Key Points
Patients with 58-66 chromosomes have 99% event-free survival and 100% overall survival in the 58951 EORTC-CLG study.
The higher the ploidy, the better the prognosis in the 58951 EORTC-CLG study.
Abstract
The aim of our study was to analyze the factors contributing to heterogeneity of prognosis in patients with hyperdiploidy>50 chromosomes (HD>50), a group of B-cell precursor acute lymphoblastic leukemia with favorable outcome. The 541 HD>50 patients registered prospectively in the 58951 European Organisation for Research and Treatment of Cancer (EORTC) Children’s Leukemia Group (CLG) trial, identified by karyotype (446 patients) and by DNA index (DI) (490 patients), had a 6-year event-free survival (EFS) of 89.0% (standard error [SE] = 1.5%) and a 6-year overall survival (OS) of 95.9% (SE = 0.9%). The strongest prognostic factor was the modal number of chromosomes (MNC): the 6-year EFS of 51-53, 54-57, and 58-66 MNC groups were 80%, 89%, and 99%, respectively (P < .0001). Ploidy assessed by DI was also a favorable factor: the higher the DI, the better the outcome. The 6-year EFS of the 3 subgroups of DI < 1.16/≥1.16-<1.24/≥1.24 were 83%, 90%, and 95%, respectively (P = .009). All usual combinations of trisomies (chromosomes 4, 10, 17, 18) were significant favorable factors but had lower EFS when MNC was lower than 58. In multivariate analysis, MNC remained the strongest factor. Consequently, the best indicator for excellent outcome was ploidy assessed by karyotype because patients with 58-66 chromosomes stood every chance of being cured (OS of 100% at 6-year follow-up) with less-intensive therapy. This trial was registered at www.clinicaltrials.gov as #NCT00003728. Registered: http://www.eortc.org/, http://clinicaltrials.gov/show/NCT00003728.
Introduction
Hyperdiploidy higher than 50 chromosomes (HD>50), also called high hyperdiploidy, has been recognized as a distinct entity of favorable outcome among B-cell precursor acute lymphoblastic leukemias (B-preALLs) since the early 1980s.1-3
In HD>50, the modal number of chromosomes (MNC) ranges from 51 to 66 or 67 chromosomes, peaks at 55 or 56, and the chromosome gains occur nonrandomly: gains of chromosomes X, 4, 6, 10, 14, 17, 18, and 21 are common within the range MNC 51-54, trisomies of chromosomes 8, 5, 11, 12 appear as of MNC 57-60 in addition to the former series, whereas gains of chromosomes 2, 3, 9, 16, and 22 are observed among the highest MNC 63-67 as evidenced in Heerema’s study.4-6
Hyperdiploidies can also be detected by flow cytometry (FCM), which measures the cellular DNA content and allows determination of the DNA index (DI).7 In HD>50, DIs range from 1.10 to 1.45, and the cutoff 1.16 corresponds to MNC 53 or 54.7,8
The first study showing that outcome is heterogeneous within HD>50 has been based on DI.7 This study has established that hyperdiploidies with DI ≥ 1.16 are the most favorable group in HD>50. Another study based on MNC has revealed that patients with 56-67 chromosomes fare better than those with 51-55.5 The outcome of HD>50 has improved over years and the overall survival of patients with DI ≥ 1.16 treated with current therapies now ranges around 95% to 96%.9-12 Other studies have focused on the favorable prognostic impact of specific trisomies or association of trisomies: +18 for the Medical Research Council group,4,13 +4,+10 for the Pediatric Oncology Group,14 +10,+17 for the Children’s Cancer Group,15 and +4,+10,+17 on a meta-analysis of the Children’s Cancer Group and Pediatric Oncology Group,16 leading the Children’s Oncology Group group to use +4,+10,+17 and +4,+10 in conjunction with other features for stratifying patients in low risk arms.17,18 However, the prognostic value of these combinations has not been validated by other protocols.4,5,13
Few studies have focused on the prognostic impact of structural abnormalities observed in HD>50. The poorer outcome initially reported by Pui et al19 has not been corroborated by other large series with long follow-ups,4,5,13 except for 2 reports relying on small series of patients.20,21
Given this absence of consensus, the current trials use different criteria for identifying patients with best prognosis. The large group of HD>50 identified by both cytogenetic analyses and DI in the 58951 European Organisation for Research and Treatment of Cancer (EORTC) Children’s Leukemia Group (CLG) protocol enabled us to reassess these prognostic factors.
Materials and methods
Patients
A total of 541 children, presenting with B-preALL and HD>50, were included in this study. They were registered in the 58951 CLG-EORTC protocol that recruited 2039 patients over a 10-year period from 1998 to 2008. Diagnosis of B-lineage ALL was established according to European Group for the Immunological Characterization of Leukemia criteria.22 Minimal residual disease (MRD) monitoring was based on polymerase chain reaction quantitation of clonospecific T-cell receptor and immunoglobulin gene rearrangements as previously described.23 All patients were treated according to the 58951 EORTC-CLG protocol, a Berlin-Frankfurt-Münster–like trial with treatment phases including induction (IA), consolidation (IB/IB′), late intensification II and maintenance, associated with chemotherapeutic central nervous system (CNS) prophylaxis without any cranial irradiation. Patients were assigned to different risk groups: very low risk (VLR), average risk (AR), and very high risk (VHR). VLR was defined as white blood cell (WBC) counts <10 × 109 per liter, HD>50 identified either by karyotype or by DI > 1.16, and no CNS or gonadal involvement. VHR criteria consisted of blast counts in peripheral blood (PB) ≥ 1 × 109 per liter at completion of the prephase (1 week of corticosteroids and intrathecal injection of methotrexate), acute undifferentiated leukemia, failure to achieve complete remission, or MRD ≥ 10−2 (≥1000 blasts/100 000 mononuclear cells) at completion of induction IA (D35). All AR patients were children without VLR or VHR characteristics subdivided in AR low (WBC < 100 × 109 per liter) and AR high (WBC ≥ 100 × 109 per liter or CNS/gonadal involvement) groups described in more detail in de Moerloose et al.24 This protocol was accepted by the EORTC Protocol Review Committee and by each local ethics committee in Belgium, France, and Portugal. In each center, informed consents were obtained from children’s parents before enrolling them in the study in accordance with the Declaration of Helsinki.
Methods
Both cytogenetics and FCM were used for identifying HD>50.
Cytogenetics.
Cytogenetic analyses were performed locally in 22 cytogenetic centers participating in the study, reviewed by N.D. (France, Portugal) and F.S. (Belgium). A successful karyotype of HD>50 required at least 3 abnormal metaphases. Poor-quality karyotypes with all chromosome gains difficult to ascertain but with a modal number clearly assessable were classified as successful karyotypes.
Fluorescence in situ hybridization (FISH) analyses were carried out with centromeric probes for testing gains of chromosomes X,4,6,10,14,17,18 and with probes used for FISH screening of t(9;22)/BCR-ABL, t(12;21)/ETV6-RUNX1, and 11q23/MLL that provided data on gains of chromosomes 9, 22, 12, 21, and 11, respectively.
Flow cytometry.
DI was measured locally according to standard techniques based on Vindelov’s guidelines25 and expressed as the DNA content ratio of blast cells vs reference diploid cells. In order to assess the validity of the DIs measured in each center (mDIs), the theoretical DIs (thDIs) were calculated when feasible, ie, when all chromosome gains were clearly identified on karyotypes.8
Criteria for eligibility.
Karyotypes presenting both a modal number ranging from 51 to 67 and a classical profile of chromosome gains of HD>50 were included. Because the objective of the study was to analyze the outcome of HD>50, patients with recurrent translocations or with Down syndrome were excluded.
DI was also used for detecting HD>50. When karyotypes were successful, DIs were retained whatever their value, but when they were not available, only DIs ranging from 1.10 to 1.45 were considered as indicative of HD>50.
When only FISH techniques could be used, a full set of probes comprising at least chromosomes X, 4, 6, 10, 14, 17, 18, 21 was required in order to include a patient in HD>50.
Statistical methods
EFS was measured from date of complete remission (CR) achieved either after IA or IB/IB′ to date of relapse or death in CR; patients who did not reach CR after IA/IB/IB′ were considered as events at time 0. The overall survival (OS) was measured from date of registration in the study to death, whatever the cause. For these 2 endpoints, the follow-up of patients without an event was censored at the last date of examination.
Survival distributions were estimated according to the Kaplan-Meier technique, and standard errors (SE) of estimates were obtained via the Greenwood formula. The Cox Proportional Hazards Model was used to determine the prognostic importance of each factor analyzed and to obtain hazard ratio (HR) estimates as well as corresponding 95% confidence intervals (CIs). For ordered 3-categorical variables (eg, MNC), either 2 binary variables were considered in the Cox model in order to evaluate the HR of a hazard rate category vs the one of the baseline (MNC 51-53) or an ordered categorical variable (eg, 0, if MNC 51-53; 1, if MNC 54-57; 2, if MNC 58-66). The tests used were the overall Wald test for the former and the Wald test for linear trend for the latter.26
Results
Frequency of HD>50
The 541 patients with HD>50 represented 26% of the 2039 patients registered in the 58 951 study and 33% of the 1651 B-preALLs. Fifteen patients were excluded from the study because they also presented either a Down syndrome (6 patients) or a recurrent translocation: 4 with t(9;22)(q34;q11), 3 with t(12;21)(p13;q22), 1 with t(1;19)(q23;p13), and 1 with an MLL rearrangement.
Number of patients according to means of identification
HD>50 was detected by a successful karyotype in 446 patients (82%), by DI in 490 (91%), by both techniques in 398 patients (74%), and by FISH only in 2 (0.4%). Karyotype was not informative in 95 patients because of cytogenetic failure (55 cases), because only normal diploid cells were detected (33 cases), or because no analyses were carried out (7 cases).
Overall results
Among 541 patients, 292 were male and 249 female with a median age of 3 years (range 0-17), peaking between 2 and 5; they had low leukocytoses (median WBC 5.6 × 109 per liter; range 0.3-176) with WBC count lower than 10 × 109 per liter in 71% of patients. They were treated according to VLR, ARlow, ARhigh, and VHR risk groups in 42%, 51%, 4%, and 3%, respectively. Most patients (97%) were good responders to prephase, and 540 out of 541 achieved complete remission: 539 after induction and 1 after consolidation. MRD at D35 (assessable in 85% of patients) was <10−3 in 419 (91.5%), 10−3<10−2 in 26 (5.7%), and ≥10−2 in 13 (2.8%). After a median 6-year follow-up, 486 (90%) of patients were in continuous complete remission (CCR), 48 (9%) relapsed, and 7 (1%) died of treatment-related toxicity. Most events occurred off therapy between the second and the sixth years of follow-up with 39 of 48 relapses in bone marrow (BM), the last one occurring at 8.5 years in BM. The 6-year event-free survival (EFS) of the cohort was 89.0% (SE = 1.5%), and the 6-year OS was 95.9% (SE = 0.9%). At the last follow-up, 519 (96%) were alive.
Cytogenetic features of HD>50
Modal numbers ranged from 51 to 66 chromosomes, peaking at 54 (77 cases), 55 (65 cases), and 56 (76 cases). The gains duly identified involved chromosome 21 in 98% of patients; chromosomes 6, 4, X, 14, 4, 18, 10, and 17 in 67% to 84% of patients; chromosomes 8, 5, 9, 11, 22, and 12 in 17% to 37% of patients; and chromosomes 15, Y, 19, 7, 16, 20, 2, 3, and 13 in 6% to 10% of cases.
Structural abnormalities were detected in 47% of good-quality karyotypes (197 of 421). The most frequent fully identified changes were trisomies 1q (n = 61), deletions 6q (n = 20), isochromosomes 7q (n = 13), deletions 12p (n = 11), deletions 9p (n = 8), isochromosomes 17q (n = 7), and 14q32 abnormalities (n = 5). Abnormalities not fully identified (markers) were reported in 86 karyotypes.
DNA index
Comparison of mDI with thDI was feasible for 368 patients. The values of mDI and thDI were identical in 92 patients (24%). In the remaining population, the median difference was 0.02 (range 0.01-0.16). There were 15 patients with DI in the normal range attributable to a failure of FCM to detect the aneuploid clone. Considering that the thDI is the expected value for a given patient, we can state that mDIs were overestimated in 139 patients (38%) and underestimated in 129 (35%). However, despite these discrepancies, 80% of patients remained in the same subgroup (the 3 subgroups defined by different outcomes: <1.16/≥1.16 to <1.24/≥1.24) even with an under- or overestimated value of DI. The percentage of DIs leading to misclassification was 10% at each cutoff (1.16 and 1.24): 5% of overclassification and 5% of underclassification. The comparison of mDI and thDI obtained for each MNC (Table 1) showed that each MNC had the same median values of mDI and thDI and that mDI equal to 1.16 corresponded to MNC 54. This comparison led us to select MNC 54 as equivalent to mDI 1.16 and MNC 58 as equivalent to 1.24.
Median DNA indexes obtained for each modal number of chromosomes
| MNC . | No. of cases . | mDI, median (range) . | thDI, median (range) . |
|---|---|---|---|
| 51 | 10 | 1.09 (1-1.17) | 1.09 (1.07-1.12) |
| 52 | 20 | 1.11 (1-1.19) | 1.11 (1.08-1.13) |
| 53 | 45 | 1.13 (1-1.19) | 1.13 (1.10-1.16) |
| 54 | 64 | 1.16 (1-1.23) | 1.16 (1.12-1.18) |
| 55 | 60 | 1.17 (1.05-1.26) | 1.17 (1.14-1.24) |
| 56 | 56 | 1.19 (0.97-1.43) | 1.19 (1.15-1.22) |
| 57 | 36 | 1.21 (1-1.36) | 1.21 (1.17-1.25) |
| 58 | 29 | 1.24 (0.96-1.59) | 1.24 (1.20-1.27) |
| 59 | 10 | 1.24 (1-1.32) | 1.24 (1.21-1.32) |
| 60 | 13 | 1.28 (0.99-1.44) | 1.28 (1.27-1.33) |
| 61 | 9 | 1.30 (1.19-1.33) | 1.30 (1.28-1.34) |
| 62 | 6 | 1.32 (1.30-1.34) | 1.32 (1.29-1.36) |
| 63 | 2 | — (1.18-1.35) | 1.345 (1.34-1.35) |
| 64 | 2 | — (1.33-1.41) | 1.36 (1.36-1.36) |
| 65 | 2 | — (1.32-1.39) | 1.365 (1.36-1.37) |
| 66 | 4 | 1.41 (1.39-1.43) | 1.41 (1.39-1.43) |
| MNC . | No. of cases . | mDI, median (range) . | thDI, median (range) . |
|---|---|---|---|
| 51 | 10 | 1.09 (1-1.17) | 1.09 (1.07-1.12) |
| 52 | 20 | 1.11 (1-1.19) | 1.11 (1.08-1.13) |
| 53 | 45 | 1.13 (1-1.19) | 1.13 (1.10-1.16) |
| 54 | 64 | 1.16 (1-1.23) | 1.16 (1.12-1.18) |
| 55 | 60 | 1.17 (1.05-1.26) | 1.17 (1.14-1.24) |
| 56 | 56 | 1.19 (0.97-1.43) | 1.19 (1.15-1.22) |
| 57 | 36 | 1.21 (1-1.36) | 1.21 (1.17-1.25) |
| 58 | 29 | 1.24 (0.96-1.59) | 1.24 (1.20-1.27) |
| 59 | 10 | 1.24 (1-1.32) | 1.24 (1.21-1.32) |
| 60 | 13 | 1.28 (0.99-1.44) | 1.28 (1.27-1.33) |
| 61 | 9 | 1.30 (1.19-1.33) | 1.30 (1.28-1.34) |
| 62 | 6 | 1.32 (1.30-1.34) | 1.32 (1.29-1.36) |
| 63 | 2 | — (1.18-1.35) | 1.345 (1.34-1.35) |
| 64 | 2 | — (1.33-1.41) | 1.36 (1.36-1.36) |
| 65 | 2 | — (1.32-1.39) | 1.365 (1.36-1.37) |
| 66 | 4 | 1.41 (1.39-1.43) | 1.41 (1.39-1.43) |
This table includes the 368 patients who had available both a measured DNA index (mDI) and a fully analyzed karyotype, allowing calculation of the theoretical DI (thDI). MNC indicates modal number of chromosomes.
Correlation between MNC and DI
Table 2, in the row labeled “Measured DNA index,” shows that the populations of patients selected by MNC and DI differed for a few patients. The example of the 101 patients included in the subgroup 58-66 MNC shows that most patients (n = 66) presented DI ≥ 1.24, but 22 had DIs lower than 1.24. Three main reasons accounted for these low values of DI. One of them was linked to the range of mDIs observed among patients with 58 chromosomes: their median mDI was 1.24, but the range was wider (from 1.21 to 1.27), which explains why patients with mDIs ranging from 1.21 to 1.23 were classified in the subgroup MNC 58-66 but in the intermediate subgroup of DI. The second reason related to the underestimation of mDI evidenced by comparison of mDI with thDI (previously explained), and the third (minor) reason corresponded to measurement errors of DI, as in the 4 patients with DI lower than 1.16 incompatible with MNC 58-66.
Characteristics and outcome of the 541 patients according to karyotype
| . | Karyotype available . | . | . | ||
|---|---|---|---|---|---|
| . | MNC 51-53 (n = 87) . | MNC 54-57 (n = 258) . | MNC 58-66 (n = 101) . | Karyotype not available (n = 95) . | Total (n = 541) . |
| Gender, n (%) | |||||
| Male | 42 (48.3) | 128 (49.6) | 63 (62.4) | 59 (62.1) | 292 (54.0) |
| Female | 45 (51.7) | 130 (50.4) | 38 (37.6) | 36 (37.9) | 249 (46.0) |
| Age | |||||
| Median, years (range) | 3 (0-17) | 4 (1-17) | 3 (1-16) | 3 (1-17) | 3 (0-17) |
| <1 , n (%) | 1 (1.1) | 0 (0) | 0 (0) | 0 (0) | 1 (0.2) |
| 1-2 , n (%) | 5 (5.7) | 27 (10.5) | 9 (8.9) | 12 (12.6) | 53 (9.8) |
| 2-5 , n (%) | 58 (66.7) | 151 (58.5) | 70 (69.3) | 58 (61.1) | 337 (62.3) |
| 6-9 , n (%) | 12 (13.8) | 45 (17.4) | 11 (10.9) | 15 (15.8) | 83 (15.3) |
| ≥10 , n (%) | 11 (12.6) | 35 (13.6) | 11 (10.9) | 10 (10.5) | 67 (12.4) |
| WBC | |||||
| Median, ×109 per liter (range) | 6.9 (1-164) | 5.4 (0.4-160) | 5.4 (0.8-100.3) | 4.7 (0.3-176.4) | 5.6 (0.3-176.4) |
| <10 × 109 per liter, n (%) | 54 (62.1) | 185 (71.7) | 70 (69.3) | 73 (76.8) | 382 (70.6) |
| 10 to <25 × 109 per liter, n (%) | 15 (17.2) | 34 (13.2) | 19 (18.8) | 14 (14.7) | 82 (15.2) |
| 25 to <100 × 109 per liter, n (%) | 17 (19.5) | 35 (13.6) | 11 (10.9) | 5 (5.3) | 68 (12.6) |
| ≥100 × 109 per liter, n (%) | 1 (1.1) | 4 (1.6) | 1 (1.0) | 3 (3.2) | 9 (1.7) |
| PB blast count | |||||
| Median, × 109/L (range) | 2.3 (0-139) | 1.3 (0-141) | 0.8 (0-92) | 0.7 (0-146) | 1.2 (0-146) |
| BM blasts | |||||
| Median, % (range) | 95 (41-99) | 94 (32-99) | 92 (27-99) | 90 (14-99) | 94 (14-99) |
| NCI risk groups, n (%) | |||||
| SR | 70 (80.5) | 203 (78.7) | 85 (84.2) | 82 (86.3) | 440 (81.3) |
| HR | 17 (19.5) | 55 (21.3) | 16 (15.8) | 13 (13.7) | 101 (18.7) |
| Trisomy/tetrasomy, n (%) | |||||
| Chromosome 4 | 46 [54]‡ | 195 [78]‡ | 87 [92]‡ | 27 [90]† | 355 [77]‡ |
| Chromosome 10 | 31 [37]‡ | 172 [69]‡ | 79 [83]‡ | 25 [81]† | 307 [67]‡ |
| Chromosome 17 | 38 [46]‡ | 170 [69]‡ | 70 [77]‡ | 23 [96]† | 301 [68]‡ |
| Chromosome 18 | 39 [48]‡ | 201 [81]‡ | 71 [79]‡ | 17 [85]† | 328 [75]‡ |
| Trisomies 4, 10 | 17 [20]‡ | 131 [52]‡ | 73 [76]‡ | 21 [64] † | 242 [52]‡ |
| Trisomies 4, 10, 17 | 6 [7]‡ | 94 [37]‡ | 54 [56]‡ | 14 [41]† | 168 [36]‡ |
| Trisomies 4, 10, 18 | 5 [6]‡ | 108 [43]‡ | 56 [58]‡ | 11 [32]† | 180 [38]‡ |
| Measured DNA index, n (%) | |||||
| <1.16 | 63 (72.4) | 58 (22.5) | 4 (4.0) | 21 (22.1) | 146 (27.0) |
| ≥1.16 to <1.24 | 14 (16.1) | 156 (60.5) | 18 (17.8) | 52 (54.7) | 240 (44.4) |
| ≥1.24 | 2 (2.3) | 17 (6.6) | 66 (65.3) | 19 (20.0) | 104 (19.2) |
| Not assessed | 8 (9.2) | 27 (10.5) | 13 (12.9) | 3 (3.2) | 51 (9.4) |
| Prephase response, n (%) | |||||
| Good | 85 (98) | 249 (96.5) | 101 (100) | 89 (93.7) | 524 (96.9) |
| Poor | 2 (2) | 9 (3.5) | 0 (0) | 6 (6.3) | 17 (3.1) |
| EORTC risk groups, n (%) | |||||
| VLR | 17 (19.5) | 124 (48.1) | 56 (55.4) | 30 (31.6) | 227 (42) |
| AR low | 65 (74.7) | 111 (43.0) | 42 (41.6) | 56 (58.9) | 274 (50.6) |
| AR high | 3 (3.4) | 14 (5.4) | 3 (3.0) | 3 (3.2) | 23 (4.3) |
| VHR | 2 (2.3) | 9 (3.5) | 0 (0) | 6 (6.3) | 17 (3.1) |
| Complete remission, n (%) | 87 (100) | 257 (99.6) | 101 (100) | 95 (100) | 540 (99.8) |
| MRD at D35, n (%) | |||||
| <10−3 | 65 [83]§ | 204 [92.7]§ | 81 [96.4]§ | 69 [94.5]§ | 419 [91.5]§ |
| ≥10−3 to <10−2 | 9 [11.5]§ | 12 [5.4]§ | 2 [2.4]§ | 3 [4.1]§ | 26 [5.7]§ |
| ≥10−2 | 4 [5.1]§* | 7 [3.2]§* | 1 [1.2]§* | 1 [1.4]§* | 13 [2.8]§ |
| Not assessed | 9 | 38 | 17 | 22 | 83 |
| EFS status, n (%) | |||||
| No CR (induction death) | 0 (0) | 1 (0.4) | 0 (0) | 0 (0) | 1 (0.2) |
| CCR | 70 (80.5) | 232 (90.0) | 100 (99.0) | 84 (88.4) | 486 (89.8) |
| Relapse | 15 (17.2) | 21 (8.1) | 1 (1.0) | 11 (11.6) | 48 (8.9) |
| BM only | 10 | 12 | 1 | 8 | 31 |
| CNS only | 1 | 4 | 0 | 0 | 5 |
| CNS combined | 2 | 1 | 0 | 2 | 5 |
| Other | 2 | 4 | 0 | 1 | 7 |
| TRM | 2 (2.3) | 4 (1.6) | 0 (0) | 0 (0) | 6 (1.1) |
| Survival, n (%) | |||||
| Alive | 78 (89.7) | 249 (96.5) | 101 (100) | 91 (95.8) | 519 (95.9) |
| Dead | 9 (10.3) | 9 (3.5) | 0 (0) | 4 (4.2) | 22 (4.1) |
| . | Karyotype available . | . | . | ||
|---|---|---|---|---|---|
| . | MNC 51-53 (n = 87) . | MNC 54-57 (n = 258) . | MNC 58-66 (n = 101) . | Karyotype not available (n = 95) . | Total (n = 541) . |
| Gender, n (%) | |||||
| Male | 42 (48.3) | 128 (49.6) | 63 (62.4) | 59 (62.1) | 292 (54.0) |
| Female | 45 (51.7) | 130 (50.4) | 38 (37.6) | 36 (37.9) | 249 (46.0) |
| Age | |||||
| Median, years (range) | 3 (0-17) | 4 (1-17) | 3 (1-16) | 3 (1-17) | 3 (0-17) |
| <1 , n (%) | 1 (1.1) | 0 (0) | 0 (0) | 0 (0) | 1 (0.2) |
| 1-2 , n (%) | 5 (5.7) | 27 (10.5) | 9 (8.9) | 12 (12.6) | 53 (9.8) |
| 2-5 , n (%) | 58 (66.7) | 151 (58.5) | 70 (69.3) | 58 (61.1) | 337 (62.3) |
| 6-9 , n (%) | 12 (13.8) | 45 (17.4) | 11 (10.9) | 15 (15.8) | 83 (15.3) |
| ≥10 , n (%) | 11 (12.6) | 35 (13.6) | 11 (10.9) | 10 (10.5) | 67 (12.4) |
| WBC | |||||
| Median, ×109 per liter (range) | 6.9 (1-164) | 5.4 (0.4-160) | 5.4 (0.8-100.3) | 4.7 (0.3-176.4) | 5.6 (0.3-176.4) |
| <10 × 109 per liter, n (%) | 54 (62.1) | 185 (71.7) | 70 (69.3) | 73 (76.8) | 382 (70.6) |
| 10 to <25 × 109 per liter, n (%) | 15 (17.2) | 34 (13.2) | 19 (18.8) | 14 (14.7) | 82 (15.2) |
| 25 to <100 × 109 per liter, n (%) | 17 (19.5) | 35 (13.6) | 11 (10.9) | 5 (5.3) | 68 (12.6) |
| ≥100 × 109 per liter, n (%) | 1 (1.1) | 4 (1.6) | 1 (1.0) | 3 (3.2) | 9 (1.7) |
| PB blast count | |||||
| Median, × 109/L (range) | 2.3 (0-139) | 1.3 (0-141) | 0.8 (0-92) | 0.7 (0-146) | 1.2 (0-146) |
| BM blasts | |||||
| Median, % (range) | 95 (41-99) | 94 (32-99) | 92 (27-99) | 90 (14-99) | 94 (14-99) |
| NCI risk groups, n (%) | |||||
| SR | 70 (80.5) | 203 (78.7) | 85 (84.2) | 82 (86.3) | 440 (81.3) |
| HR | 17 (19.5) | 55 (21.3) | 16 (15.8) | 13 (13.7) | 101 (18.7) |
| Trisomy/tetrasomy, n (%) | |||||
| Chromosome 4 | 46 [54]‡ | 195 [78]‡ | 87 [92]‡ | 27 [90]† | 355 [77]‡ |
| Chromosome 10 | 31 [37]‡ | 172 [69]‡ | 79 [83]‡ | 25 [81]† | 307 [67]‡ |
| Chromosome 17 | 38 [46]‡ | 170 [69]‡ | 70 [77]‡ | 23 [96]† | 301 [68]‡ |
| Chromosome 18 | 39 [48]‡ | 201 [81]‡ | 71 [79]‡ | 17 [85]† | 328 [75]‡ |
| Trisomies 4, 10 | 17 [20]‡ | 131 [52]‡ | 73 [76]‡ | 21 [64] † | 242 [52]‡ |
| Trisomies 4, 10, 17 | 6 [7]‡ | 94 [37]‡ | 54 [56]‡ | 14 [41]† | 168 [36]‡ |
| Trisomies 4, 10, 18 | 5 [6]‡ | 108 [43]‡ | 56 [58]‡ | 11 [32]† | 180 [38]‡ |
| Measured DNA index, n (%) | |||||
| <1.16 | 63 (72.4) | 58 (22.5) | 4 (4.0) | 21 (22.1) | 146 (27.0) |
| ≥1.16 to <1.24 | 14 (16.1) | 156 (60.5) | 18 (17.8) | 52 (54.7) | 240 (44.4) |
| ≥1.24 | 2 (2.3) | 17 (6.6) | 66 (65.3) | 19 (20.0) | 104 (19.2) |
| Not assessed | 8 (9.2) | 27 (10.5) | 13 (12.9) | 3 (3.2) | 51 (9.4) |
| Prephase response, n (%) | |||||
| Good | 85 (98) | 249 (96.5) | 101 (100) | 89 (93.7) | 524 (96.9) |
| Poor | 2 (2) | 9 (3.5) | 0 (0) | 6 (6.3) | 17 (3.1) |
| EORTC risk groups, n (%) | |||||
| VLR | 17 (19.5) | 124 (48.1) | 56 (55.4) | 30 (31.6) | 227 (42) |
| AR low | 65 (74.7) | 111 (43.0) | 42 (41.6) | 56 (58.9) | 274 (50.6) |
| AR high | 3 (3.4) | 14 (5.4) | 3 (3.0) | 3 (3.2) | 23 (4.3) |
| VHR | 2 (2.3) | 9 (3.5) | 0 (0) | 6 (6.3) | 17 (3.1) |
| Complete remission, n (%) | 87 (100) | 257 (99.6) | 101 (100) | 95 (100) | 540 (99.8) |
| MRD at D35, n (%) | |||||
| <10−3 | 65 [83]§ | 204 [92.7]§ | 81 [96.4]§ | 69 [94.5]§ | 419 [91.5]§ |
| ≥10−3 to <10−2 | 9 [11.5]§ | 12 [5.4]§ | 2 [2.4]§ | 3 [4.1]§ | 26 [5.7]§ |
| ≥10−2 | 4 [5.1]§* | 7 [3.2]§* | 1 [1.2]§* | 1 [1.4]§* | 13 [2.8]§ |
| Not assessed | 9 | 38 | 17 | 22 | 83 |
| EFS status, n (%) | |||||
| No CR (induction death) | 0 (0) | 1 (0.4) | 0 (0) | 0 (0) | 1 (0.2) |
| CCR | 70 (80.5) | 232 (90.0) | 100 (99.0) | 84 (88.4) | 486 (89.8) |
| Relapse | 15 (17.2) | 21 (8.1) | 1 (1.0) | 11 (11.6) | 48 (8.9) |
| BM only | 10 | 12 | 1 | 8 | 31 |
| CNS only | 1 | 4 | 0 | 0 | 5 |
| CNS combined | 2 | 1 | 0 | 2 | 5 |
| Other | 2 | 4 | 0 | 1 | 7 |
| TRM | 2 (2.3) | 4 (1.6) | 0 (0) | 0 (0) | 6 (1.1) |
| Survival, n (%) | |||||
| Alive | 78 (89.7) | 249 (96.5) | 101 (100) | 91 (95.8) | 519 (95.9) |
| Dead | 9 (10.3) | 9 (3.5) | 0 (0) | 4 (4.2) | 22 (4.1) |
TRM, treatment-related mortality.
Percentages calculated on cases assessed by FISH.
Percentages calculated on cases with gains assessable (either on karyotype or on FISH or both).
Data obtained from patients with MRD assessable.
Patients switched to VHR: 3 in the 51-53 group, 5 in the 54-57 group, 1 in the 58-66 group, and 1 in the karyotype not available group.
Outcome
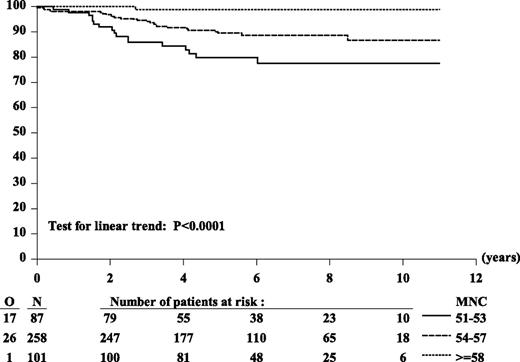
Outcome was assessed according to MNC in the 446 patients with a successful karyotype. This group is representative of the entire cohort as shown by their outcomes, 6-year EFS of 89.4% (SE = 1.6%) and 6-year OS of 96.2% (SE = 0.9%), comparable to those of the 541 HD>50. Used as a continuous variable, MNC appeared to be of prognostic importance for EFS: the higher the MNC, the lower the risk of an event (HR for each increment in MNC: 0.82, P = .003). With a cut-point at MNC 55 (MNC 51-54 vs MNC 55-66 chromosomes), the HR was 0.52 (P = .054), whereas with a cut-point of 54, it was 0.36 (P = .0006). Six-year EFS of the 87 patients with 51 to 53 chromosomes was 80%, whereas that of the 359 patients with 54 to 66 chromosomes was 92%. In the latter group, we further looked for an additional cut-point for MNC in order to define 2 subgroups comprising a sufficient number of patients with a distinct outcome. The 6-year EFS of the subgroups MNC 54-57 chromosomes (258 patients) and MNC 58-66 chromosomes (101 patients) were 89% (95% CI: 84% to 92%) and 99% (95% CI: 93% to 100%), respectively (Figure 1 and Table 2). Their OS were 97% (95% CI: 94% to 99%) and 100%, respectively.
Kaplan-Meier curves of EFS according to MNC. Six-year EFS estimates: 80% for MNC 51-54, 89% for MNC 54-57, 99% for MNC 58-66. HR MNC 58-66 vs MNC 51-54: 0.49; HR (MNC 54-57 vs MNC 51-54): 0.04 Wald test of heterogeneity: P = .0025. Considered as 3-ordered categorical variable (MNC 58-66 vs MNC 54-57 vs MNC 51-54), the estimated HR was 0.36 (95% CI, 0.22 to 0.58); Wald test for linear trend: P < .0001.
Kaplan-Meier curves of EFS according to MNC. Six-year EFS estimates: 80% for MNC 51-54, 89% for MNC 54-57, 99% for MNC 58-66. HR MNC 58-66 vs MNC 51-54: 0.49; HR (MNC 54-57 vs MNC 51-54): 0.04 Wald test of heterogeneity: P = .0025. Considered as 3-ordered categorical variable (MNC 58-66 vs MNC 54-57 vs MNC 51-54), the estimated HR was 0.36 (95% CI, 0.22 to 0.58); Wald test for linear trend: P < .0001.
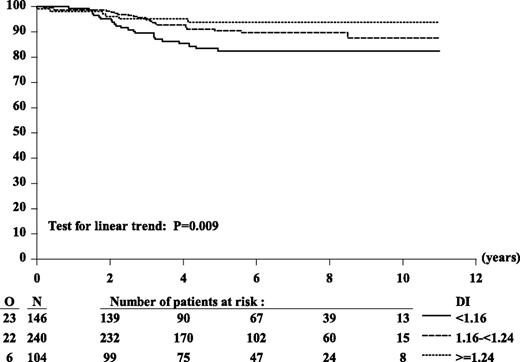
Outcome according to DI showed that patients with DI ≥ 1.16 fared better (6-year EFS: 91%) than did those with DI < 1.16 (6-year EFS: 82%) (HR: 0.49; P = .01). To analyze outcome as we did for MNC, we subdivided patients into 3 groups with 2 cutoffs: 1.16 and 1.24, equivalent to MNC 54 and 58, respectively. Patients with DI ≥ 1.24 had the best outcome (6-year EFS: 94%) compared with those with DI ≥ 1.16 to <1.24 (6-year EFS: 90%) and those with DI < 1.16 (6-year EFS: 82%) (Figure 2 and supplemental Table 1). To compare the prognostic importance of MNC and DI, we then tested only patients who had available both MNC and DI (n = 398). Analyses performed on these 398 patients showed results similar to those obtained from 490 patients with available DI.
Kaplan-Meier curves of EFS according to DI. Six-year EFS estimates: 82% (DI < 1.16), 90% (DI 1.16 to <1.24), 94% (DI ≥ 1.24). HR DI 1.16 to <1.24 vs DI < 1.16: 0.56; HR (DI ≥ 1.24 vs DI < 1.16): 0.35; Wald test of heterogeneity: P = .03. Considered as 3-ordered categorical variable (DI < 1.16 vs DI 1.16 to <1.24 vs DI ≥ 1.24), the estimated HR was 0.58 (95% CI, 0.38 to 0.87); Wald test for linear trend: P = .009.
Kaplan-Meier curves of EFS according to DI. Six-year EFS estimates: 82% (DI < 1.16), 90% (DI 1.16 to <1.24), 94% (DI ≥ 1.24). HR DI 1.16 to <1.24 vs DI < 1.16: 0.56; HR (DI ≥ 1.24 vs DI < 1.16): 0.35; Wald test of heterogeneity: P = .03. Considered as 3-ordered categorical variable (DI < 1.16 vs DI 1.16 to <1.24 vs DI ≥ 1.24), the estimated HR was 0.58 (95% CI, 0.38 to 0.87); Wald test for linear trend: P = .009.
Outcome was also tested according to presence of trisomies or combination of trisomies. Only trisomies 4 (6-year EFS: 92%; P = .025) and 18 (6-year EFS: 91%; P = .012) proved to be significant favorable factors in our study and their association (+4,+18) showed a stronger significance for good outcome (6-year EFS: 93%; P = .002). In order to compare our findings with those in other protocols, we studied the outcome of patients with double trisomies (DT) +4,+10 and triple trisomies (TT) +4,+10,+17, although isolated trisomies 10 or 17 were not of prognostic importance in our series. The 242 patients with DT fared better than did those without (6-year EFS of DT: 94% vs non-DT: 84%; P = .003), and the 168 patients with TT fared better than those without (6-year EFS of TT: 95% vs non-TT: 86%; P = .005). However, favorable outcomes were also obtained with other combinations of trisomies, and +4,+10,+18 (EFS: 95%; P = .003; HR: 0.31) proved to be similar to +4,+10,+17 (EFS: 95%; P = .006; HR: 0.34).
To compare the prognoses of these combinations of trisomies with that of our best group of MNC (MNC 58-66), we tested their outcome when they were not associated with MNC 58-66. These hierarchical variables showed that any combination of trisomies had a less favorable outcome when found in karyotypes with MNC lower than 58. The DT +4,+18 had 100% EFS in the group MNC 58-66 but only 91% when MNC ranged from 51 to 57 chromosomes (MNC 51-57), whereas the DT +4,+10 showed 100% EFS when associated with MNC 58-66 and 91% EFS for MNC 51-57. Similar results were found in all combinations of TT (EFS of +4,+10,+17: 100% when associated with MNC 58-66 and 93% for MNC 51-57) (EFS of TT +4,+10,+18: 100% when associated with MNC 58-66 and 92% for MNC 51-57). Likewise, the 4 trisomies together +4,+10,+17,+18 had a lower EFS (93%) in the MNC 51-57 group in comparison with their EFS in the MNC 58-66 group (100%).
Other factors did not appear to be of prognostic importance: gender, age, WBC, presence/absence of structural abnormalities, response to the prephase, and risk groups (supplemental Table 2). This might be due to the low number of events or to the impact of AR and VHR treatments on the outcome of the non-VLR group.
Multivariate analysis
MNC and DI were highly correlated (Table 2); therefore by including these 2 variables in a Cox model, a collinearity phenomenon occurred: the relative prognostic importance of MNC remained highly significant, whereas that of DI was no longer significant, the corresponding HR being even greater than 1 (Table 3, model 1).
Results of multivariate Cox model either in patients (N = 398) with karyotype and DNA index assessed (model 1) or in all patients (N = 446) with a karyotype assessed (models 2, 3, and 4)
| . | HR (95% CI) . | P value* . |
|---|---|---|
| Model 1 (number of EFS events = 40) | ||
| MNC: 54-57 vs 51-53 | 0.42 (0.21, 0.88) | .02 |
| MNC: 58-66 vs 51-53 | 0.04 (0.004, 0.37) | .004 |
| DI: 0 = DI <1.16; 1 = DI 1.16 to <1.24; 2 = DI ≥ 1.24 | 1.14 (0.62, 2.07) | .68 |
| Model 2 | ||
| MNC: 54-57 vs 51-53 | 0.50 (0.27, 0.93) | .03 |
| MNC: 58-66 vs 51-53 | 0.04 (0.006, 0.33) | .003 |
| WBC: 10 to <25 vs <10 × 109 per liter | 1.41 (0.66, 2.98) | .38 |
| WBC: ≥25 vs <10 × 109 per liter | 0.86 (0.36, 2.07) | .73 |
| Model 3 | ||
| MNC: 54-57 vs 51-53 | 0.50 (0.26, 0.94) | .03 |
| MNC: 58-66 vs 51-53 | 0.05 (0.006, 0.36) | .003 |
| EORTC risk group: AR low vs VLR | 0.96 (0.49, 1.88) | .91 |
| EORTC risk group: AR high vs VLR | 0.48 (0.064, 3.65) | .48 |
| EORTC risk group: VHR vs VLR | 1.97 (0.712, 5.45) | .19 |
| Model 4 | ||
| MNC: 54-57 vs 51-53 | 0.50 (0.27, 0.94) | .03 |
| MNC: 58-66 vs 51-53 | 0.05 (0.006, 0.35) | .003 |
| MRD: 10−3 to <10−2 vs undetectable | 1.73 (0.60, 4.97) | .31 |
| MRD: ≥10−2 vs undetectable | 4.03 (1.40, 11.6) | .01 |
| MRD: unknown vs undetectable | 2.39 (1.12, 11.4) | .02 |
| . | HR (95% CI) . | P value* . |
|---|---|---|
| Model 1 (number of EFS events = 40) | ||
| MNC: 54-57 vs 51-53 | 0.42 (0.21, 0.88) | .02 |
| MNC: 58-66 vs 51-53 | 0.04 (0.004, 0.37) | .004 |
| DI: 0 = DI <1.16; 1 = DI 1.16 to <1.24; 2 = DI ≥ 1.24 | 1.14 (0.62, 2.07) | .68 |
| Model 2 | ||
| MNC: 54-57 vs 51-53 | 0.50 (0.27, 0.93) | .03 |
| MNC: 58-66 vs 51-53 | 0.04 (0.006, 0.33) | .003 |
| WBC: 10 to <25 vs <10 × 109 per liter | 1.41 (0.66, 2.98) | .38 |
| WBC: ≥25 vs <10 × 109 per liter | 0.86 (0.36, 2.07) | .73 |
| Model 3 | ||
| MNC: 54-57 vs 51-53 | 0.50 (0.26, 0.94) | .03 |
| MNC: 58-66 vs 51-53 | 0.05 (0.006, 0.36) | .003 |
| EORTC risk group: AR low vs VLR | 0.96 (0.49, 1.88) | .91 |
| EORTC risk group: AR high vs VLR | 0.48 (0.064, 3.65) | .48 |
| EORTC risk group: VHR vs VLR | 1.97 (0.712, 5.45) | .19 |
| Model 4 | ||
| MNC: 54-57 vs 51-53 | 0.50 (0.27, 0.94) | .03 |
| MNC: 58-66 vs 51-53 | 0.05 (0.006, 0.35) | .003 |
| MRD: 10−3 to <10−2 vs undetectable | 1.73 (0.60, 4.97) | .31 |
| MRD: ≥10−2 vs undetectable | 4.03 (1.40, 11.6) | .01 |
| MRD: unknown vs undetectable | 2.39 (1.12, 11.4) | .02 |
Wald test.
In the whole 446 series, MNC remained of strong prognostic importance after adjustment by initial WBC or by EORTC risk groups, which were highly correlated with MNC (Tables 2 and 3). Similar results were obtained when VHR risk group also included patients switched to a VHR treatment after induction (data not shown).
The prognostic importance of MNC adjusted by MRD level evaluated on D35 remained highly significant (Table 3, model 4).
Discussion
The 541 patients in our series of HD>50 presented the standard features expected for this entity. Their frequency (26% in the 58951 trial and 33% of B-preALLs) is in agreement with other large series of childhood ALLs reported in literature (reviewed by Paulsson et al27 ) as well as the slight predominance of males (54%), the young age of patients, and the low leukocytoses. The 6-year EFS of 89.0% and the 6-year OS of 95.9% found in our series of HD>50 treated between 1998 and 2008 are in keeping with the overall improvement reported by other protocols during similar periods of time.9-13,18 Improvement in outcome, obtained not by new medication but by an optimal use of existing drugs, is observed for all cytogenetic groups, as demonstrated by the outcome of patients with t(12;21)/ETV6-RUNX1 in our 58 951 protocol (6-year EFS: 91.6%; 6-year OS: 96%).
Heterogeneity of prognosis within the HD>50 entity has already been emphasized by several authors, but the outstanding outcome of patients with the highest MNC, found in our series, has not been reported to date: we here show that patients with 58 to 66 chromosomes stand every chance of being cured (6-year EFS: 99% and 6-year OS: 100%).
The group of 101 patients with MNC 58-66 that had an excellent outcome comprised 23% of patients with a successful karyotype and 18% of the entire cohort. It is worth noting that this group consisted of more males than females (sex ratio: 1.65), had fewer peripheral blast cells at diagnosis, and was most frequently treated with less-intensive therapy. The favorable impact of increased MNC was further demonstrated by the outcome of the subgroup of 53 patients with MNC ≥ 60 chromosomes who had a 6-year EFS of 100%, no positive MRD at D35, and no relapses. The male preponderance was even stronger in this small group (sex ratio: 1.8) as well as the frequent VLR stratification (64% of patients). In the MNC 58-66 group, the 20 patients with potential adverse risk factors were all in CCR at 6-year follow-up. They included 16 National Cancer Institute high-risk patients with undetectable MRD at D35 (14 assessable) treated according to VLR, ARlow, and ARhigh in 6, 9, and 1 patient, respectively, 3 patients with detectable MRD (1 with MRD ≥ 10−2 and 2 with MRD 10−3 to <10−2), and 1 with WBC > 100 × 109 per liter. These 20 patients were all good responders to prephase. The only patient who relapsed was a 4-year-old girl with WBC 4.8 × 109 per liter, undetectable MRD after IA, 58 chromosomes on karyotype, a standard profile of gains, and ikaros family zinc finger 1 deletion, which may have affected her prognosis.
Unlike in previous reports,4,13-16 in our study the number of chromosomes was more significant than any specific trisomies or any combination thereof. Nevertheless, our results do not contradict these reports because we found more favorable prognoses among patients with +18, with double trisomies +4,+10 or triple trisomies +4,+10,+17 as in the MRC and Children’s Oncology Group series, respectively. Our data differed by showing that other combinations of trisomies (+4,+18 and +4,+10,+18) were also significant favorable factors and that any association of trisomies fared worse when karyotypes had fewer than 58 chromosomes, thus demonstrating that MNC had a stronger prognostic impact and that the good outcomes of specific trisomies were mostly due to their association with MNC higher than 58. If the prognostic impact of these trisomies had been stronger than MNC, they would have conferred the same favorable prognosis independently of the MNC. Given the sequential and nonrandom gains that occur in HD>50, any combination of trisomies are more likely to occur when MNC rises and almost all chromosomes contribute to trisomies in the MNC 58-66 group.
Patients with the best prognosis were also identified by DI assessed by FCM. Patients with DI ≥ 1.24 (DI equivalent to MNC ≥ 58) fared the best. Despite the strong correlation between MNC and DI, DI proved to be less accurate than MNC for identifying the subgroup of patients with an excellent outcome because individual patients with 58 or 59 chromosomes may have a DI lower than 1.24 (because 1.24 is a median value), and also because DI may be underestimated as evidenced by the comparison between thDI and mDI. Despite these technical drawbacks, our results show that the subgroup of patients with the best outcomes can also be identified by DI (DI ≥ 1.24).
Different factors may have contributed to the excellent results obtained in patients with the highest ploidy and to the emergence of the subgroup MNC 58-66. One of them is excluding HD>50 with t(9;22) and MLL rearrangement that confers adverse risk. Likewise, our series did not contain near-triploidy/duplication of hypodiploidy 30-39 chromosomes, a rare entity in childhood ALL associated with a dismal outcome that may be difficult to single out when the hyperdiploid clone is preponderant and the hypodiploid nonproliferative.28 However, the main reason probably lies in the overall improvement of outcome obtained with current therapies. This is exemplified in our EORTC trials by the improved outcome obtained with the 58 951 protocol (reported in this study) compared with that of the previous decade (58 881 protocol), in which HD>50 (selected on the same criteria as in 58 951) had lower EFS and OS (6-year EFS and OS: 81.7% and 88.6%, respectively). In the 58 881 protocol, MNC had an impact on outcome (P = .0002), but there was no difference in outcome between the 54-57 and 58-66 MNC groups (6-year EFS: 85%) (supplemental Figure 1). These poorer results may be explained by a less-effective Asparaginase received by some patients during induction or reintensification,29 which had a significant impact on HD>50 as shown by the better outcomes obtained with E. coli Asparaginase (supplemental Figure 2 and supplemental Table 3) and the improved EFS of all MNC groups when only those patients who received the most effective Asparaginase were considered (supplemental Figure 3). The other therapeutic changes initiated in the 58 951 study that may account for the overall improvement obtained were the stopping of intravenous administration of 6-Mercaptopurine during maintenance, which proved to be detrimental to outcome30 (supplemental Figure 4); therapy intensification (AR high) for patients with WBC ≥ 100 × 109 per liter or extramedullary involvement that might explain why WBC counts were no longer of prognostic value in the 58 951 study; the shift to VHR for patients who had MRD ≥ 10−2 at D35 and an increased number of intrathecal chemotherapy administrations that decreased the risk of CNS relapse and possibly of nonCNS relapse as well. However, it must be emphasized that the 58-66 MNC group with excellent outcome was little affected by 2 of these changes (MRD and intensification for high WBC) because this group was closely linked to good risk factors and therefore frequently treated in the VLR or AR-low arm. Moreover, the de-escalation of therapy applied to VLR patients contributed to the low number of toxic deaths (1% of patients) observed in the 58 951 protocol. Nevertheless, the excellent outcome reported here for patients with the highest MNC and DI, established on retrospective analysis of data and optimized cut-points, requires confirmation on an independent cohort of patients treated with current therapies, especially the cut-point 58 chromosomes/DI:1.24 reported for the first time. If data on 58-66 chromosomes are validated, they call for further treatment de-escalation.
Our results are in agreement with previous reports on HD>50 with low ploidy level5,7 because patients with 51 to 53 chromosomes and DI < 1.16 had the lowest EFS and OS, although they were mainly stratified in the AR arms in the 58 951 protocol. These results confirm that patients with low ploidy level, even though they belong to HD>50 and have a relatively good outcome (in comparison with nonhyperdiploid ALLs) should not be submitted to de-escalation. Further studies are needed for clarifying why there are such heterogeneous prognoses among patients with HD>50.
In conclusion, in a large series of patients with HD>50, our results showed that ploidy has the strongest prognostic value and that its impact has not been neutralized by improved therapies of the last decades. In our study, the best indicator for excellent outcome was ploidy assessed with karyotype. Ploidy assessed by DI was also a good indicator, although less significant. MNC has only been provided so far by conventional karyotype but when microarray genomic profiling becomes cost effective and widespread among cytogenetic laboratories, chromosome gains will also be inferred with higher accuracy from copy number aberrations. DI obtained by CMF is and will remain a useful supplement of karyotype or copy number aberrations for verifying the presence of aneuploid clones.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mariana Titorov for editorial assistance, Agnès Chassevent for advice in interpreting DNA index data, Emmanuelle Delabesse for collecting cytogenetic data, and the EORTC HQ Data Management Department members (Séraphine Rossi, Lies Meirlaen, Liv Meert, Aurélie Dubois, Christine Waterkeyn, Alessandra Busato, Isabel VandeVelde, and Gabriel Solbu) for their support of this trial as well as Dr. Francisco Bautista (EORTC-CLG Fellow).
A complete list of contributors appears in the Appendix.
This study was supported by the EORTC Charitable Trust Foundation, Vlaamse Liga Tegen Kanker, Belgian Federation Against Cancer (nonprofit organization), TéléVie, and Kinderkankerfonds, Belgium.
Authorship
Contribution: N.D. reviewed cytogenetic data, designed research, and wrote the article; S.S. performed statistical analyses and analyzed and interpreted results; F.S. reviewed and provided cytogenetic data; H.C. and S.G. reviewed the MRD and immunophenotypic data, respectively; M.B. performed molecular analyses; G.P., K.Y., B.N., A.U., P.L., A.F., P.R., Y.B., and Y.B. included patients in the trial and provided clinical data; N.D., F.S., M.P.P., C.G., M.T., and P.H. provided cytogenetic data; M.K. was the clinical research physician at the EORTC Headquarters; Y. Benoit and Y. Bertrand are chairman and co-chairman of the EORTC-CLG, respectively.
Conflict-of-Interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicole Dastugue, Hematology Laboratory, Purpan Hospital, Place Baylac, 31059, Toulouse cedex, France; e-mail: dastugue.n@chu-toulouse.fr.
Appendix
EORTC-CLG members who contributed to this study are:
Lyon, France: Y. Bertrand, N. Philippe, K. Kebaili, S. Girard, M.P. Pagès; Toulouse, France: G. Plat, A. Robert, E. Delabesse, A. Chassevent, N. Dastugue; Paris, France: H. Cavé, K. Yakouben, D. Adjaoud, E. Vilmer, J.H. Dalle, B. Lescoeur, G. Sterkers, A. Aboura; Gent, Belgium: Y. Benoit, B. De Moerloose, C. Dhooge, G. Laureys, J. Philippé, F. Speleman, N. Van Roy; Lille, France: B. Nelken, F. Mazingue, M. Fournier, J.L. Laï, N. Grardel; Leuven, Belgium: A. Uyttebroeck, M. Renard, N. Boeckx, P. Vandenberghe, L. Michaux, A. Hagemeijer; Strasbourg, France: P. Lutz, M.P. Gaub, A. Eischen, L. Mauvieux, M. Lessard, C. Gervais; Brussels, Belgium: H. Reine Fabiola, A. Ferster, C. Devalck, B. Cantiniaux, P. Heimann; Porto, Portugal: L. Norton, M.R. Teixeira, V. Costa, S. Borges, C. Correia, L. Torres, S. Lisboa, J. Vieira, G. Martins, C. Palmeira; Besançon, France: P. Rohrlich, E. Plouvier, V. Laithier, M.A. Collonge-Rame, F. Garnache-Ottou; Reims, France: M. Munzer, I. Luquet; Caen, France: P. Boutard, O. Minckes, V. Salaun, G. Plessis; Nice, France: N. Sirvent, A. Deville, M. Poirée, C. Soler, S. Raynaud; Grenoble, France: D. Plantaz, C. Lefebvre, D. Leroux; Liège, Belgium: C. Hoyoux, M.F. Dresse, P. Philippet, C. Herens; Montpellier, France: G. Margueritte, S. Taviaux; Poitiers, France: F. Millot, F. Brizard; Brussels UZ, Belgium: M. Bakkus, A. Van Damme, J. Van der Werff, J. Otten; Nantes, France: F. Mechinaud, P. Talmant; Angers, France: X. Rialland, L. Baranger.