzi; NMRI × C57/Bl6). The animals were boosted 4 times during a period of 4 weeks before serum samples were taken and tested for binding to hapten 1 conjugated to bovine serum albumin by Luminex (LiquiChip; QIAGEN, Hilden Germany). After 3 weeks and additional booster immunizations, 2 mice with the greatest titers were sacrificed and their spleen cells isolated for fusion and hybridoma generation according to standard techniques.17 Hybridomas were selected in complete Dulbecco’s modified Eagle medium 10% fetal calf serum with 1× HAT supplement for 10-14 days. Hybridoma supernatants were screened for specific binding by Luminex and then subcloned by limiting dilution. Monoclonal antibodies were purified with the use of Mab Select (GE Healthcare, Munich, Germany) according to the manufacturer’s instructions.
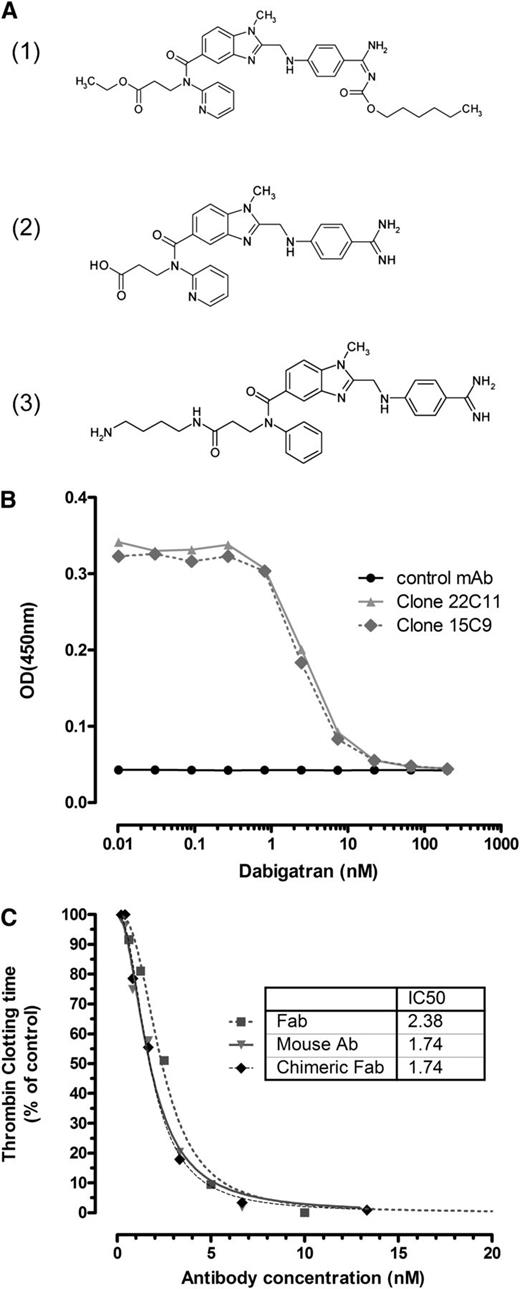
aDabi-Fab binds to dabigatran and reverses its effect in vitro. (A) The orally available double prodrug dabigatran etexilate (1) is converted into the active form dabigatran (2), which is able to bind reversibly to the catalytic site of thrombin. The hapten (3) was used as immunogen for antibody generation. (B) Immobilized mouse antibodies were analyzed for binding to a hapten-peroxidase conjugate, which was competed off by increasing concentrations of soluble dabigatran. As a representative example, 2 dabigatran-specific antibodies together with a negative control, mAb, are shown. Complete inhibition is observed at a dabigatran concentration >10nM. (C) The prolonged clotting time with addition of dabigatran (7nM) in vitro in a thrombin clotting assay was reversed in a concentration-dependent manner by the mouse antibody (clone 22), the chimeric Fab, and the humanized Fab, aDabi-Fab without a loss in potency. Data are represented as mean of 3 determinations.
aDabi-Fab binds to dabigatran and reverses its effect in vitro. (A) The orally available double prodrug dabigatran etexilate (1) is converted into the active form dabigatran (2), which is able to bind reversibly to the catalytic site of thrombin. The hapten (3) was used as immunogen for antibody generation. (B) Immobilized mouse antibodies were analyzed for binding to a hapten-peroxidase conjugate, which was competed off by increasing concentrations of soluble dabigatran. As a representative example, 2 dabigatran-specific antibodies together with a negative control, mAb, are shown. Complete inhibition is observed at a dabigatran concentration >10nM. (C) The prolonged clotting time with addition of dabigatran (7nM) in vitro in a thrombin clotting assay was reversed in a concentration-dependent manner by the mouse antibody (clone 22), the chimeric Fab, and the humanized Fab, aDabi-Fab without a loss in potency. Data are represented as mean of 3 determinations.
ELISA and affinity measurements
Specific binding of purified antibodies to dabigatran was analyzed in a competition enzyme-linked immunosorbent assay (ELISA), in which antibodies at a concentration of 1 µg/mL phosphate-buffered saline were immobilized onto microtiter plates (MaxiSorp; Nunc, Langenselbold, Germany). After blocking the wells with 3% bovine serum albumin in phosphate-buffered saline, hapten-peroxidase conjugate was added in the absence or presence of increasing concentrations of dabigatran and incubated for 1 hour. Bound peroxidase conjugate was detected with the use of One-step-Turbo reagent (Pierce, Rockford, IL) according to the manufacturer’s instructions. The affinities and binding kinetics of the antibodies and Fab molecules were determined with Kinexa technology (Sapidyne Instruments, Boise, ID). A constant concentration of Fab or antibody was incubated with various concentrations of dabigatran for 15 hours until equilibrium was reached. The concentration of free antibody was determined by capturing unbound Fab on a dabigatran-biotin conjugate coupled to NeutrAvidin beads. The captured mouse Fab was detected with an antimouse IgG (Fab-specific) F(ab′)2 fragment labeled with fluorescein isothiocyanate. The captured humanized Fab was detected with an antihuman IgG conjugated with Cy5. The dissociation constants were calculated by the use of a 1:1 binding model.
Humanization and production of Fabs
The variable regions from the heavy and light chains of the mouse antibodies were cloned and sequenced with the use of standard methods. The sequences were confirmed by protein analysis with mass spectrometry and N-terminal sequencing of the antibodies. DNA constructs encoding chimeric Fabs, comprising the specific mouse variable regions and human IgG constant regions, were generated to confirm the function of the lead antibodies to ensure that the correct v-region sequence had been obtained. The variable region of the Fab was then humanized through a design and screening process. Human framework sequences were selected for each of the mouse leads on the basis of the framework homology, complementarity determining region (CDR) structure, conserved canonical residues, conserved interface packing residues, and other parameters. A combinatorial framework library was made in instances in which human and murine residues were varied in such a way that in any given position there could be either a human or murine residue. Such a library was made for those amino acids that were different between human germline and murine antibody. The Fab library variants were expressed individually from an Escherichia coli expression system and screened via ELISA for binding to biotin-dabigatran. Only the Fabs that showed better or equal binding compared with the chimeric parent Fab were selected. Gene sequences encoding the humanized variable regions of the heavy and light chain were synthesized de novo on the basis of the amino acid sequences and subcloned into Fab expression format within pcDNA3.1 mammalian expression vectors. The Fab heavy and light chain constructs were cotransfected into CHO-DG44 cells, and the cleared culture supernatants were purified by the use of a two-step chromatographic process.
Thrombin time assay
Dabigatran (7nM final concentration) was added to pooled human plasma (3.2% sodium citrate, n = 10 volunteers). Bovine thrombin (0.4 U/mL; Siemens Healthcare Diagnostics, Marburg, Germany) was added to initiate clotting, and dabigatran prolonged clotting in this assay from 63 (0%) to 105 seconds (100%). Increasing concentrations of the antibodies or fragments were then added to plasma containing dabigatran and incubated for 5 minutes at 37°C. Clotting was performed on a CL4 coagulometer (Behnk Elektronik, Norderstedt, Germany). Time required to clot was measured and recorded and calculated at percentage of control to allow comparisons across different antibodies. Each assay was performed in triplicate.
SPR analysis
FV, FXIII, protein C, von Willebrand factor, S-2238, 10% pooled plasma (n = 6 individuals), dabigatran, and peptides representing known thrombin cleavage sites in FV, FVIII, FXIII, protein C, fibrinogen, and PAR1 were tested for binding to aDabi-Fab. The Fab was immobilized on a CM5 chip at pH 4 via EDC-NHS coupling. Surface plasmon resonance (SPR) analysis was performed in HBS-T buffer (10mM HEPES, 150mM NaCl, 0.05% Tween-20, pH 7.4) at 25°C with a constant flow rate of 30 µL/min. All thrombin substrates were injected at physiological concentrations (1µM for S-2238 and dabigatran) for 60 seconds followed by a dissociation time of 120 seconds.
Assays measuring thrombotic potential of aDabi-Fab
Three separate assays examining the thrombotic potential of aDabi-Fab were tested. (1) Increasing concentrations of aDabi-Fab were added to citrated prothrombin-depleted plasma (Hyphen BioMed, Neuville sur Oise, France) and incubated for 1 minute at 37°C. Clotting was determined after resupplementing with the minimal prothrombin concentration (12.5 µg/mL) required to restore clotting on a coagulometer (same as mentioned previously) after recalcification. Each clotting time was performed in triplicate. (2) Human-citrated plasma (n = 3) containing increasing concentrations of either aDabi-Fab or 0.1-5 pM human thrombin (Sigma-Aldrich) was recalcified and incubated for 120 minutes at 37°C under continued stirring. The reaction was stopped by adding ice-cold bentonite from the fibrinopeptide A (FPA) ELISA kit (Imunoclone FPA; American Diagnostica, Stamford, CT) to each well, and FPA generation was measured according to manufacturer’s instructions. (3) Increasing concentrations of aDabi-Fab or a coagulation factor concentrate Feiba (Baxter AG, Vienna, Austria) were added to plasma. Thrombin generation was performed by use of calibrated automated thrombinography with a Fluoroskan Ascent plate reader (Thermo Labsystems OY, Helsinki, Finland). Thrombin generation was initiated with the provided reagent containing 5pM tissue factor (Thrombinoscope BV, Maastricht, The Netherlands). Thrombin generation curves were calculated by use of the manufacturer’s software, from which lag time (minutes), peak thrombin concentration, and endogenous thrombin potential (ETP, area under the thrombin-time integral, nM × seconds) were calculated. The ETP reflects the total amount of thrombin generated as a function of time.
Platelet aggregation studies
We performed platelet aggregation in human platelet-rich plasma (PRP) by using light transmission aggregometry according to Born.18 Free-flowing whole blood (60 mL) was obtained from an antecubital vein via an 18-gauge needle from 3 healthy human volunteers. Blood was collected in tubes containing 3.13% sodium citrate (1 in 10 dilution with whole blood). Blood was centrifuged at 200g to obtain PRP. Samples (300 µL of PRP) were placed in a 6-channel aggregometer, equilibrated for 5 minutes at 37°C, and calibrated with platelet-poor plasma from same individual (0-1 Volts). Photometric tracings were digitally recorded over the course of 5 minutes after the addition of agonist, and curves were evaluated as area under the curve (AUC) over this time interval. Each PRP sample was incubated with 5mM CaCl2 and 3 mg/mL aDabi-Fab (final concentration). SFLLRN (1-5µM final concentration, LOXO [1mM], Dossenheim, Germany) was used as a positive control.
Reversal of anticoagulant activity in vivo
All experiments were performed with approval from the regional German animal ethics board. Male Wistar rats (∼300 g) were anesthetized with pentobarbital as a bolus (60 mg/kg intraperitoneally) and a continuous infusion for maintenance anesthesia (20 mg/kg/h intraperitoneally); internal body temperature was maintained with a heating pad. The carotid artery was isolated and cannulated to draw blood for sampling and the right jugular vein to administer the substances. Dabigatran (active substance) was infused as a 0.3-µmol/kg bolus + 0.1 µmol/kg/h infusion over the course of 50 minutes via the jugular vein; control animals received the respective vehicle in same volume and infusion rate. After the first 20 minutes of dabigatran infusion, aDabi-Fab (equimolar dose) was injected as a single bolus at t = 0. Blood samples were collected in sodium citrate solution (final concentration 0.313%). Neutralization of anticoagulant effects of intravenous dabigatran by the Fab in rats ex vivo was measured by activated partial thromboplastin time (aPTT) and thrombin time (TT) at serial time points (via carotid artery) after bolus injection.
Whole-blood TT (3 U/mL) was measured in a coagulometer CL4 (Behnk Elektronik) with the use of bovine thrombin reagent (Siemens). Blood (50 µL) was prewarmed to 37°C, 100 µL of thrombin reagent was added, and the time to the onset of blood clotting was determined in seconds. Whole-blood aPTT was measured with PTT reagent (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. Blood (50 µL) was prewarmed to 37°C, and 50 µL of PTT reagent was added, followed by calcium chloride (50 µL of 0.025 mol/L; Siemens) after a 3-minute incubation. The time to the onset of blood clotting was determined in seconds.
Protein crystallography
Detailed information on complex formation, crystallization, and crystallography is compiled in the supplemental data; see the Blood Web site.
Results
Antibody screening and engineering
Mice were immunized with dabigatran-derived haptens coupled to carrier proteins for the generation of dabigatran-specific monoclonal antibodies. Isolated antibodies were screened for binding to the immunogen and for specific binding to soluble dabigatran in a competitive ELISA (Figure 1B). Antibodies showed binding to a hapten-peroxidase conjugate and could be competed off by increasing the concentrations of dabigatran. To minimize the immunogenicity, a combinatorial library of humanized Fabs was constructed and potent Fabs were selected by binding affinity.
The functional inhibition of dabigatran activity was tested in a modified TT assay. Clotting time was prolonged from 60 to 100 seconds in the presence of 7nM dabigatran. When the dabigatran-specific antibodies were added in increasing concentrations, the anticoagulant activity of dabigatran was reversed in a concentration-dependent manner (Figure 1C). The parental mouse antibodies, mouse Fabs, and humanized Fab all showed similar inhibitory activity with half maximal inhibitory concentration (ie, IC50) values between 2 and 4nM. Clotting times were reduced to baseline at antibody concentrations >10nM, indicating complete neutralization of dabigatran at equimolar concentrations of dabigatran and aDabi-Fab. The antibody alone in the absence of dabigatran had no effect on the coagulation time. Similarly, a negative control Fab, not specific for dabigatran, did not reverse the anticoagulant effect of dabigatran.
The affinities of mouse and humanized Fabs for dabigatran were measured in solution. The parental mouse Fab had a binding affinity (KD) of 32pM, this was improved with the humanized aDabi-Fab, which displayed increased binding with a KD of 2.1pM (Table 1). The high affinity of aDabi-Fab correlated with an extremely slow off-rate of ∼0.7 × 10−6 s−1, almost 200-fold lower compared with the parental mouse Fab. To better understand the molecular mechanism of the dabigatran-specific Fab, structural studies were performed.
Affinities of parental mouse Fab 15c9 and humanized aDabi-Fab
| . | Format . | KD, pM . | ka, ×105 M−1s−1 . | kd, ×10−6 s−1 . |
|---|---|---|---|---|
| aDabi-Fab | Humanized | 2.1 ± 0.6 | 3.4 ± 0.4 | 0.7 ± 0.2 |
| Fab 15c9 | Mouse | 32 ± 11 | 42 ± 16 | 134 ± 69 |
| . | Format . | KD, pM . | ka, ×105 M−1s−1 . | kd, ×10−6 s−1 . |
|---|---|---|---|---|
| aDabi-Fab | Humanized | 2.1 ± 0.6 | 3.4 ± 0.4 | 0.7 ± 0.2 |
| Fab 15c9 | Mouse | 32 ± 11 | 42 ± 16 | 134 ± 69 |
ka, association rate constant; KD, binding affinity; kd, dissociation rate constant.
KD, ka, and calculated kd are shown mean ± SD of at least 3 individual measurements.
Overall structure of the complex
The crystal structure of aDabi-Fab in complex with dabigatran was solved at 1.7 Å resolution. Two crystallographically independent Fab:dabigatran complexes are located in each asymmetric unit of the crystal. The Fab is composed of two variable domains (VLκ and VH) and two constant domains (CLκ and CHIgG1). These domains show the typical immunoglobulin fold.19 In the sections to follow, Fab chains H + L and A + B and will be named Fab 1 and Fab 2, respectively, and the residue numbering used is according to Johnson and Wu.20 A detailed description of overall structural features can be found in the supplemental materials. The description of structural features refers to Fab 1 because this is the protomer that is completely defined by electron density. If the respective values differ significantly for Fab 2, this will be mentioned explicitly.
Dabigatran binding site
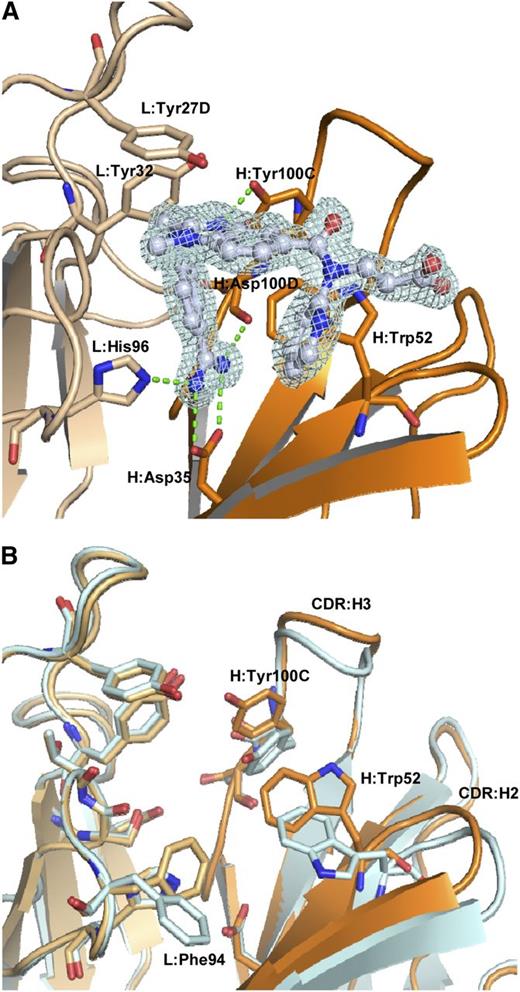
The dabigatran binding site of the aDabi-Fab is located in a concave region at the interface of the variable domains (Figure 2A). The tendency to form concave paratopes has been reported for antibodies recognizing small antigens and haptens.21 All CDR loops except L2 are involved in dabigatran binding. The binding is mediated by hydrophobic interactions, H-bonds, and a salt bridge. The benzamidine moiety of dabigatran inserts into a cavity formed by heavy and light chain, whereas the benzimidazole, carboxamide, and pyridine moieties are partially exposed to solvent. The propionic acid moiety of the hapten is almost completely exposed to solvent. The most prominent interaction is conveyed via the benzamidine moiety of dabigatran. It extends deeply into a cavity that lies at the interface of heavy and light chain. The amidine group of dabigatran forms a bidentate salt bridge to H:Asp35 in CDR:H1 with nitrogen-oxygen distances of 2.9 Å and 3.0 Å, respectively. This interaction is additionally stabilized: one of the nitrogen atoms forms a hydrogen bond to Nε of L:His96 (2.9 Å) in CDR:L3; the other acts as a hydrogen donor for the backbone carbonyl oxygen of H:Asp100D (3.2 Å) in CDR:H3. These polar interactions are expected to contribute strongly to the free energy of binding because they reside in an otherwise-hydrophobic environment provided by the side chains of L:Phe94, H:Ile33, H:Val50, and H:Phe100F, which provide efficient solvent shielding. An additional hydrogen bond is formed between H:Tyr100C, located in CDR:H3, and the aldimine nitrogen of the benzimidazole moiety of dabigatran. Here the hydroxyl group of H:Tyr100C acts as hydrogen donor with an oxygen-nitrogen distance of 2.65 Å and a NHO-angle of 160°, indicating a fairly strong hydrogen bond.22 H:Trp52 in CDR:H2 and L:Tyr27D contribute to the binding by aromatic π-interactions with the benzamidine ring, the pyridine ring of the hapten and the benzimidazole moiety, respectively. L:Tyr32 displays a N-H/π interaction with the amine nitrogen of dabigatran. In this nonclassical hydrogen bond, the π-electrons of the phenyl ring act as H-bond acceptor.23
Binding mode of aDabi-Fab and conformational differences between apo and holo structure. (A) Binding site of aDabi-Fab in complex with dabigatran (the respective electron density of dabigatran is shown in pale cyan, contoured at 1.5 sigma). The benzamidine group of dabigatran (carbon atoms colored in gray) extends into a cavity formed by the interface of light (light ochre) and heavy chain (dark ocre). It forms a bidentate salt bridge to H:Asp35 and additional hydrogen bonds to L:His96 and H:Asp100D, respectively (indicated as green dotted lines). The amine nitrogen of dabigatran forms a nonclassical H-bond to L:Tyr32. L:Tyr27D interacts with the benzimidazole moiety of dabigatran via parallel displaced π-stacking. H:Tyr100C forms a hydrogen bond to the aldimine nitrogen. H:Trp52 forms two T-shaped aromatic interactions with the benzamidine and pyridine moiety of dabigatran, respectively. (B) Superposition of the binding site of aDabi-Fab in its apo conformation and bound to dabigatran (color scheme as in Figure 4, the apo structure is colored in pale cyan). The orientation of the sidechain of H:Tyr100C changes by 3.2 Å (hydroxyl oxygen) upon binding of dabigatran, which causes the apex of CDR:H3 to increase its relative distance to the binding site by 2.1 Å (measured from the Cα of H:Tyr100). About the same distance CDR:H2 moves toward the binding site (measured from the Cα of H:Gly55) and the H:Trp52 and L:Phe94 adopt different side chain conformations. This concerted movement opens the binding site of dabigatran.
Binding mode of aDabi-Fab and conformational differences between apo and holo structure. (A) Binding site of aDabi-Fab in complex with dabigatran (the respective electron density of dabigatran is shown in pale cyan, contoured at 1.5 sigma). The benzamidine group of dabigatran (carbon atoms colored in gray) extends into a cavity formed by the interface of light (light ochre) and heavy chain (dark ocre). It forms a bidentate salt bridge to H:Asp35 and additional hydrogen bonds to L:His96 and H:Asp100D, respectively (indicated as green dotted lines). The amine nitrogen of dabigatran forms a nonclassical H-bond to L:Tyr32. L:Tyr27D interacts with the benzimidazole moiety of dabigatran via parallel displaced π-stacking. H:Tyr100C forms a hydrogen bond to the aldimine nitrogen. H:Trp52 forms two T-shaped aromatic interactions with the benzamidine and pyridine moiety of dabigatran, respectively. (B) Superposition of the binding site of aDabi-Fab in its apo conformation and bound to dabigatran (color scheme as in Figure 4, the apo structure is colored in pale cyan). The orientation of the sidechain of H:Tyr100C changes by 3.2 Å (hydroxyl oxygen) upon binding of dabigatran, which causes the apex of CDR:H3 to increase its relative distance to the binding site by 2.1 Å (measured from the Cα of H:Tyr100). About the same distance CDR:H2 moves toward the binding site (measured from the Cα of H:Gly55) and the H:Trp52 and L:Phe94 adopt different side chain conformations. This concerted movement opens the binding site of dabigatran.
Conformational differences between apo and holo structure
The crystal structure of the uncomplexed Fab was solved at 1.9 Å resolution with one protomer in the asymmetric unit. The CDR loops adopt similar conformations as observed in the complex structure. Noteworthy, the binding site for dabigatran is occupied by two glycerol molecules. Because these originate from the cryo buffer, one can assume the binding site to be occupied by water molecules under physiological conditions. The comparison of the free Fab with the dabigatran complex reveals no major conformational change of the protein backbone at the binding site of the hapten but some major side chain reorientations (Figure 2B). The side chain orientations of H:Tyr100C, H:Trp52, and L:Phe94 occlude the dabigatran pocket in the apo form. In the apo structure, H:Trp52 is involved in a crystal contact. However, we also observed this conformer in a costructure of aDabi-Fab with benzamidine that crystallized in a different spacegroup (data not shown). Thus, the apo structure seems to be a preferred low-energy conformation of the Fab. In molecular dynamics simulations (data not shown) we observed both apo and bound conformations of the aDabi-Fab and conclude that both are prevalent conformers in solution.
Binding to thrombin substrates
The binding pattern of aDabi-Fab displays structural similarities to thrombin regarding its interactions with dabigatran. Therefore, binding of aDabi-Fab to thrombin substrates was analyzed by SPR (Figure 3A). None of the tested substrates, including S-2238, factors V, VIII, XIII, fibrinogen, von Willebrand factor, protease-activated receptor-1 (PAR-1), or protein C, bound to aDabi-Fab at physiological concentrations.
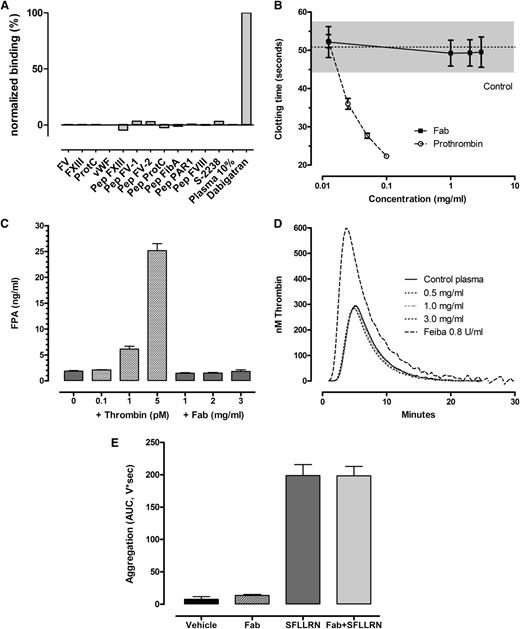
aDabi-Fab does not induce clotting activity (A) Lack of binding of thrombin substrates to aDabi-Fab in SPR. (B) Lack of thrombotic activity of aDabi-Fab in prothrombin-deficient plasma when supplemented with small amounts of prothrombin. Data are expressed as mean ± SD, n = 3. (C) Lack of effect of Fab on fibrinogen conversion to fibrin measured as FPA generation in plasma in vitro. Data shown as mean of triplicate determinations. (D) Lack of effect of aDabi-Fab to induce further thrombin generation in plasma, in contrast to Feiba, which elevated thrombin generation. Data mean of 5 determinations. (E) Effect of aDabi-Fab (3 mg/mL) and SFLLRN (2µM) or a combination on platelet aggregation in human platelet rich plasma. Data expressed as mean ± SD, n = 3.
aDabi-Fab does not induce clotting activity (A) Lack of binding of thrombin substrates to aDabi-Fab in SPR. (B) Lack of thrombotic activity of aDabi-Fab in prothrombin-deficient plasma when supplemented with small amounts of prothrombin. Data are expressed as mean ± SD, n = 3. (C) Lack of effect of Fab on fibrinogen conversion to fibrin measured as FPA generation in plasma in vitro. Data shown as mean of triplicate determinations. (D) Lack of effect of aDabi-Fab to induce further thrombin generation in plasma, in contrast to Feiba, which elevated thrombin generation. Data mean of 5 determinations. (E) Effect of aDabi-Fab (3 mg/mL) and SFLLRN (2µM) or a combination on platelet aggregation in human platelet rich plasma. Data expressed as mean ± SD, n = 3.
Functional clotting assays
Several assays were performed to test whether the structural similarities of aDabi-Fab and thrombin result in thrombin-like activity in plasma. When undiluted normal plasma (containing ∼100 µg/mL prothrombin) was added to prothrombin-depleted plasma, the clotting time was 22.3 ± 0.8 seconds upon recalcification. The minimal dilution of normal plasma to induce clotting upon recalcification was ∼12.5 µg/mL (1:8 dilution), which resulted in an approximate doubling of the clotting time to 51.9 ± 5.3 seconds. Adding concentrations of aDabi-Fab up to 3 mg/mL with 12.5 µg/mL prothrombin did not shorten the clotting time, demonstrating the lack of procoagulant activity of the Fab (Figure 3B). In further studies we examined the ability of aDabi-Fab to convert fibrinogen to fibrin by measuring the release of FPA. Figure 3C illustrates that there was no increase in FPA when aDabi-Fab was incubated with recalcified plasma; in contrast, adding small amounts of thrombin (0.1-5 pM) resulted in increased FPA levels in recalcified plasma. In addition, when the Fab was added to plasma (3 mg/mL) there was no increased thrombin generation as indicated by ETP. In contrast, when Feiba was added in increasing concentrations, there was an elevation in peak thrombin generation and the ETP (Figure 3D).
Platelet aggregation studies
Potential effects of aDabi-Fab on platelet activation were tested by adding 3 mg/mL Fab to recalcified PRP as a potential agonist to stimulate platelet aggregation via PAR-1. SFLLRN (1-5µM final concentration) was used as a positive control. Platelet aggregation over 5 minutes (AUC) increased with increasing concentrations of SFLLRN, and 2µM was the minimum concentration to achieve maximum aggregation (198.9 ± 29.1, AUC, n = 3, Figure 3E). The addition of aDabi-Fab alone had no effect on platelet aggregation; similarly, incubating aDabi-Fab with platelets before the addition of SFLLRN did not enhance or decrease the platelet aggregation response as compared with SFLLRN alone.
In vivo reversal of anticoagulation
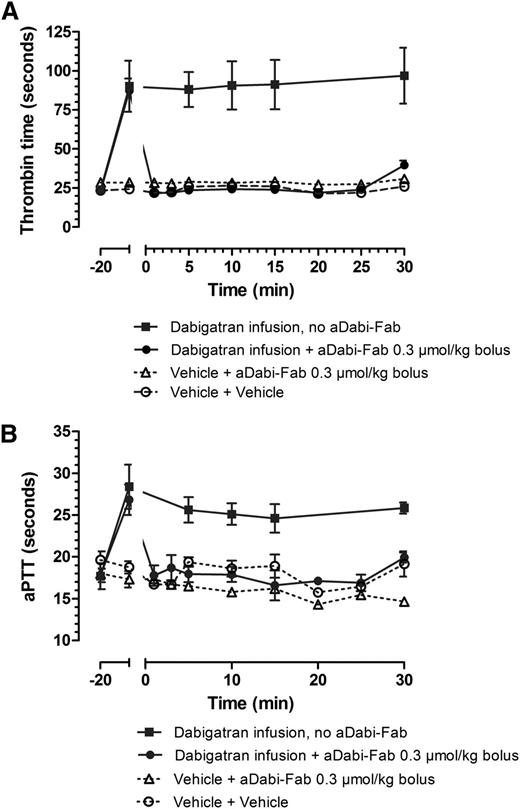
After the first 20 minutes of infusion in rats, dabigatran was present in plasma levels of ∼200 ng/mL, and the infusion was continued during the entire subsequent 30 minutes to maintain these levels, both in control and aDabi-Fab treatment groups. Dabigatran alone (squares) prolonged TT ∼4-fold over control (Figure 4A) and aPTT ∼2-fold over control (Figure 4B). The addition of a single bolus injection of aDabi-Fab completely reversed the prolonged anticoagulant activity within 1 minute of injection, and this reversal was maintained during the course of ∼25 minutes despite continual infusion of further dabigatran. A slight elevation of anticoagulation reappearing at 25 minutes after the single bolus of aDabi-Fab probably represents saturation of aDabi-Fab.
aDabi-Fab reverses the anticoagulative effects of dabigatran in vivo. The effect of a continual infusion of dabigatran (squares, solid line) given as a bolus loading dose (0.3 µmol/kg) and continual infusion (0.1 µmol/kg/h). aDabi-Fab (circles, solid line, 0.3 µmol/kg) was given as a single bolus at t = 0. Thrombin time (A) and aPTT (B) were measured at different time points over 30 minutes after the injection of aDabi-Fab. Data represented as mean ± SE, n = 4-6 per group.
aDabi-Fab reverses the anticoagulative effects of dabigatran in vivo. The effect of a continual infusion of dabigatran (squares, solid line) given as a bolus loading dose (0.3 µmol/kg) and continual infusion (0.1 µmol/kg/h). aDabi-Fab (circles, solid line, 0.3 µmol/kg) was given as a single bolus at t = 0. Thrombin time (A) and aPTT (B) were measured at different time points over 30 minutes after the injection of aDabi-Fab. Data represented as mean ± SE, n = 4-6 per group.
Discussion
To generate a specific neutralizing reagent for dabigatran, we generated a humanized Fab and studied its pharmacological activity in vitro and in vivo. On the basis of the crystal structure of the Fab in complex with dabigatran, the molecular binding mechanism was analyzed in detail and compared with the dabigatran thrombin interaction.
Crystal structure explains high affinity
The extraordinarily high affinity of 2.1pM of aDabi-Fab for dabigatran, which is ∼350 times greater than for thrombin, can be qualitatively explained by the structural data. Both dabigatran and aDabi-Fab adopt low-energy conformations in the complex, as inferred from statistical conformer analysis of structures with motifs homologous to dabigatran in the CSD (ie, Cambridge structural database) and molecular dynamics simulation of the Fab, respectively. Molecular dynamics simulations suggested the existence of both crystallographically observed aDabi-Fab conformations (apo and holo) in solution. The binding thus most probably follows a “conformational selection” mechanism as also reported for thrombin.24 A tight network of interactions, including numerous hydrogen bonds to all buried polar atoms of dabigatran, contributes to the high binding affinity.
aDabi-Fab mimics structural features of thrombin
When we compared the structure of thrombin:dabigatran complex2 with the structure we present here, it becomes obvious that dabigatran adopts distinct bound conformations. When superimposing the benzimidazole scaffold of dabigatran, we found that the carboxamide and carboxylate group as well as the pyridine ring are very similarly positioned (root-mean-square deviation 0.35 Å), whereas the benzamidine moiety is twisted by 66° because of a difference in torsion angles around the exocyclic bond at the benzimidazole.
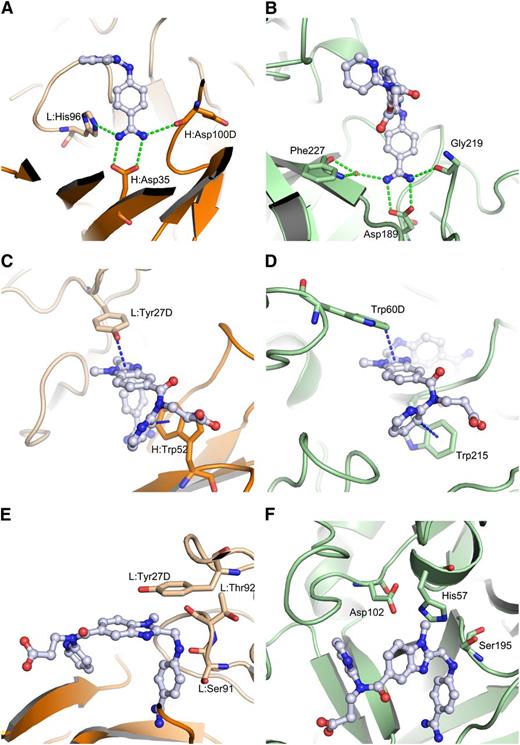
Despite the differences in bound conformations, the way thrombin and aDabi-Fab bind dabigatran are strikingly similar (Figure 5). Most notably, a salt bridge anchors the benzamidine moiety of dabigatran in a hydrophobic cavity in both proteins. In the thrombin structure, this salt bridge is formed by Asp189 in the S1-pocket and additional hydrogen bonds are formed by the backbone carbonyl oxygen of Gly219 and a water molecule with the amidine. In the Fab, a salt bridge is formed by H:Asp35, and a similar hydrogen bonding pattern is formed by the ε-nitrogen of L:His96 and the backbone carbonyl oxygen of H:Asp100D. When both structures are superimposed on the benzimidazole moiety of dabigatran, additional similarities become obvious. Thrombin interacts with the benzimidazole moiety of dabigatran via parallel displaced π-stacking with Trp60D (3.5 Å distance of closest atoms) in the S2-pocket, whereas the Fab uses L:Tyr27D for the same purpose. In both crystal structures the pyridine ring of dabigatran displays a T-shaped π-interaction with a tryptophane. In the S4-pocket of thrombin this is Trp215 (3.3 Å distance), in aDabi-Fab it is H:Trp52. Notably, the indole moieties occupy the same space, but they are orientationally inverted due to the position of the main chains from which the side chains protrude. In the thrombin structure the tryptophane protrudes from a strand that is close to the benzamidine group, whereas in the Fab it is positioned in a strand that is closer to the propionic acid moiety of dabigatran. Interestingly, the N-methyl group on the benzimidazole scaffold that forms hydrophobic interactions within the S2-pocket of thrombin is not involved in binding to the Fab.
Comparison of binding modes of dabigatran to Fab and thrombin. aDabi-Fab mimics structural features of thrombin to bind dabigatran (same color scheme as in Figure 3, thrombin is colored in pale green, H-bonds and aromatic interactions are indicated as dotted lines in green and blue, respectively). The dabigatran complex structure of aDabi-Fab and thrombin is displayed in the left and right panels, respectively. (A-B) The benzamidine moiety of dabigatran binds into a cavity of the proteins and forms a bidentate salt bridge to H:Asp35 and Asp189, respectively. The salt bridge is further stabilized by a conserved hydrogen bonding pattern with L:His96 and H:Asp100D or with a water molecule and Gly219. (C-D) The benzimidazole moiety of dabigatran interacts by π-stacking with L:Tyr27D and Trp60D, respectively. The pyridine ring of dabigatran interacts with a T-shaped π-stacking with H:Trp52 and Trp215, respectively. (E-F) The catalytic triad of thrombin is formed by His57, Asp102, and Ser195. The Fab contains in the same region an arrangement of L:Tyr27D, L:Ser91, and L:Thr92 that is not able to convey proteolytic activity.
Comparison of binding modes of dabigatran to Fab and thrombin. aDabi-Fab mimics structural features of thrombin to bind dabigatran (same color scheme as in Figure 3, thrombin is colored in pale green, H-bonds and aromatic interactions are indicated as dotted lines in green and blue, respectively). The dabigatran complex structure of aDabi-Fab and thrombin is displayed in the left and right panels, respectively. (A-B) The benzamidine moiety of dabigatran binds into a cavity of the proteins and forms a bidentate salt bridge to H:Asp35 and Asp189, respectively. The salt bridge is further stabilized by a conserved hydrogen bonding pattern with L:His96 and H:Asp100D or with a water molecule and Gly219. (C-D) The benzimidazole moiety of dabigatran interacts by π-stacking with L:Tyr27D and Trp60D, respectively. The pyridine ring of dabigatran interacts with a T-shaped π-stacking with H:Trp52 and Trp215, respectively. (E-F) The catalytic triad of thrombin is formed by His57, Asp102, and Ser195. The Fab contains in the same region an arrangement of L:Tyr27D, L:Ser91, and L:Thr92 that is not able to convey proteolytic activity.
aDabi-Fab does not functionally mimic thrombin
The structural similarities of aDabi-Fab to thrombin and the potent binding to dabigatran may suggest possible thrombin-like enzymatic activity of the antidote. However, inspection of the binding site of aDabi-Fab unsurprisingly shows the absence of a thrombin-like catalytic triad. Thrombin is a pleiotropic molecule with multiple potential substrates. We did not observe binding for a variety of thrombin substrates to the Fab in SPR, including factors V, VIII, XIII, fibrinogen, von Willebrand factor, PAR-1, protein C, or the thrombin-specific substrate S-2238. In addition, we demonstrate that the Fab alone does not affect clotting in a diluted thrombin time assay. It does not shorten the clotting time in a system in which prothrombin was limited and aDabi-Fab was the only protein that could potentially exert any thrombin-like activity. It also does not convert the main substrate of thrombin, fibrinogen, into fibrin as measured by FPA release and it does not mimic or enhance the thrombin-mediated feedback of coagulation, resulting in elevated thrombin generation. It does not interfere with PAR-1 activation by SFLRRN or activate platelet aggregation alone. These functional results, taken together, therefore demonstrate that although there are similarities in the molecular recognition of dabigatran between aDabi-Fab and thrombin, these do not result in any prothrombotic activity of the Fab.
The aDabi-Fab demonstrates potent and immediate reversal of dabigatran anticoagulant activity as measured by TT and aPTT in rats ex vivo. Steady-state dabigatran levels of ∼200 ng/mL were neutralized within 1 minute of the injection of an intravenous bolus of aDabi-Fab. This neutralization was maintained over the course of 25 minutes despite the continual infusion of further dabigatran over this time, thus demonstrating the effective in vivo inhibition of dabigatran anticoagulant activity by aDabi-Fab in equimolar doses.
In conclusion, aDabi-Fab displays major interactions with dabigatran that are also present in its costructure with thrombin. Thus, aDabi-Fab mimics thrombin in its molecular recognition and binding of dabigatran. However, aDabi-Fab does not exhibit specific thrombin-like enzymatic activity, but rather was neutral in all coagulation assays. Additional hydrogen bonding and hydrophobic interactions allow for binding of dabigatran with a very high affinity of 2.1 pM, that is ∼350 times stronger than the 700 pM affinity of thrombin.25 On the basis of its extremely high affinity aDabi-Fab is able to neutralize dabigatran activity rapidly and completely. To our knowledge, this is the first report on a specific antidote for a next-generation anticoagulant. Further development of this antidote is currently ongoing and if successfully translated into a clinical product, this antibody fragment will be a valuable tool in treating bleeding in life-threatening, emergency situations.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Anita Bloching, Michael Ritter, Nikolai Roosz, Johanna Schurer, and Monika Kink-Eiband for technical assistance. Sanjaya Singh and Alisa Waterman were instrumental advisors and coordinators of antidote generation and humanization activities.
Authorship
Contribution: F.S. and K.C. designed and performed research and wrote the manuscript; J.v.R. and T.L. designed research and wrote the manuscript; J.P. and H.N. designed research; and C.N. and E.S. performed research.
Conflict-of-interest disclosure: All authors are employees of Boehringer Ingelheim.
Correspondence: Tobias Litzenburger, New Biological Entity Discovery, Boehringer Ingelheim GmbH & Co. KG, Birkendorfer Straße 65, 88397 Biberach, Germany; e-mail: tobias.litzenburger@boehringer-ingelheim.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal