Key Points
Ponatinib exhibits activity against AC220-resistant FLT3-ITD/F691 gatekeeper mutations.
Ponatinib is highly ineffective against FLT3-ITD activation loop mutations, particularly at the D835 residue.
Abstract
Secondary point mutations in the Fms-like tyrosine kinase 3 (FLT3) tyrosine kinase domain (KD) are common causes of acquired clinical resistance to the FLT3 inhibitors AC220 (quizartinib) and sorafenib. Ponatinib (AP24534) is a multikinase inhibitor with in vitro and clinical activity in tyrosine kinase inhibitor (TKI)-resistant chronic myeloid leukemia, irrespective of BCR-ABL KD mutation. Ponatinib has demonstrated early clinical efficacy in chemotherapy-resistant acute myeloid leukemia (AML) patients with internal tandem duplication (ITD) mutations in FLT3. We assessed the in vitro activity of ponatinib against clinically relevant FLT3-ITD mutant isoforms that confer resistance to AC220 or sorafenib. Substitution of the FLT3 “gatekeeper” phenylalanine with leucine (F691L) conferred mild resistance to ponatinib, but substitutions at the FLT3 activation loop (AL) residue D835 conferred a high degree of resistance. Saturation mutagenesis of FLT3-ITD exclusively identified FLT3 AL mutations at positions D835, D839, and Y842. The switch control inhibitor DCC-2036 was similarly inactive against FLT3 AL mutations. On the basis of its in vitro activity against FLT3 TKI-resistant F691 substitutions, further clinical evaluation of ponatinib in TKI-naïve and select TKI-resistant FLT3-ITD+ AML patients is warranted. Alternative strategies will be required for patients with TKI-resistant FLT3-ITD D835 mutations.
Introduction
Activating mutations in FLT3 are detected in approximately 30% of adult acute myeloid leukemia (AML) cases. Particularly common are internal tandem duplication (ITD) events in the juxtamembrane domain, which are associated with poor prognosis.1,2 The collective clinical experience with early FLT3 tyrosine kinase inhibitors (TKIs) in AML was disappointing; responses were largely limited to transient reductions in peripheral blood blasts, whereas bone marrow responses were exceedingly rare.3-5 These studies suggested that FLT3-ITD may represent a “passenger” rather than a “driver” lesion. An alternative explanation is that “first-generation” FLT3 TKIs failed to achieve sufficiently potent target inhibition in the leukemic cells of patients. Recently, a phase II study of the second-generation FLT3/KIT inhibitor AC220 (quizartinib) demonstrated a composite complete remission rate of 44% to 54% in relapsed and chemotherapy-refractory AML.6,7 In addition, anecdotal achievement of complete remission in FLT3-ITD+ AML patients treated with the multikinase inhibitor sorafenib on a compassionate use basis has been reported.8 The validity of FLT3-ITD as a therapeutic target in human AML was definitively demonstrated through translational studies that identified the evolution of AC220 resistance–conferring FLT3-ITD kinase domain (KD) mutations at the time of acquired resistance in 8/8 FLT3-ITD+ patients analyzed.9 This finding suggests that, as with chronic myeloid leukemia (CML), secondary mutation in the target KD is likely to represent a common mechanism of acquired resistance to clinically active TKIs and will pose a substantial barrier to response durability. In further support of this concept, KD mutations have been reported to be associated with acquired resistance to sorafenib10 and the multikinase inhibitor PKC41211 in FLT3-ITD+ AML patients.
Clinically relevant AC220 resistance–conferring mutations have thus far been restricted to 2 residues in the FLT3 KD, the “gatekeeper” residue F691 (F691L), and the activation loop (AL) residue D835 (D835V/Y/F). An in vitro mutagenesis screen identified mutations at a third AL residue, Y842 (Y842C/H), as also capable of causing substantial resistance to AC220 in vitro.9 Notably, mutations at all 3 of these residues confer in vitro cross-resistance to sorafenib.9,12 Substitutions at gatekeeper residues such as FLT3-ITD/F691 have been well-documented to confer resistance to kinase inhibitors in other malignancies, including EGFR-mutated non–small-cell lung cancer, BCR-ABL+ acute lymphoblastic leukemia, and CML.13,14 Analogs of the FLT3-ITD/D835V AL mutation have also proven problematic for a number of kinase inhibitors. Substitutions at the analogous residue (D816) in KIT, commonly associated with systemic mastocytosis, results in pathological kinase activation and confers a high degree of intrinsic resistance to imatinib and other KIT inhibitors.15,16 Mutations at D835 in FLT3-ITD have also been implicated recently in clinical resistance to sorafenib in FLT3-ITD+ AML patients.10
Although AC220 appears to harbor substantial clinical activity in FLT3-ITD+ AML, its clinical development has been complicated by toxicities including QT prolongation and myelosuppression. Clinical trials are currently exploring lower AC220 doses for retention of antileukemic activity and improved safety. Ponatinib (AP24534) is a potent inhibitor of several kinases, including ABL and FLT3, that has demonstrated in vitro activity against all drug-resistant BCR-ABL KD mutants, including the gatekeeper T315I and AL H396P mutations.17,18 Ponatinib is well-tolerated; has demonstrated significant clinical activity in TKI-resistant CML cases, including in patients with the BCR-ABL/T315I mutation19 ; and was recently approved by the US Food and Drug Administration for the treatment of CML and Ph+ acute lymphoblastic leukemia patients with resistance or intolerance to prior TKI therapy. In addition, ponatinib has demonstrated clinical activity in FLT3-ITD+ AML patients in limited phase I experience. Specifically, 2 of 7 TKI-naïve FLT3-ITD+ AML patients achieved complete remission with incomplete recovery of blood counts (CRi) on 45 mg daily ponatinib therapy.20 Ponatinib is orally administered and is not associated with appreciable QT prolongation. Because it retains activity against all TKI-resistant BCR-ABL mutants, ponatinib may be similarly effective against all FLT3-ITD KD substitutions. No studies have yet assessed the activity of ponatinib against clinically detected FLT3-ITD KD mutants. We therefore sought to test the activity of ponatinib against FLT3-ITD KD mutants that have been documented to confer clinical resistance to AC220, and to prospectively identify secondary FLT3-ITD KD mutations that can confer resistance to ponatinib in vitro, which may also confer acquired clinical resistance to this agent in FLT3-ITD+ AML patients.
Material and methods
Inhibitors
Ponatinib was a gift of ARIAD Pharmaceuticals (Cambridge, MA). AC220 and DCC-2036 were purchased from Selleckchem (Houston, TX). Sorafenib was purchased from LC Laboratories (Woburn, MA).
DNA Constructs, mutagenesis, and resistance screen
Random mutagenesis was performed as previously described.9 Cells were selected in 40 nM ponatinib in soft agar. After 10 to 21 days, visible colonies were plucked and expanded in 10 nM ponatinib.
Sequencing and alignments
Sequencing was performed from amplified genomic DNA from colonies expanded from soft agar as previously described.9
Generation of mutants
Mutations isolated in the screen were engineered into pMSCVpuroFLT3-ITD by QuikChange mutagenesis (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s recommendations.
Cell lines
Stable Ba/F3 lines were generated by retroviral spinfection with the appropriate mutated plasmid as previously described.9
Cell viability assay
Exponentially growing cells (5 × 103 cells per well) were plated in each well of a 96-well plate with 0.1 mL of RPMI 1640 + 10% fetal calf serum containing the appropriate concentration of drug (ponatinib, AC220, sorafenib, DCC-2036) in triplicate. Cells were allowed to expand for 2 days, and cellular proliferation was assessed using CellTiter-Glo reagent (Promega, Madison, WI) according to the manufacturer’s recommendation on a SpectraMax M3 microplate reader using SpectraMax Pro Software (Molecular Devices, Sunnyvale, CA). The value at varying concentrations of drug was normalized to the median value in the no-drug sample for each mutant and a mean value was calculated. Numerical IC50 values were generated using nonlinear best-fit regression analysis using Prism 5 software (GraphPad, San Diego, CA).
Immunoblotting
Exponentially growing Ba/F3 cells stably expressing mutant isoforms, along with a native FLT3-ITD control, were plated in RPMI medium 1640 + 10% fetal calf serum supplemented with kinase inhibitor at the indicated concentration. After a 90-minute incubation, the cells were washed in phosphate-buffered saline and lysed in buffer (50 mM HEPES, pH 7.4, 10% glycerol, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2) supplemented with protease and phosphatase inhibitors. The lysate was clarified by centrifugation and quantitated by BCA assay (Thermo Scientific, Rockford, IL). Protein was subjected to sodium dodecyl sulfate polyacrylamide electrophoresis and transferred to nitrocellulose membranes. Immunoblotting was performed using anti–phospho-FLT3, anti–phospho-STAT5, anti-STAT5, anti–phospho-S6, anti-S6 (Cell Signaling, Beverly, MA), and anti-FLT3 S18 antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Plasma inhibitory assay
Plasma inhibitory assay was performed as previously described21 with some modifications. Briefly, 7 × 106 parental Molm14 cells and Molm14 cells harboring the indicated mutations were resuspended in 1 mL of plasma from either a healthy control or steady-state plasma from a ponatinib-treated patient. Plasma was obtained from healthy volunteers and from CML patients treated on the phase I study of ponatinib (NCT00660920) at UCSF. All samples were collected under the UCSF institutional review board–approved cell-banking protocol (CC#112514). Informed consent was obtained in accordance with the Declaration of Helsinki. Cells were incubated at 37°C for 2 hours. FLT3 was immunoprecipitated from 400 μg of total protein using anti-FLT3 S18 antibody and used for Western immunoblotting. Remaining lysate was normalized and used for Western immunoblotting as described previously.
Model of ponatinib bound to FLT3
A homology model of FLT3 was built based on the crystal structure of KIT bound with imatinib (PDB code: 1T46) by using the protein structure prediction program Prime (Schrodinger). A docking grid file was generated from this model and ponatinib was docked into the FLT3 model using Glide SP (Schrodinger) with postdocking minimization. The docked poses of ponatinib with highest scores revealed similar binding modes, hence the model of FLT3 bound to ponatinib with the best binding score was chosen for further analysis.
Results
Activity of ponatinib against AC220 resistance-conferring FLT3-ITD KD mutants
We first sought to assess the in vitro activity of ponatinib against clinically relevant AC220 resistance–conferring FLT3-ITD KD mutants F691L and D835V/Y/F. Ponatinib potently inhibited the proliferation of Ba/F3 cells stably expressing native FLT3-ITD, with an inhibitory concentration 50 (IC50) of 3 nM, and retained activity, albeit moderately reduced, against the clinically relevant F691L mutant with an IC50 of 52 nM (Table 1 and Figure 1A). Consistent with a recent report,22 FLT3-ITD/F691I (analogous to BCR-ABL/T315I), is sensitive to ponatinib at concentrations equivalent to those required to inhibit native FLT3-ITD (Table 1 and Figure 1). Although the F691I mutant has been shown to confer in vitro resistance to a number of FLT3 TKIs,12 it has not been implicated in clinical resistance to AC220 or any other agent to date. Notably, ponatinib was substantially less active against Ba/F3 cells expressing FLT3-ITD AL mutants D835V/Y/F in both cell-based proliferation and biochemical assays of phosphorylation of FLT3 and its downstream target, STAT5 (Table 1 and Figure 1B). Derivatives of the human FLT3-ITD+ cell line Molm14 harboring the F691I, F691L, or D835Y mutations, which were established through selection in escalating concentrations of AC220, recapitulated these findings in cell proliferation assays (supplemental Figure 1).
IC50 values of Ba/F3 cells expressing FLT3-ITD and mutant isoforms in the presence of AC220, sorafenib, ponatinib, or DCC-2036
| Mutation . | IC50 for cellular proliferation (nM) . | |||
|---|---|---|---|---|
| AC220 . | Sorafenib . | Ponatinib . | DCC-2036 . | |
| Parental + IL3 | >1000 | 3761 | >1000 | >5000 |
| ITD alone | 0.13 | 1.3 | 3.0 | 19 |
| ITD + F691L | 102 | 1189 | 52 | 49 |
| ITD + F691I | 122 | 648 | 4.2 | 51 |
| ITD + D835V | 120 | 2209 | 349 | 255 |
| ITD + D835Y | 28 | 675 | 284 | 315 |
| ITD + D835F | 166 | 2374 | 414 | 1417 |
| ITD + D835H | 5.5 | 164 | 211 | 148 |
| ITD + D839G | 1.9 | 112 | 54 | 51 |
| ITD + Y842C | 33 | 469 | 229 | 47 |
| ITD + Y842H | 18 | 260 | 80 | 30 |
| Mutation . | IC50 for cellular proliferation (nM) . | |||
|---|---|---|---|---|
| AC220 . | Sorafenib . | Ponatinib . | DCC-2036 . | |
| Parental + IL3 | >1000 | 3761 | >1000 | >5000 |
| ITD alone | 0.13 | 1.3 | 3.0 | 19 |
| ITD + F691L | 102 | 1189 | 52 | 49 |
| ITD + F691I | 122 | 648 | 4.2 | 51 |
| ITD + D835V | 120 | 2209 | 349 | 255 |
| ITD + D835Y | 28 | 675 | 284 | 315 |
| ITD + D835F | 166 | 2374 | 414 | 1417 |
| ITD + D835H | 5.5 | 164 | 211 | 148 |
| ITD + D839G | 1.9 | 112 | 54 | 51 |
| ITD + Y842C | 33 | 469 | 229 | 47 |
| ITD + Y842H | 18 | 260 | 80 | 30 |
Activity of ponatinib against AC220 resistance–conferring FLT3-ITD KD mutations. (A) Relative proliferation of Ba/F3 populations stably expressing FLT3-ITD mutant isoforms after 48 hours in various concentrations of ponatinib (error bars represent SD of triplicates from the same experiment). (B) Western blot analysis using anti–phospho-FLT3, anti–phospho-STAT5, anti-FLT3, and anti-STAT5 antibody performed on lysates from IL-3–independent Ba/F3 populations expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed to ponatinib at the indicated concentrations for 90 minutes.
Activity of ponatinib against AC220 resistance–conferring FLT3-ITD KD mutations. (A) Relative proliferation of Ba/F3 populations stably expressing FLT3-ITD mutant isoforms after 48 hours in various concentrations of ponatinib (error bars represent SD of triplicates from the same experiment). (B) Western blot analysis using anti–phospho-FLT3, anti–phospho-STAT5, anti-FLT3, and anti-STAT5 antibody performed on lysates from IL-3–independent Ba/F3 populations expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed to ponatinib at the indicated concentrations for 90 minutes.
An in vitro mutagenesis screen of FLT3-ITD in a clinically achievable concentration of ponatinib identifies mutations restricted to the FLT3-ITD AL
To delineate the spectrum of FLT3-ITD KD mutations that may potentially cause clinical resistance to ponatinib in an unbiased manner, we conducted an in vitro saturation mutagenesis screen of FLT3-ITD similar to the assay we recently used to successfully identify clinically relevant AC220-resistant KD mutations in FLT3-ITD.9 Resistant clones were selected in 40 nM ponatinib, which is less than the trough concentrations safely achieved in patients in a phase I study19 and is identical to the concentration used in a similar study of BCR-ABL.17 A screen of 50 independently derived clones revealed mutations at 3 AL residues (D835 [n = 31], D839 [n = 3], and Y842 [n = 16]), which confer varying degrees of resistance to ponatinib (Figure 2A). D835V, D835Y, Y842C, and Y842H substitutions identified in this screen were also isolated in a previous screen using 20 nM AC220. Although D835V and Y mutants are highly resistant to both AC220 and ponatinib, Y842 substitutions confer intermediate resistance to these inhibitors (Table 1 and Figure 2B-C) and have not been identified in clinical samples to date. The D835H mutation was also recovered with ponatinib and was found to confer moderate resistance to ponatinib but relatively minimal resistance to AC220 (Table 1). In addition, at low frequency, the D839G mutation was identified, which conferred mild resistance to both drugs (Table 1 and Figure 2B-C). The clinically relevant F691L mutation, which was notably the most common substitution (>50% of resistant clones) identified in our previous saturation mutagenesis screen for AC220 resistance,9 was not recovered in 40 nM ponatinib, even though D839G and F691L appear to confer a similar mild degree of relative resistance to ponatinib in vitro (Table 1 and Figure 2B-C).
Mutation screen of FLT3-ITD identifies AL KD mutations that confer resistance to ponatinib. (A) Numbers of independent ponatinib-resistant Ba/F3/FLT3-ITD subpopulations with amino acid substitution at the indicated residue obtained from a saturation mutagenesis assay (N = 50 clones). (B) Relative proliferation of Ba/F3 populations stably expressing ponatinib-resistant FLT3-ITD mutant isoforms after 48 hours in various concentrations of ponatinib (error bars represent SD of triplicates from the same experiment). Data shown are representative of multiple experiments. (C) Western blot analysis using anti–phospho-FLT3, anti–phospho-STAT5, anti-FLT3, and anti-STAT5 antibody performed on lysates from IL-3–independent Ba/F3 populations expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed to ponatinib at the indicated concentrations for 90 minutes.
Mutation screen of FLT3-ITD identifies AL KD mutations that confer resistance to ponatinib. (A) Numbers of independent ponatinib-resistant Ba/F3/FLT3-ITD subpopulations with amino acid substitution at the indicated residue obtained from a saturation mutagenesis assay (N = 50 clones). (B) Relative proliferation of Ba/F3 populations stably expressing ponatinib-resistant FLT3-ITD mutant isoforms after 48 hours in various concentrations of ponatinib (error bars represent SD of triplicates from the same experiment). Data shown are representative of multiple experiments. (C) Western blot analysis using anti–phospho-FLT3, anti–phospho-STAT5, anti-FLT3, and anti-STAT5 antibody performed on lysates from IL-3–independent Ba/F3 populations expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed to ponatinib at the indicated concentrations for 90 minutes.
Compound FLT3-ITD/F691L-AL mutations confer increased in vitro resistance to ponatinib
In anticipation that ponatinib may be clinically useful in patients who develop resistance to AC220 as a result of acquired F691L mutations (provided adequate drug levels can be attained in vivo to inhibit this mutant), we sought to determine whether FLT3 AL mutations identified in our mutagenesis screen could impart increased relative resistance to ponatinib when found in conjunction with FLT3-ITD/F691L. We created Ba/F3 cell lines containing FLT3-ITD/F691L and substitutions at FLT3 AL residues (D835, D839, and Y842) on the same allele, and determined the IC50 values for proliferation compared with FLT3-ITD and FLT3-ITD/F691L alone. All compound F691L-AL mutants assessed conferred a high degree of resistance to ponatinib (Table 2), including substitutions that conferred only mild to moderate resistance to ponatinib in the setting of FLT3-ITD alone. These data suggest that compound FLT3-ITD/F691L AL mutations (and perhaps other compound mutations as well) will likely cause acquired clinical resistance to ponatinib in some FLT3-ITD+ AML patients who have developed resistance to prior TKI therapy.
IC50 values of Ba/F3 cells expressing FLT3-ITD/F691L mutant isoforms in the presence of ponatinib
| Mutation . | IC50 for cellular proliferation (nM) . |
|---|---|
| Parental + IL3 | >1000 |
| ITD alone | 2.5 |
| ITD + F691L | 61 |
| ITD + F691L/D835V | 632 |
| ITD + F691L/D835Y | 409 |
| ITD + F691L/D835F | 742 |
| ITD + F691L/D835H | 300 |
| ITD + F691L/D839G | 393 |
| ITD + F691L/Y842C | 344 |
| ITD + F691L/Y842H | 379 |
| Mutation . | IC50 for cellular proliferation (nM) . |
|---|---|
| Parental + IL3 | >1000 |
| ITD alone | 2.5 |
| ITD + F691L | 61 |
| ITD + F691L/D835V | 632 |
| ITD + F691L/D835Y | 409 |
| ITD + F691L/D835F | 742 |
| ITD + F691L/D835H | 300 |
| ITD + F691L/D839G | 393 |
| ITD + F691L/Y842C | 344 |
| ITD + F691L/Y842H | 379 |
Modeling of FLT3-ponatinib interactions
To understand the potential structural effects of secondary FLT3 KD mutations on ponatinib binding, we used a docking model of ponatinib bound to FLT3, based on the crystal structure of the close homolog KIT bound with imatinib.23 In this model, ponatinib is shown to bind to the inactive form of FLT3 in a DFG-out binding mode and occupies three major pockets, including the ATP pocket, the hydrophobic pocket located behind the gatekeeper residue, and the extended hydrophobic pocket near the DFG motif. We first sought to understand the effects of mutations at the F691 residue on ponatinib’s activity from a structural perspective. As illustrated in Table 1, ponatinib inhibits native and F691I mutant-FLT3 with similar potencies but has reduced activity against the F691L gatekeeper mutant. The gatekeeper residue F691 in native FLT3 does not block the hydrophobic pocket located near the residue. This is demonstrated in both the crystal structure of auto-inhibited FLT324 and in our homology model, where the phenylalanine adopts a similar conformation and is folded back from the ATP binding site, forming the bottom of this hydrophobic pocket. No conformational change of F691 appears to be necessary to accommodate ponatinib binding to FLT3 where the side chain of F691 and the 2-methylphenyl substituent of ponatinib are oriented in a parallel fashion and make strong hydrophobic interactions (Figure 3A). When the F691 residue is mutated to isoleucine, the side chain of isoleucine can easily adopt a similar conformation to that of F691, particularly in the hydrophobic pocket, without introducing significant protein structural changes or obstructing inhibitor binding (Figure 3A-B). Conversely, mutation of F691 to leucine is expected to have a more direct steric influence on ponatinib binding because of the presence of a terminal sp3 carbon-branched side chain in leucine compared with the flat phenylalanine side chain (Figure 3A,C). Indeed, minor steric clashes with inhibitor are observed in the F691L, but not the F691I, model of FLT3 bound with ponatinib (Figure 3B-C). Furthermore, because the hydrophobic nature of the side chain is maintained in this phenylalanine-to-leucine substitution, reinforcement of the hydrophobic spine with resultant stabilization of the active conformation that leads to autoactivation, as reported in the case of T315I ABL,25 is unlikely to play a dominant role in influencing kinase activity or binding of ponatinib (as a type II kinase inhibitor) to F691L mutants in FLT3. Importantly, we observed that the FLT3 F691L mutant is not significantly autophosphorylated when transiently expressed in 293T cells without a co-occurring ITD, and hence is unactivated (supplemental Figure 2), supporting the notion that the effect of the F691L mutation on the activity of ponatinib is primarily steric in nature.
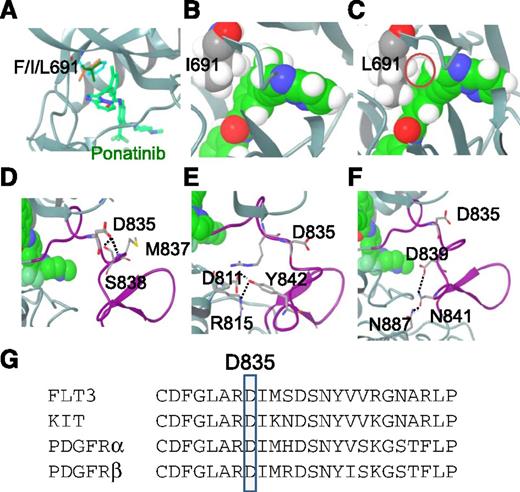
Modeling of FLT3-ponatinib interactions. (A) Docking model of FLT3 bound to ponatinib: gatekeeper residue F691 (cyan), I691 (brown), and L691 (green). Although F691 and I691 are compatible with ponatinib binding, L691 appears to be more intruding into the inhibitor binding pocket. (B) Space-filling model of F691I (gray) FLT3 bound with ponatinib (green). Hydrogen atoms are shown in white. (C) Space-filling model of F691L FLT3 bound with ponatinib. The minor steric clash between L691, particularly the hydrogen atom of the side chain, and ponatinib is highlighted with a red circle. (D) Model of ponatinib bound to FLT3/D835. D835 is located on the AL as shown in the purple segment. D835 makes hydrogen bond interactions with the side chain of S838 and main chain of M837, as represented by the black dashed line. (E) Model of ponatinib bound to FLT3/Y842. Y842 is located in the AL shown in purple. It makes hydrogen bonds with the side chains of D811 and R815. (F) Model of ponatinib bound to FLT3/D839. D839 forms part of the hydrogen bond network involving D839, N841, and N887, as represented by the black dashed lines. (G) AL amino-acid sequence alignment of FLT3, KIT, and PDGFR-α and -β, showing homology of FLT3/D835, KIT/D816, and PDGFR-α/D842, -β/D850.
Modeling of FLT3-ponatinib interactions. (A) Docking model of FLT3 bound to ponatinib: gatekeeper residue F691 (cyan), I691 (brown), and L691 (green). Although F691 and I691 are compatible with ponatinib binding, L691 appears to be more intruding into the inhibitor binding pocket. (B) Space-filling model of F691I (gray) FLT3 bound with ponatinib (green). Hydrogen atoms are shown in white. (C) Space-filling model of F691L FLT3 bound with ponatinib. The minor steric clash between L691, particularly the hydrogen atom of the side chain, and ponatinib is highlighted with a red circle. (D) Model of ponatinib bound to FLT3/D835. D835 is located on the AL as shown in the purple segment. D835 makes hydrogen bond interactions with the side chain of S838 and main chain of M837, as represented by the black dashed line. (E) Model of ponatinib bound to FLT3/Y842. Y842 is located in the AL shown in purple. It makes hydrogen bonds with the side chains of D811 and R815. (F) Model of ponatinib bound to FLT3/D839. D839 forms part of the hydrogen bond network involving D839, N841, and N887, as represented by the black dashed lines. (G) AL amino-acid sequence alignment of FLT3, KIT, and PDGFR-α and -β, showing homology of FLT3/D835, KIT/D816, and PDGFR-α/D842, -β/D850.
The other class of ponatinib-resistant mutations occurs on certain amino acids located in the AL of FLT3, including D835, Y842, and D839. D835 is located 4 residues C-terminal to the DFG motif in the AL of FLT3. In both the crystal structure of auto-inhibited FLT3 and the model of FLT3 bound with ponatinib, D835 plays a critical role in stabilizing the DFG-out inactive conformation of the AL. D835 forms bifurcated hydrogen bond interactions with the side chain of S838 and the main chain of M837, both of which are located on the AL C-terminal to D835 (Figure 3D). Mutation of D835 to tyrosine, valine, phenylalanine, or histidine would result in loss of the hydrogen bond interactions, helping shift the equilibrium of FLT3 toward an activated state, which explains why mutations at this residue are activating mutations.26 With respect to inhibitor binding, there are no direct molecular interactions between residue D835 and ponatinib; however, ponatinib binding is predicted to be dependent on a DFG-out conformation of the FLT3 KD. The interactions of D835 with its neighboring residues located on the AL in the native FLT3 help stabilize this DFG-out inactive conformation, whereas the D to V or D to Y/F/H mutations all destabilize this conformation through loss of hydrogen bond capability, thereby decreasing the binding affinity of ponatinib to FLT3. Furthermore, D835 is solvent exposed with limited molecular packing interactions with other residues on the AL. This may explain why all D835 mutations result in approximately the same reduction in potency against ponatinib (Table 1). Interestingly, the homologous aspartic acid residues in KIT and PDGFR-α or β (Figure 3G) appear to play a similar role, as shown in the crystal structures of KIT and other similar biochemical assays.23 Mutation at this position often leads to activation of these kinases and reduced binding affinity to kinase inhibitors.27,28
Y842 is also located in the AL of FLT3, 7 residues C-terminal to D835. In the model and crystal structure of auto-inhibited FLT3, Y842 makes bifurcated hydrogen bonds to D811 and is hydrogen-bonded to R815 (Figure 3E). In addition, Y842 also makes extensive hydrophobic packing interactions with neighboring residues, including D839, R834, I836, and P851. Similar to Y842, D839 is located in the AL and is involved in a hydrogen bond network that stabilizes the conformation of the AL. As shown in Figure 3F, D839 is hydrogen-bonded to the neighboring residue N841 of the AL, which in turn makes an additional hydrogen bond to N887 from an α helix in the vicinity of the AL. Both Y842 and D839 therefore play a similar role to D835 in stabilizing the DFG-out inactive form of FLT3. Mutation of Y842 to C or H, or D839 to G, would result in loss of these molecular interactions, leading to the destabilization of the inactive form of FLT3 and reduced binding affinity to ponatinib. Consistent with the idea that these substitutions are activating, each mutation appears to exhibit some level of autophosphorylation when transiently expressed in 293T cells in the absence of an ITD (supplemental Figure 2), and have all been reported to occur de novo in AML patients.29,30 In contrast to the solvent exposed D835, Y842 is mostly buried inside protein; hence, the Y842H mutation may preserve more molecular packing interactions with its neighboring residues than Y842C, resulting in less destabilization of the inactive form of FLT3 and a smaller effect on the inhibitory activity of ponatinib (Table 1).
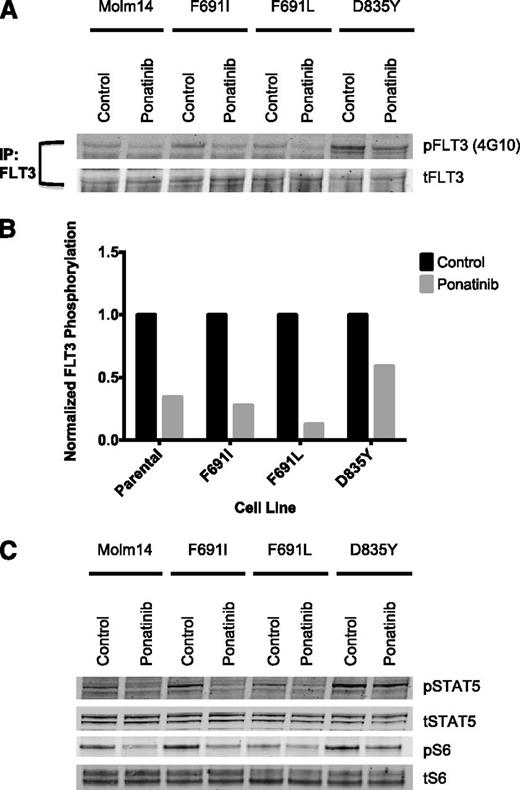
Plasma isolated from ponatinib-treated CML patients inhibits signaling of FLT3-ITD and FLT3-ITD/F691 mutants
To test whether clinically achieved plasma concentrations of ponatinib are sufficient to biochemically inhibit FLT3 autophosphorylation and downstream signaling of a gatekeeper mutant, we performed a plasma inhibitory assay using plasma obtained at steady state from a CML patient treated with the clinically approved 45-mg daily dose of ponatinib. Western blot demonstrates equivalent inhibition of phosphorylated FLT3 in parental Molm14 cells and in AC220-resistant Molm14 cells harboring F691I or F691L mutations upon exposure to ponatinib-containing plasma (Figure 4A-B), but reduced inhibition in cells expressing the ponatinib-resistant D835Y mutation. Plasma from this patient was also able to inhibit downstream phosphorylation of STAT5, a putative direct substrate of FLT3-ITD,31 and ribosomal S6, a clinically relevant biomarker of FLT3 inhibition in patients (Figure 4C).32
Plasma inhibitory assay shows ponatinib is active against FLT3-ITD and FLT3-ITD/F691 mutants at clinically achievable plasma concentrations. (A) Western blot analysis for phosphotyrosine and total FLT3 performed after immunoprecipitation using anti-FLT3 antibody on lysates prepared from Molm14 cells expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed for 120 minutes to healthy control and steady-state plasma obtained from a patient treated with 45 mg of ponatinib per day. (B) Quantitation of FLT3 autophosphorylation after immunoprecipitation from Molm14 cells expressing the indicated FLT3 mutant isoforms. (C) Western blot analysis using anti–phospho-S6, anti–phospho-STAT5, anti-S6, and anti-STAT5 antibody performed on whole-cell lysates prepared from Molm14 cells expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed for 120 minutes to normal control and steady-state plasma obtained from a patient treated with 45 mg ponatinib per day.
Plasma inhibitory assay shows ponatinib is active against FLT3-ITD and FLT3-ITD/F691 mutants at clinically achievable plasma concentrations. (A) Western blot analysis for phosphotyrosine and total FLT3 performed after immunoprecipitation using anti-FLT3 antibody on lysates prepared from Molm14 cells expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed for 120 minutes to healthy control and steady-state plasma obtained from a patient treated with 45 mg of ponatinib per day. (B) Quantitation of FLT3 autophosphorylation after immunoprecipitation from Molm14 cells expressing the indicated FLT3 mutant isoforms. (C) Western blot analysis using anti–phospho-S6, anti–phospho-STAT5, anti-S6, and anti-STAT5 antibody performed on whole-cell lysates prepared from Molm14 cells expressing the FLT3-ITD mutant isoforms indicated. Cells were exposed for 120 minutes to normal control and steady-state plasma obtained from a patient treated with 45 mg ponatinib per day.
Switch control inhibitor DCC-2036 is inactive against FLT3 AL mutations
DCC-2036 is an investigational first-in-class switch control inhibitor of multiple kinases, including ABL and FLT3. The ability of DCC-2036 to bind the transiently formed switch-control pocket of ABL allows it to retain activity against BCR-ABL/T315I by avoiding direct contact with the T315 residue.33 DCC-2036 also demonstrates the ability to bind both phosphorylated and nonphosphorylated ABL, suggesting it may be able to overcome some of the resistance caused by activating FLT3 AL mutations. To determine whether DCC-2036, with its unique mode of binding and demonstrated in vitro activity against the BCR-ABL/T315I mutation, would retain activity against AC220 and ponatinib-resistant FLT3 KD mutations, we assessed its activity against our panel of drug-resistant FLT3-ITD KD mutations. DCC-2036 was able to inhibit the proliferation of Ba/F3 cells containing the FLT3 gatekeeper F691L and F691I mutations at IC50 concentrations (49 nM and 51 nM), only slightly higher than that required to inhibit FLT3-ITD alone (19 nM), and retained activity against select FLT3 AL mutations (D839G, Y842C/H) at similar drug concentrations. However, mutations at D835 were highly resistant to DCC-2036 (Table 1). In general, DCC-2036 was the least potent of all tested inhibitors against FLT3-ITD (Table 1), suggesting it may be less likely to be effective in AML patients at clinically achievable doses.
Discussion
Recent clinical experience with AC220 and sorafenib in FLT3-ITD+ AML has demonstrated the ability of FLT3 TKIs to achieve complete responses, even in the setting of chemotherapy-resistant disease. Unfortunately, relapses typically occur rapidly and are commonly caused by the acquisition of drug-resistant mutations in the KD that presumably negatively affect drug binding. Although it is anticipated that FLT3-independent (“off-target”) mechanisms will also confer acquired resistance to clinically active FLT3 TKIs, experience to date strongly suggests that potent next-generation FLT3 inhibitors that retain activity against AC220 resistance–conferring mutants hold considerable therapeutic promise.
Here we confirm that the multikinase inhibitor ponatinib, which is clinically active in CML and has demonstrated clinical activity in FLT3-ITD+ AML in limited phase I experience, retains in vitro activity against the pan-resistant FLT3-ITD/F691I gatekeeper mutant, analogous to BCR-ABL/T315I. Moreover, we demonstrate that ponatinib largely retains activity in vitro (at clinically achievable drug concentrations) against the clinically relevant FLT3-ITD/F691L mutant that was noted to evolve in 3 of 8 analyzed AML patients with acquired resistance to AC220. Notably, F691L was the most commonly detected FLT3-ITD mutation recovered from an in vitro saturation mutagenesis screen using AC2209 and has been implicated in in vitro resistance to other FLT3 inhibitors currently in clinical development.34 Encouragingly, an in vitro saturation mutagenesis screen of FLT3-ITD using a concentration of ponatinib (40 nM) identical to that used in a previously reported screen for ponatinib-resistant BCR-ABL mutations,17 and below that achieved clinically (cmax:145 nM, trough:64 nM) in patients treated on a phase I study with the recommended phase II dose of 45 mg per day,19 identified only FLT3 AL substitutions as a cause of ponatinib resistance, including two mutants with IC50s for proliferation that lie within the range of clinically achievable concentrations (D839G and Y842H). The relative preservation of activity of ponatinib against the F691L mutant warrants clinical investigation of this agent in patients with TKI-resistant AML who harbor this substitution. However, only clinical and translational studies of patients treated with ponatinib can definitively assess the potential clinical activity of this agent in AML patients with F691L mutations. Notably, AL substitutions at the D835 residue in FLT3-ITD, which are commonly associated with acquired clinical resistance to AC220 and sorafenib, confer a high degree of cross-resistance to ponatinib in vitro and are thus predicted to represent an important cause of clinical resistance to this agent and others, including the switch-control inhibitor DCC-2036. Ponatinib is presumed to bind to FLT3 in an inactive, DFG-out conformation (“type II inhibitor”), similar to the manner in which it is known to bind to ABL.17 It is predicted that a “type I” TKI, capable of binding to FLT3 in an active, DFG-in conformation, will be required to adequately inhibit the kinase activity of FLT3-ITD D835 mutants.
In light of its preclinical and early clinical promise and favorable toxicity profile, ponatinib may represent a useful second-line TKI in AML patients with acquired resistance to AC220 or sorafenib as a consequence of secondary KD mutation at F691, and thus warrants more formal investigation in TKI-pretreated as well as TKI-naïve FLT3-ITD+ AML patients. Further categorization of the spectrum of resistance-conferring mutations to clinically active FLT3 inhibitors through translational studies promises to facilitate a more personalized approach toward patients with FLT3-ITD+ AML treated with TKIs. Although ponatinib appears to be active against all TKI-resistant BCR-ABL mutants, it is clearly ineffective against multiple FLT3 AL mutants in vitro, and a combination of potent FLT3 TKIs may therefore be required to adequately suppress all resistance-conferring mutants and substantially improve response duration. To that end, ponatinib may represent a cornerstone upon which an effective FLT3 TKI combination strategy can be built.
Presented in abstract form at the 53rd annual meeting of the American Society of Hematology, San Diego, CA, December 6, 2011.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Art and Alison Kern and the Edward S. Ageno family for their support; and Drs David Dalgarno, Victor Rivera, and Joseph Gozgit for their comments and suggestions on the manuscript.
This work was supported by grants from the National Cancer Institute (1RO1 CA166616-01) (N.P.S.), the National Institutes of Health T-32 Molecular Mechanisms of Cancer (C.C.S. and E.A.L.), and the Leukemia and Lymphoma Society (C.C.S. and N.P.S.).
Authorship
Contribution: C.C.S., E.A.L., and X.Z. designed experiments, performed research, analyzed data, and wrote the manuscript. N.P.S. designed experiments, analyzed data, and wrote the manuscript. L.E.D., K.C.L., W.K.S., and S.S. performed experiments and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare the following competing financial interests. C.C.S (research funding, Plexxikon); N.P.S. (research funding associated with the conduct of clinical trials, ARIAD Pharmaceuticals, Ambit Biosciences, Plexxikon); and X.Z. is an employee of and holds equity interest in ARIAD Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Neil P. Shah, M.D., Ph.D., 505 Parnassus Ave, Suite M1286, Box 1270, San Francisco, CA 94143; e-mail: nshah@medicine.ucsf.edu.