Key Points
Hdac1 and, to a lesser extent, Hdac2 behave as oncosuppressors during tumor initiation, but they work as oncogenes in tumor maintenance.
Class I HDAC inhibitors (VPA) accelerate tumorigenesis in murine models of leukemia, which suggests caution in their clinical use.
Abstract
Aberrant recruitment of histone deacetylases (HDACs) by the oncogenic fusion protein PML-RAR is involved in the pathogenesis of acute promyelocytic leukemia (APL). PML-RAR, however, is not sufficient to induce disease in mice but requires additional oncogenic lesions during the preleukemic phase. Here, we show that knock-down of Hdac1 and Hdac2 dramatically accelerates leukemogenesis in transgenic preleukemic mice. These events are not restricted to APL because lymphomagenesis driven by deletion of p53 or, to a lesser extent, by c-myc overexpression, was also accelerated by Hdac1 knock-down. In the preleukemic phase of APL, Hdac1 counteracts the activity of PML-RAR in (1) blocking differentiation; (2) impairing genomic stability; and (3) increasing self-renewal in hematopoietic progenitors, as all of these events are affected by the reduction in Hdac1 levels. This led to an expansion of a subpopulation of PML-RAR–expressing cells that is the major source of leukemic stem cells in the full leukemic stage. Remarkably, short-term treatment of preleukemic mice with an HDAC inhibitor accelerated leukemogenesis. In contrast, knock-down of Hdac1 in APL mice led to enhanced survival duration of the leukemic animals. Thus, Hdac1 has a dual role in tumorigenesis: oncosuppressive in the early stages, and oncogenic in established tumor cells.
Introduction
Epigenetic alterations are considered as relevant as genetic mutations in tumorigenesis.1 Small molecules that inhibit the activity of epigenetic enzymes have the potential to revert those changes and have entered clinical use, validating the concept of epigenetic therapy.2 As an example, histone deacetylase inhibitors (HDACi) that target multiple HDACs with nonredundant functional roles in normal cells and tumor cells3,4 are effective for the treatment of cutaneous T-cell lymphoma.2 Epigenetic drugs, however, have single-agent efficacy only in selected hematologic malignancies, underlying the need for optimized drugs and treatment strategies.2 Initial genetic dissection of the role of specific HDACs in murine models has revealed that inactivation of Hdac1 and Hdac2 results in hematopoietic defects, and gene deletion in established tumor cells results in loss of proliferation and/or decreased survival duration.5,6 Indeed, the development of class I (HDACs 1, 2, 3, and 8)–selective HDACi such as the US Food and Drug Administration–approved compound romidepsin, supports the proposed pro-oncogenic role of class I HDACs.7,8
APL is caused by the fusion protein promyelocytic leukemia-retinoic acid receptor (PML-RAR),9 and expression of PML-RAR in murine hematopoietic progenitors leads to APL that reflects the human disease.10 Before full leukemic transformation, PML-RAR induces a “preleukemic” stage without an overtly dramatic phenotype that leads to substantial alterations in the self-renewing properties of the preleukemic cells.11,12 Therefore, it is assumed that PML-RAR is necessary but not sufficient to cause APL, and additional genetic hits, largely unknown, must occur for the clonal expansion of leukemic blasts.
Different HDAC-containing complexes associate with PML-RAR. These complexes (such as NURD and SMRT/NCoR) include class I HDACs.13 Through its association with HDACs, PML-RAR is able to transcriptionally silence its target genes and regulate the stability of the oncosuppressor p53.14 In view of these observations, we functionally assessed the role of individual HDACs, in this case Hdac1/2, in the development and progression of APL.
Methods
Mice
Transgenic mCGPR/PR mice were generously provided by T.J. Ley (Washington University, St Louis, MO) and backcrossed into the C57BL/6J strain. C57BL/6J p53−/− mice were obtained from the Jackson Laboratory. Eμ-myc transgenic mice were provided by Dr Jerry Adams (Walter and Eliza Hall Institute, Melbourne, Australia).15 For lineage-negative (lin−) transplantation experiments, lethally irradiated C57BL/6J mice, 12 to 14 weeks old, were inoculated intravenously with 300 000 green fluorescence protein (GFP+)–transduced lin– cells, together with 500 000 spleen cells obtained from a wild-type (WT) mouse.16 For leukemia transplantation, GFP+-transduced leukemic cells were injected intravenously (200 000 cells/mouse) in syngeneic recipient mice. APL leukemias used for Hdac1 knockdown experiments shown in Figure 5 have been described previously.16 For the generation of Eμ-myc lymphomas knocked down for Hdac1 and Hdac2, fetal livers were harvested from E13.5-E15.5 Eμ-myc embryos (in the C57BL/6/CD45.2 strain) and were transduced with pLMSshRNAmiR30.SCR, pLMSshRNAmiR30.Hdac1 or pLMSshRNAmiR30Hdac2 constructs as detailed previously.17,18 Irradiated recipient PTPRCA/CD45.1 mice19 were injected with unsorted fetal liver cells (1 × 106 GFP+/mouse) and were euthanized once lymphoma developed, at which point the lymph nodes, spleen, and thymus were harvested and were stored in liquid nitrogen for further analysis. Animals were checked periodically for clinical signs of disease (by inspection, and periodic blood analysis using the AcT™ 5diff Beckman Coulter, Brea, CA). Long-term reconstitution of the hematopoietic compartment was assessed by scoring the percentage of GFP+ cells in peripheral blood at least 4 months after transplantation. Mice were euthanized by CO2 inhalation when they became detectably ill. The survival rate was calculated using the Kaplan-Meier method. All procedures involving animals were done in accordance with national and international laws and policies.
Purification of lin− cells and infection procedures
Lin− cells from WT C57BL/6J or transgenic mCGPR/PR mice and APL blasts from leukemic animals were obtained and maintained as previously described.16 Cells were infected using the viral supernatants (see below) and then were sorted by GFP positivity (FACSAria, Becton Dickinson, Franklin Lakes, NJ16 ).
Eμ-myc fetal liver cells were grown in high-glucose Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, penicillin (100 U/mL)/streptomycin (100 mg/mL), 0.1 mM of L-asparagine, 50 mM of 2-mercaptoethanol, recombinant IL-3 (2ng/mL), IL-6 (2ng/mL), and stem cell factor (SCF; 10 ng/mL). All cytokines came from PeproTech.
Retroviral shRNA constructs and virus production
ShRNA-based retroviral plasmids (used for the experiments with preleukemic, leukemic, and p53 null cells) were generated by ligating synthetic oligonucleotides targeting the indicated mRNAs into a modified pRETRO-SUPER vector20 in which the cDNA for puromycin selection had been replaced with that encoding for enhanced GFP. The shRNA sequences (supplemental Table 4) are in the format: target sequence sense (underlined)–loop-target sequence antisense (underlined).
The pRETRO-SUPER vectors and package plasmid pCL-Eco were cotransfected into packaging ecotropic Phoenix cells, and the viral supernatants were produced as previously described.16
Targeting sequences for shRNA-miR30 constructs (used for the experiments with Eµ-myc cells) were identified using the previously described DSIR algorithm.21 The top-ranked short hairpin RNAs (shRNAs) were used to create 10 miR30 sequences (97mer, Sigma-Aldrich) and these were cloned into the pLMS vector after generation of approximately 110bp shRNA-miR30’s by amplification of 97mers using 5′mir30-XhoI (CAGAAGGCTCGAGAAGGTATATTGCTGTTGACAGTGAGCG) and 5′miR30-EcoRI (CTAAAGTAGCCCCTTGAATTCCGAGGCAGTAGGCA) primers. The miRNA oligomer sequences (supplemental Table 4) are in the format: mir-30 context-sense (underlined)–loop-antisense (underlined). Viral supernatants were produced as previously described.17,18 Knock-downregulation of gene expression can be associated with experimental artifacts, because of off-target effects.22 We can exclude that off-target effects confound the interpretation of our results, for the following reasons: (1) The use of different sequences to interfere with Hdac1 expression, both in vivo and in vitro, gave identical results. (2) The in vitro Hdac1 knock-down phenotype (potentiation of differentiation block and enhancement of colony-forming ability) was rescued by re-expressing a noninterferable Hdac1 protein in lin− cells. (3) The in vivo pharmacologic inhibition of HDACs recapitulates the observed Hdac1 knock-down phenotype.
Cell lines
Human APL NB4 cells were grown in RPMI supplemented with 10% fetal bovine serum, 2 mM of L-glutamine, and antibiotics. Phoenix cells were purchased from ATCC and were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM of L-glutamine, and antibiotics. Cultures were maintained in a humidified tissue culture incubator at 37°C in 5% CO2.
Immunoblots and antibodies
Whole-cell extracts were obtained by lysis in sodium dodecyl sulfate (SDS) buffer (50 mM of Tris HCl, 10% glycerol, 2% SDS). Proteins were separated by SDS-polyacrylamide gel electrophoresis, blotted onto polyvinylidene difluoride membrane, and probed with the indicated antibodies. For determination of p53 status of Eμ-myc lymphomas, 1.5 × 106 Fluorescence-activated cell-sorting (FACS) GFP+ tumor cells were exposed to 10 Gy of irradiation and incubated at 37°C in 10% CO2 for 1 hour alongside nonirradiated controls and then were lysed. The antibodies used in the study were anti-Hdac1 (ab7028), anti-Hdac2, (ab7029), anti-HDAC3 (ab7030), anti-H4-Ac K5 (ab51997), and anti-3H (ab1791) (Abcam, Cambridge, UK); anti-RARα (sc-551) (Biotechnology Inc, Santa Cruz, CA); anti-acetylated histone 3H and H4 were detected using a homemade antibody23 ; anti-p53 (Novacastra, NCL-p53-505), anti-p19ARF (Santa Cruz sc32748), anti-Hsp90 (Stressgen ADI-SPA-830), and anti-β-actin (Sigma Aldrich).
Differentiation, serial replating, and cytogenetic assays
A total of 5000 sorted lin− cells were plated in methylcellulose medium (MethoCult SF M3236, Stem Cell Technology, Vancouver, BC, Canada) containing 15% fetal calf serum, 2 ng/mL of IL-3, 2 ng/mL of IL-6, 50 ng/mL of SCF, 60 ng/mL of G-CSF, and 20 ng/mL of GM-CSF (Peprotech, Rocky Hill, NJ). After 7 to 10 days of culture, colonies were scored and the cells were used for immunolabeling, serial replating experiments, and morphologic analysis.16
Analysis of mitotic chromosome spreads
Lin− cells were plated in methylcellulose medium as previously described. After 4 days, cells were harvested and resuspended in RPMI medium containing 10% fetal calf serum, 20 ng/mL of IL-3, 20 ng/mL of IL-6, 100 ng/mL of SCF, and 0.1μg/mL of colcemid (Gibco; Life Technologies Europe, Monza, Italy). After 4 hours, cells were washed and resuspended in hypotonic buffer (0,075M KCl), incubated for 1 hour at 37°C, and then fixed with methanol/acetone (3:1). Cells were spotted on glass slides and stained with Giemsa (Sigma-Aldrich, St Louis, MO). Metaphases were analyzed by optical microscopy.
In vivo bromodeoxyuridine (BrdU) incorporation
BrdU incorporation was analyzed by FACS analysis (BD, FACSCantoII BD, FACSDiva Software V6.1.1, Becton Dickinson, Franklin Lakes, NJ) after 4 hours of a single intraperitoneal injection of BrdU (40 mg/Kg) using the FITC BrdU Flow Kit (Becton Dickinson).
Immunophenotyping and histopathologic analysis
FACS analysis (BD, FACSCantoII BD, FACSDiva Software V6.1.1) was performed on splenocytes, and bone marrow cells were derived from transplanted mice. Cells derived from healthy mice were used as a control. The antibodies used for the immunophenotyping were Ly-6A/E (SCA1) PE-Cy5.5 conjugated, CD117 (C-Kit) APC conjugated, anti-mouse CD45.2 FITC conjugated, CD11b (MAC1), Ly-6G (Gr-1), CD3e, and CD45R (B220) all PE-Cy7 conjugated (eBioscience, San Diego, CA). Blood smears and cytospins were stained using the May-Grünwald-Giemsa method (Sigma-Aldrich).
Pharmacologic treatments
Preleukemic 6- to 8-week-old mCGPR/PR mice were intraperitoneally injected with valproic acid (VPA; Sigma) dissolved in phosphate-buffered saline (PBS) at a dose of 400 mg/kg twice a day for 14 days. For differentiation analysis, 12 hours after the last injection, the mice were euthanized and bone marrow cells were analyzed by FACS for surface differentiation markers or plated in methylcellulose as described. For in vivo experiments, the mice were followed until leukemia development. For APL mice treatment, 2*10^5 APL cells were injected intravenously into nonirradiated congenic-recipient mice (C57Bl/6-Ly5.1) and then treated with VPA (intraperitoneally at the dose of 400 mg/kg twice a day for 4 weeks) when the blast cells (Ly5.2) in peripheral blood reached 5% to 10%.24
Quantitative polymerase chain reaction
Total RNA was purified using RNeasy Mini Kit (QIAGEN, Valencia, CA), quantified and reverse transcribed. From 5 to 10 ng of cDNA were used to perform quantitative polymerase chain reaction using SYBR Green Reaction Mix (Perkin Elmer, Boston, MA). mRNA levels were normalized against GAPDH mRNA. The sequences of the primers used in this study are listed in supplemental Table 4.
Statistical analysis
Statistical analyses were performed using the unpaired sample 2-tailed Student t test (GraphPad software) unless otherwise specified. Statistical analysis of the Kaplan-Meier survival curves was done using the log-rank test (Prism 4.0 software).
Results
Hdac1/Hdac2 knock-down accelerates APL development
As a model system for PML-RAR–driven APL, we used the well-characterized mCGPR/PR mouse model.12 Lin− cells enriched in hematopoietic stem cells and progenitors from mCGPR/PR mice16 were transduced with GFP-tagged retroviral vectors targeting Hdac1 or the control (CTRL) firefly luciferase transcripts. FACS-purified GFP+/lin− cells were then analyzed in vitro and in vivo (supplemental Figure 1). RNA and protein analysis confirmed a strong reduction of Hdac1 by the targeting shRNA constructs (Figure 1A, supplemental Figure 2), with a compensatory increase in the expression of Hdac2 but not Hdac3 (Figure 1A) as previously reported.6,25 In addition, we observed increased acetylation of histone H4 at lysine 5, that is preferentially deacetylated by Hdac1,25,26 whereas global histone acetylation remained relatively unchanged (Figure 1A). Knock-down of Hdac1 had no effect on the expression of PML-RAR (supplemental Figure 3 and 4).
Hdac1 knock-down accelerates APL development. (A) Immunoblot analysis of Hdac1, 2, and 3 expression, and histone 3H/H4 acetylation in GFP+ lin− cells derived from mCGPR/PR mice and transduced with the indicated retroviral vectors. Total histone 3H served as a loading control. (B) Leukemia-free survival curves of mice transplanted with the indicated GFP+ lin− cells. HDAC1-KDA and HDAC1-KDB indicate 2 different shRNA against Hdac1 (supplemental Table 4). CTRL vs Hdac1: P < .0001. (C) Hdac1, Hdac2, and Hdac3 expression in bone marrows of leukemic mice. Vinculin was used as a loading control. (D) Cytologic analysis of APL blasts from the spleens of moribund animals. Original magnification ×1000, May Grünwald-Giemsa staining, Olympus BX51. (E) Representative immunophenotype of leukemic cells derived from the spleens of moribund animals.
Hdac1 knock-down accelerates APL development. (A) Immunoblot analysis of Hdac1, 2, and 3 expression, and histone 3H/H4 acetylation in GFP+ lin− cells derived from mCGPR/PR mice and transduced with the indicated retroviral vectors. Total histone 3H served as a loading control. (B) Leukemia-free survival curves of mice transplanted with the indicated GFP+ lin− cells. HDAC1-KDA and HDAC1-KDB indicate 2 different shRNA against Hdac1 (supplemental Table 4). CTRL vs Hdac1: P < .0001. (C) Hdac1, Hdac2, and Hdac3 expression in bone marrows of leukemic mice. Vinculin was used as a loading control. (D) Cytologic analysis of APL blasts from the spleens of moribund animals. Original magnification ×1000, May Grünwald-Giemsa staining, Olympus BX51. (E) Representative immunophenotype of leukemic cells derived from the spleens of moribund animals.
Reconstitution of irradiated syngeneic recipient mice with GFP+/lin−/Hdac1 knockdown (Hdac1-kd) cells led to a dramatic shortening of leukemia onset compared with controls (Figures 1B, supplemental Table 1A), and knock-down of Hdac1 was maintained in leukemias derived from these mice (Figures 1C, supplemental Figure 5). Interestingly, the upregulation of Hdac2 observed in freshly transduced cells was not maintained in leukemic blasts (Figure 1C). CTRL and Hdac1-kd leukemias were essentially indistinguishable macroscopically, histologically, and cytologically (Figure 1D, supplemental Figure 6A-B). In contrast, immunophenotypic analysis revealed a greater proportion of immature Gr-1+/c-Kit+/Hdac1-kd cells (Figure 1E, supplemental Table 2). Transplantation of primary Hdac1-kd or CTRL APLs into syngeneic recipients resulted in the development of secondary leukemias at similar rates (supplemental Figure 7), indicating that Hdac1 knock-down mainly affects the preleukemic stage.
Similar studies were performed using shRNA vectors targeting Hdac2. Western blot analysis of transduced GFP+/lin−/Hdac2-kd cells confirmed decreased Hdac2 expression with limited concomitant increase in Hdac1 levels, suggesting that the modality of regulation of Hdac1 and Hdac2 differs (supplemental Figure 8A). Knock-down of Hdac2 led to a significant acceleration of APL development (supplemental Figure 8B), although less pronounced than Hdac1 knock-down (supplemental Figure 8C), and APL cells from these mice maintained decreased Hdac2 expression (supplemental Figure 8D). Interestingly, although Hdac1 levels were generally unchanged, in a single case (n. 10), we observed a strong reduction of Hdac1 levels in leukemic cells, hinting at a potentially cooperating oncosuppressive role for Hdac1 and Hdac2 (supplemental Figure 8D).
Expression analysis of a collection of primary human APL samples and normal CD34+ progenitor cells showed that Hdac1 and Hdac2 transcript levels were reduced in APL (supplemental Figure 9A-B). To further validate this observation, we surveyed the expression levels of Hdac1 and Hdac2 in primary human AML samples from available gene-profiling datasets.27 Interestingly, Hdac1 mRNA levels were selectively reduced in primary APL samples, compared with other AML subtypes, and relative to normal progenitor CD34+ cells and normal promyelocytes (supplemental Figure 9C). Hdac2 levels were also down-regulated compared with normal CD34+ cells in several AML subtypes, although during normal myeloid differentiation, there is a strong decrease in expression in promyelocytes (supplemental Figure 9D). Collectively, these results support a potential oncosuppressor role, at least of Hdac1, in human APL.
Hdac1 behaves as a general oncosuppressor
To determine if accelerated tumorigenesis after knock-down of Hdac 1 or 2 was restricted to APL, we performed additional knock-down experiments on 2 different murine lymphoma models. We firstly examined the role of Hdac1 and 2 on spontaneous lymphomagenesis occurring in p53−/− mice.28 After transduction with the Hdac1- or 2-targeting vectors, lin− cells derived from p53−/− mice were transplanted into syngeneic, lethally irradiated mice (Figure 2A-B). Thymic lymphomas developed in all transplanted animals, and knock-down of Hdac1 significantly accelerated lymphomagenesis (Figure 2B left panel, supplemental Table 1B). Hdac2 knock-down, in contrast, did not result in a significant change in survival of the mice (Figure 2B right panel, supplemental Table 1B), suggesting that the oncosuppressive functions of Hdac1 and Hdac2 may be context dependent.
Hdac1/2 knock-down accelerates lymphomagenesis. (A-B) Lin− cells from p53−/− mice transduced with the control vector (CTRL) or vectors carrying an shRNA against Hdac1 or Hdac2 were sorted for GFP positivity and injected into lethally irradiated recipient WT mice. (A) Immunoblot analysis of cells transduced with the indicated retroviral vectors. The arrow indicates HDAC2. The asterisk indicates HDAC1 residual staining from the previous immunoblotting assay. Hdac3 served as a loading control. (B) Leukemia-free survival curves of mice transplanted with p53 null lin− cells transduced with the control vector (CTRL) or a vector carrying an shRNA against Hdac1 (left panel, P < .05) or Hdac2 (right panel, p=ns). (C-D) Eμ-myc fetal liver progenitor cells were transduced with the control vector (CTRL) or vectors carrying an shRNA against Hdac1 or Hdac2 and injected into irradiated recipient PTPRCA mice. (C) Kaplan-Meier survival curves are shown; hdac1 P < .05, hdac2 p=ns. (D) Western blots were performed on FACS-sorted GFP+ cell lysates from a control spontaneous Eμ-myc lymphoma (lane 1) and the indicated primary lymphomas with knockdown of HDAC1 (lanes 2 and 3) or HDAC2 (lanes 4-6) and were analyzed for the expression of the indicated HDACs. B-actin was used as a loading control.
Hdac1/2 knock-down accelerates lymphomagenesis. (A-B) Lin− cells from p53−/− mice transduced with the control vector (CTRL) or vectors carrying an shRNA against Hdac1 or Hdac2 were sorted for GFP positivity and injected into lethally irradiated recipient WT mice. (A) Immunoblot analysis of cells transduced with the indicated retroviral vectors. The arrow indicates HDAC2. The asterisk indicates HDAC1 residual staining from the previous immunoblotting assay. Hdac3 served as a loading control. (B) Leukemia-free survival curves of mice transplanted with p53 null lin− cells transduced with the control vector (CTRL) or a vector carrying an shRNA against Hdac1 (left panel, P < .05) or Hdac2 (right panel, p=ns). (C-D) Eμ-myc fetal liver progenitor cells were transduced with the control vector (CTRL) or vectors carrying an shRNA against Hdac1 or Hdac2 and injected into irradiated recipient PTPRCA mice. (C) Kaplan-Meier survival curves are shown; hdac1 P < .05, hdac2 p=ns. (D) Western blots were performed on FACS-sorted GFP+ cell lysates from a control spontaneous Eμ-myc lymphoma (lane 1) and the indicated primary lymphomas with knockdown of HDAC1 (lanes 2 and 3) or HDAC2 (lanes 4-6) and were analyzed for the expression of the indicated HDACs. B-actin was used as a loading control.
We then investigated a role for Hdac1 and 2 in the context of c-myc overexpression using the Eμ-myc mouse.15 Eμ-myc fetal liver hematopoietic progenitor cells were transduced with retroviruses expressing shRNAs targeting Hdac 1 or 2. Unsorted cells were transplanted into irradiated recipient mice, and tumor development was monitored (Figure 2C, supplemental Table 1C). Consistent with the data outlined above, knock-down of Hdac1 hastened the rate of myc-induced lymphomagenesis, whereas the effects of Hdac2 knock-down were considerably more modest and did not achieve statistical significance (Figure 2C, supplemental Table 1C). GFP+ tumors isolated from Eμ-myc/Hdac1-kd and Eμ-myc/Hdac2-kd mice showed robust and specific knock-down of the targeted protein (Figure 2D). Moreover, we tested these tumors for maintenance of a p53 response and observed the p53 pathway to be functionally intact as demonstrated by γ-irradiation–induced upregulation of p53 and constitutively low expression of p19ARF, which is used as a surrogate marker for WT p53 expression in this model29 (supplemental Figure 10). Finally, sequence analysis of p53 revealed it to be WT in all Eμ-myc/Hdac1-kd and Eμ-myc/Hdac2-kd tumors (data not shown). These data suggest that Eμ-myc tumors with loss of Hdac1 or 2 develop in a manner that maintains expression of WT p53. Taken together, these data extend our initial findings in APL and indicate a more general oncosuppressive role for Hdac1.
Hdac1 knock-down enhances differentiation block and genomic instability of PML-RAR–expressing cells
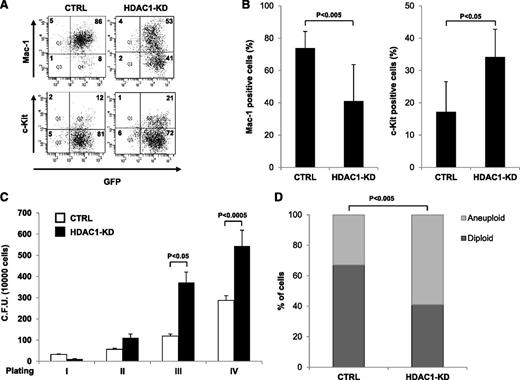
APL is characterized by a block in myeloid differentiation and alteration of genomic stability.30,31 Both events are supposedly driven by the direct action of PML-RAR, and at least in part, this is because of its association with Hdacs,32 although Hdacs can affect both biological processes independently of PML-RAR.33,34 Therefore, we investigated whether Hdac1 knock-down affected either or both of these processes in the preleukemic setting to accelerate PML-RAR–driven APL. As previously shown,16,35 expression of PML-RAR in the preleukemic phase caused a modest defect on differentiation in vitro (supplemental Figure 11), and Hdac1 knock-down greatly augmented this effect with a strong decrease of Mac-1+ cells and an increase of c-Kit+, immature cells (Figure 3A-B). Interestingly, this phenomenon was PML-RAR dependent because Hdac1 knock-down in WT lin− cells did not significantly affect differentiation (supplemental Figure 12). PML-RAR expression results in an enhanced proliferative ability of lin− cells,35 and consistent with the observed potentiation of the differentiation block, Hdac1 knock-down led to a further increase in the proliferative potential of PML-RAR–expressing lin− cells, as demonstrated by the higher number of colonies recovered in serial replating experiments (Figure 3C). We next determined the effect of Hdac1 knock-down on genomic stability. Colony-forming assays with GFP+/lin− cells confirmed previous studies36 demonstrating that PML-RAR expression enhanced genome instability compared with WT cells (supplemental Figure 13). Hdac1 knock-down amplified this phenomenon with more than 50% of metaphases being aneuploid after in vitro plating (Figure 3D).
Hdac1 knock-down enhances the myeloid differentiation block and genomic instability imposed by PML-RAR in lin− cells derived from mCGPR/PR mice. Lin− cells were transduced with the indicated vectors, sorted for GFP positivity, and plated in methylcellulose-containing medium. (A-B) Analysis of the expression of the indicated differentiation markers in control and Hdac1 knock-down preleukemic cells. A representative dot plot (A) and the statistical analysis of 3 independent experiments (B) are shown. (C) Analysis of the proliferative potential of control and Hdac1 knock-down preleukemic cells by a serial replating assay. (D) Plot of the percentages of aneuploid and diploid cells in metaphase spreads from CTRL (n = 82) and HDAC1-KD (n = 76) cells. The statistical analysis was performed with the Fisher exact test (GraphPad software).
Hdac1 knock-down enhances the myeloid differentiation block and genomic instability imposed by PML-RAR in lin− cells derived from mCGPR/PR mice. Lin− cells were transduced with the indicated vectors, sorted for GFP positivity, and plated in methylcellulose-containing medium. (A-B) Analysis of the expression of the indicated differentiation markers in control and Hdac1 knock-down preleukemic cells. A representative dot plot (A) and the statistical analysis of 3 independent experiments (B) are shown. (C) Analysis of the proliferative potential of control and Hdac1 knock-down preleukemic cells by a serial replating assay. (D) Plot of the percentages of aneuploid and diploid cells in metaphase spreads from CTRL (n = 82) and HDAC1-KD (n = 76) cells. The statistical analysis was performed with the Fisher exact test (GraphPad software).
To confirm the specificity of the Hdac1 knock-down effects, PML-RAR–expressing lin−/Hdac1-kd cells were superinfected with a retrovirus expressing Hdac1 cDNA that could not be targeted by the RNAi knock-down constructs (supplemental Figure 14A). Strikingly, expression of nontargetable Hdac1 essentially abolished the differentiation block observed on Hdac1 knock-down, confirming that the observed phenotype was the result of the selective loss of the protein (supplemental Figure 14B-C). Taken together, these results indicate that downregulation of Hdac1 potentiates the effects of PML-RAR on differentiation and genomic stability of hematopoietic progenitors, consistent with the observed acceleration of leukemogenesis in vivo.
Hdac1 knock-down primes the expansion of the prospective leukemic-initiating cell compartment at the preleukemic stage
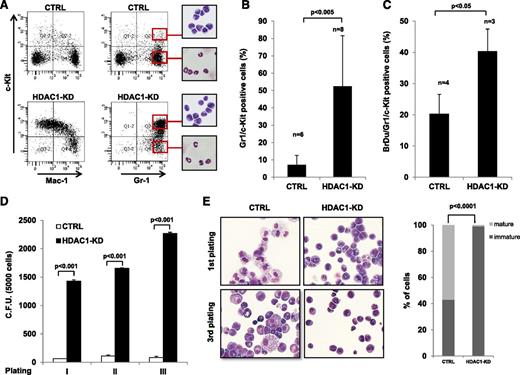
To determine the effect of Hdac1 knock-down on the differentiation of preleukemic myeloid cells in vivo, irradiated recipient mice were reconstituted with PML-RAR–expressing GFP+/lin−/Hdac1-kd and GFP+/lin−/CTRL cells, and bone marrows from transplanted mice were analyzed at different time points during the preleukemic phase (supplemental Figure 15). Knock-down of Hdac1 caused a dramatic expansion of immature Gr-1+/c-Kit+ cells and a decrease in expression of Mac-1 (Figure 4A-B). As a consequence of the expansion of Gr-1+/c-Kit+/Mac-1low cells, we observed an imbalance in lineage distribution after knock-down of Hdac1 in preleukemic APL mice, with a shift toward the myeloid lineage at the expense of the lymphoid compartment (Figure 4A, supplemental Table 3). Consistent with their expansion, the proliferative ability in vivo of PML-RAR–expressing Gr-1+/c-Kit+/Hdac1-kd cells was strongly enhanced compared with CTRL cells as assessed by BrdU incorporation (Figure 4C). This effect was confirmed in vitro, as shown by the increase in colony-forming cells in serial methylcellulose assays (Figure 4D). In addition, when cultured in methylcellulose supplemented with a prodifferentiation cytokine cocktail, PML-RAR–expressing Gr-1+/c-Kit+/Hdac1-kd cells maintained their undifferentiated phenotype which lasted for several platings, whereas PML-RAR–expressing CTRL cells showed an activated differentiation program (Figure 4E). Intriguingly, in frankly leukemic mice, it has been shown that the leukemic stem cell compartment is enriched in Gr-1+/c-Kit+ cells.37,38 Therefore, we transplanted PML-RAR–expressing Gr-1+/c-Kit+/Hdac1-kd cells from preleukemic mice into secondary recipient mice, which developed APL (supplemental Figure 16). Taken together, these results show that during the preleukemic stage, Hdac1 functions to prevent the expansion of a PML-RAR–expressing cell subpopulation prototypic of the leukemic stem cell compartment in the leukemic stage, which is able to give rise to APL.
Hdac1 knock-down primes the expansion of Gr-1+/c-Kit+ cells in vivo. Lin− cells derived from mCGPR/PR transgenic mice were transduced with the indicated vectors, and sorted GFP+ cells were injected into lethally irradiated mice. Mice were euthanized 30 days after transplantation, and their phenotype was analyzed. (A) A representative dot plot showing the levels of the indicated differentiation markers in bone marrow cells. Insets show the cytologic analysis of the sorted subpopulations (red squares). (B) Plot of the levels of Gr-1+/c-Kit+ cells in the indicated murine bone marrow samples. (C) Mice transplanted with the indicated cells were injected with BrdU. In the graph, the levels of BrdU incorporation are shown. (D) The indicated Gr-1+/c-Kit+ cells were plated in methylcellulose medium and were scored for colony-forming ability. In the graph, the mean and standard deviation of 3 independent experiments are shown. (E) Morphologic analysis of the indicated Gr-1+/c-Kit+ cells, harvested after plating in methylcellulose medium. Left panels, representative cytospins; right panel, percentage of mature and immature cells, original magnification ×1000, May Grünwald-Giemsa staining, Olympus BX51. Statistical analysis was performed with the Fisher exact test.
Hdac1 knock-down primes the expansion of Gr-1+/c-Kit+ cells in vivo. Lin− cells derived from mCGPR/PR transgenic mice were transduced with the indicated vectors, and sorted GFP+ cells were injected into lethally irradiated mice. Mice were euthanized 30 days after transplantation, and their phenotype was analyzed. (A) A representative dot plot showing the levels of the indicated differentiation markers in bone marrow cells. Insets show the cytologic analysis of the sorted subpopulations (red squares). (B) Plot of the levels of Gr-1+/c-Kit+ cells in the indicated murine bone marrow samples. (C) Mice transplanted with the indicated cells were injected with BrdU. In the graph, the levels of BrdU incorporation are shown. (D) The indicated Gr-1+/c-Kit+ cells were plated in methylcellulose medium and were scored for colony-forming ability. In the graph, the mean and standard deviation of 3 independent experiments are shown. (E) Morphologic analysis of the indicated Gr-1+/c-Kit+ cells, harvested after plating in methylcellulose medium. Left panels, representative cytospins; right panel, percentage of mature and immature cells, original magnification ×1000, May Grünwald-Giemsa staining, Olympus BX51. Statistical analysis was performed with the Fisher exact test.
Hdac1 knock-down has an antitumor activity on APL cells at the leukemic stage
Our results are apparently at odds with the known antitumor effects of HDACi, shown in several contexts, including leukemias expressing the PML-RAR oncoprotein.30 Therefore, we knocked down Hdac1 (or luciferase as a control: CTRL) in frankly leukemic APL cells16 (Figure 5A-B) and observed concomitant induction of Hdac2 expression and increased histone acetylation (Figure 5B). GFP+/Hdac1-kd and GFP+/CTRL APL cells were transplanted into syngeneic recipient mice, and in contrast to the accelerated tumorigenesis observed after knock-down of Hdac1 in preleukemic cells, Hdac1 depletion caused increased overall survival duration in the recipient mice (Figure 5C). Consistent with this result and as previously observed,24,30 treatment with the class I HDACi VPA39,40 caused increased overall survival duration in frankly leukemic APL mice (Figure 5D and supplemental Figure 17).
These results suggest that Hdac1 activity plays opposing roles at different stages of APL development: it antagonizes the oncogenic activity of PML-RAR at the preleukemic stage but favors the growth of the leukemic mass in the overt leukemic state.
Pharmacologic inhibition of class I HDACs mimics the phenotype of Hdac1 knock-down
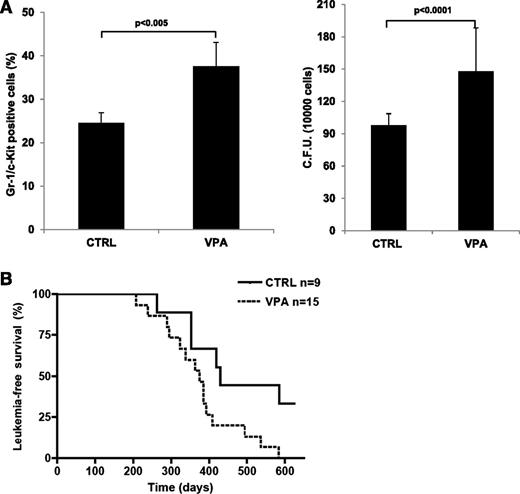
We then determined if the biological effect of Hdac1 knock-down in PML-RAR–expressing preleukemic cells could be phenocopied by inhibiting HDACs pharmacologically. Strikingly, short-term treatment with VPA doubled the number of Gr-1+/c-Kit+ cells in the bone marrow of preleukemic mice (Figure 6A, left panel). Bone marrow cells derived from VPA-treated mice gave rise to a higher number of colonies than cells derived from untreated mice (Figure 6A, right panel). Importantly, leukemia developed in VPA-treated mice within a shorter period compared with such development in untreated mice (Figure 6C, supplemental Table 1D). Taken together, these results show that pharmacologic inhibition of HDAC enzymatic activity in preleukemic mice mimics the oncogenic effects of Hdac1 knock-down.
Hdac1 knock-down in APL blasts increases mice survival duration. (A-C) APL blasts from 129sv mice were transduced with the indicated vectors, sorted for GFP positivity, and transplanted in wt mice. (A) Analysis of Hdac1 mRNA levels in APL blasts transduced with the indicated retroviral vectors. Values are normalized against GAPDH and referred to CTRL. The graph represents the average and standard deviation of 3 independent experiments. (B) Immunoblot analysis of the expression of Hdac1, 2, and 3 and histone 3H acetylation in GFP+ APL blasts transduced as indicated. Histone 3H served as a loading control. (C) Leukemia-free survival curves of mice transplanted with APL blasts transduced with the indicated vectors and then were sorted for GFP expression (CTRL vs HDAC1-KDA or CTRL vs HDAC1-KDB: P < .05). (D) Leukemia-free survival curves of C57Bl/6-Ly5.1 recipient mice transplanted with APL cells derived from the mCGPR/PR leukemic mouse #1.VPA treatment was started when the blast cells in peripheral blood reached 5% to 10%. CTRL vs VPA: P < .001. Day 0 indicates the start of the treatment.
Hdac1 knock-down in APL blasts increases mice survival duration. (A-C) APL blasts from 129sv mice were transduced with the indicated vectors, sorted for GFP positivity, and transplanted in wt mice. (A) Analysis of Hdac1 mRNA levels in APL blasts transduced with the indicated retroviral vectors. Values are normalized against GAPDH and referred to CTRL. The graph represents the average and standard deviation of 3 independent experiments. (B) Immunoblot analysis of the expression of Hdac1, 2, and 3 and histone 3H acetylation in GFP+ APL blasts transduced as indicated. Histone 3H served as a loading control. (C) Leukemia-free survival curves of mice transplanted with APL blasts transduced with the indicated vectors and then were sorted for GFP expression (CTRL vs HDAC1-KDA or CTRL vs HDAC1-KDB: P < .05). (D) Leukemia-free survival curves of C57Bl/6-Ly5.1 recipient mice transplanted with APL cells derived from the mCGPR/PR leukemic mouse #1.VPA treatment was started when the blast cells in peripheral blood reached 5% to 10%. CTRL vs VPA: P < .001. Day 0 indicates the start of the treatment.
VPA treatment mimics the phenotype of Hdac1 knock-down in vivo. mCGPR/PR mice were treated with VPA or PBS (CTRL) for 14 days. Then, (A) 12 hours after the last injection, mice were euthanized and bone marrows were collected and analyzed for the expression of Gr-1 and c-Kit (left panel), or colony-forming ability (right panel), or (B) followed for leukemia development. In the graph, leukemia-free survival curves from VPA- and PBS-treated mice (P < .05).
VPA treatment mimics the phenotype of Hdac1 knock-down in vivo. mCGPR/PR mice were treated with VPA or PBS (CTRL) for 14 days. Then, (A) 12 hours after the last injection, mice were euthanized and bone marrows were collected and analyzed for the expression of Gr-1 and c-Kit (left panel), or colony-forming ability (right panel), or (B) followed for leukemia development. In the graph, leukemia-free survival curves from VPA- and PBS-treated mice (P < .05).
Discussion
Here, we show in different mouse tumor models (APL, p53−/−, and myc-driven lymphomas) that a single putative barrier to full transformation is surprisingly provided by class I Hdacs 1 and 2. Knock-down of Hdac1 potentiated both the block of cellular differentiation and the increased genomic instability mediated by PML-RAR in hematopoietic progenitors. Either or both biological deregulations could be sufficient to cooperate with the tumor-promoting activities of oncoproteins such as PML-RAR and Myc and would provide a functional explanation for the observed increase in frequency of transformation to full leukemia in Hdac1-deficient cells. Additionally, and potentially linked to those events, we observed in Hdac1-deficient preleukemic cells a dramatic expansion of a subpopulation of myeloid precursor cells (Gr1+ and c-Kit+ cells) that represents the major source of leukemic stem cells in the full leukemic stage.37,38 We hypothesize that in preleukemia this subpopulation acts as a reservoir of target cells for full leukemic transformation: in the presence of decreased levels of Hdac1, expansion of Gr1+/c-Kit+ cells leads to an increase in the number of candidate target cells and of the probability of transformation, thus accelerating leukemogenesis. These results are reminiscent of the role of histone H3K9 methylation in the tumor suppression of Ras and myc-driven lymphoma development in transgenic mice41 and support an important role for heterochromatin as a tumor-suppressive mechanism.41-43 Consistent with the notion that HDACs can function as tumor suppressors, Hdac1 and Hdac2 mRNA levels were found to be selectively reduced in human AML samples, suggesting that this function may be important in human leukemogenesis.
HDACs have been widely considered to have tumor-promoting and/or sustaining roles, especially given the impressive antitumor responses mediated by HDACi in certain hematologic malignancies.7 Indeed, combined genetic deletion of Hdac1 and Hdac2 results in the activation of a senescent program, and/or mitotic catastrophe and death of transformed cells.5,6 Here, we showed that knock-down of Hdac1 in transplanted PML-RAR–expressing leukemia cells prolonged the survival time of recipient mice, supporting the view that Hdac1 has oncogenic activity in established tumor cells. As a potential explanation to reconcile the opposing roles of Hdac1 unraveled in APL, we speculate that Hdac1 may have distinct functions in different subpopulations of leukemia: indeed, VPA treatment induces differentiation and death of APL blasts, sparing the leukemic stem cell compartment.24
Short-term in vivo treatment of preleukemic mice with the class I HDACi VPA phenocopied the effect of Hdac1 knock-down during PML-RAR–mediated leukemogenesis with expansion of the Gr1+/c-Kit+ population and accelerated disease. Intriguingly, this phenomenon was not observed after treatment of premalignant Eμ-myc mice with VPA, indicating that a tumor-type or oncogene-specific effect may be at play (data not shown). Nevertheless, these results are somewhat unsettling concerning the growing clinical use of HDACi. The broad inhibition of multiple HDACs by most available HDACi has been proposed as a potential explanation for the lack of sustained efficacy of these agents in most patients with cancer. Our data suggest that HDAC inhibition may block intrinsic antitumor functions of HDACs and imply that more studies are needed to systematically dissect the role of individual HDACs, at different stages of tumorigenesis, in different tumor compartments and in different tumor cell types.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Simona Ronzoni, Anna Sciullo, Ivan Muradore, Mario Faretta, and Gabriele Bucci for technical assistance; Daniela Bossi, Chiara Ronchini for helpful discussions; and Simona Citro for the HDAC1 expression plasmid.
Work in S.M.’s laboratory is supported by AIRC (Italian Association for Cancer Research), FIRC (Italian Foundation for Cancer Research), CNR (National Research Council), Flagship Project Epigen, MIUR and MIS (Ministry of Health and Research), and European Community (FP7 Blueprint and 4D Projects).
R.W.J. is a Principal Research Fellow of the National Health and Medical Research Council of Australia (NHMRC) and is supported by NHMRC program and project grants, Cancer Council Victoria, The Leukemia Foundation of Australia, Victorian Breast Cancer Research Consortium, and the Victorian Cancer Agency.
Authorship
Contribution: F.S. and O.A.B. designed and performed experiments and assisted in writing the manuscript. G.M.M. and L.C. performed experiments and provided discussion. R.D.Z., I.P., S.S., I.B., and L.F. provided discussion and technical advice. L.A., P.G.P., and S.C. provided significant discussion. S.M. and R.W.J. designed experiments, provided discussion, and assisted in writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Saverio Minucci, Department of Experimental Oncology, European Institute of Oncology, IFOM-IEO Campus, via Adamello 16, 20139 Milan, Italy; e-mail: saverio.minucci@ieo.eu; Ricky Johnstone, Peter MacCallum Cancer Centre, St. Andrews Place, East Melbourne 3002, Victoria, Australia; e-mail: ricky.johnstone@petermac.org.
References
Author notes
F.S. and O.A.B. contributed equally to this study.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal