In this issue of Blood, Vegliante et al establish for the first time the oncogenic role and mechanisms of SOX11 in mantle cell lymphoma.1
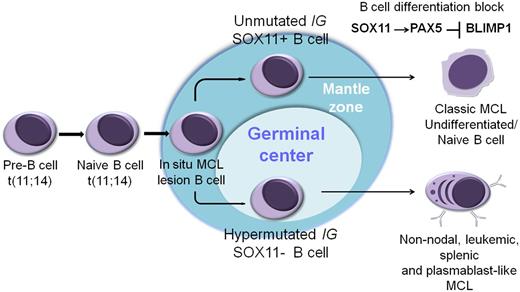
Hypothetical models of SOX11-possitive vs SOX11-negative MCL. The naive B cell carrying the t(11;14) colonizes the mantle zone of the lymphoid follicle and generates an in situ MCL lesion. Most MCLs evolve from these cells in the marginal zone with no or limited IGHV somatic mutations and SOX11 expression. SOX11 overexpression in conventional MCL may block the cells in a mature B-cell stage, preventing their further differentiation thought the SOX11-PAX5-PRDM1/BLIMP regulatory axis. Alternatively, some cells with the t(11;14) may enter the germinal center, undergo IGHV somatic hypermutation, and lack expression of SOX11. SOX11 may modulate the mature B-cell and early plasma cell differentiation program in MCL.
Hypothetical models of SOX11-possitive vs SOX11-negative MCL. The naive B cell carrying the t(11;14) colonizes the mantle zone of the lymphoid follicle and generates an in situ MCL lesion. Most MCLs evolve from these cells in the marginal zone with no or limited IGHV somatic mutations and SOX11 expression. SOX11 overexpression in conventional MCL may block the cells in a mature B-cell stage, preventing their further differentiation thought the SOX11-PAX5-PRDM1/BLIMP regulatory axis. Alternatively, some cells with the t(11;14) may enter the germinal center, undergo IGHV somatic hypermutation, and lack expression of SOX11. SOX11 may modulate the mature B-cell and early plasma cell differentiation program in MCL.
Mantle cell lymphomas (MCL) are CD5-positive mature B-cell lymphoid tumors derived from antigen-naive pregerminal center B cells located in the mantle zone surrounding normal germinal center follicles.2,3 The t(11;14)(q13;q32), a chromosomal rearrangement driving overexpression of the cyclin D1 gene, is a hallmark of this disease.2 Still, the full spectrum of genetic lesions involved in the pathogenesis of MCL remains to be established. MCLs are typically aggressive tumors associated with poor prognosis. However, recent studies have identified a distinct clinical group of t(11;14)(q13;q32)-positive MCL cases that show an indolent clinical course and prolonged survival.4-6 Notably, smoldering MCLs show hypermutated immunoglobulin genes indicating that they originate from postgerminal center B cells. In addition, they characteristically lack expression of SOX11, a transcription factor aberrantly and universally expressed at high levels in aggressive classic MCL cells. Most notably, and in contrast with its high levels of expression in classic MCL, SOX11 is not expressed in lymphoid progenitors and mature B-cell populations.
Based on these observations, Vegliante and coworkers proposed that aberrant expression of SOX11 could play an oncogenic role in the pathogenesis of classic MCL. The underlying hypothesis is that SOX11 could sit atop of an oncogenic transcriptional network controlling critical effector target genes and pathways responsible for B-cell transformation and the aggressive clinical course typically associated with SOX11-high MCLs. Should this premise hold true, deciphering the structure of the SOX11-controlled oncogenic network in MCL could identify new therapeutic targets for the treatment of this disease.
Toward this goal, these investigators first addressed the identification of SOX11 direct target genes via chromatin immunoprecipitation microarray analyses using a promoter array platform. These experiments uncovered over 1000 promoter sequences occupied by SOX11 in MCL cells. In addition, and to establish the specific role of SOX11 in the control of gene expression in MCL, they analyzed the gene expression changes associated with SOX11 small hairpin RNA knockdown. Despite the complexity of the data and the large number of genes controlled by SOX11 identified, 2 major findings stood out from these analyses. First, SOX11 knockdown in MCL cells results in upregulation of gene expression signatures associated with plasma cell differentiation while suppressing the genetic programs characteristic of B cells. In addition, PAX5, a key transcription factor strictly required for the establishment of B-cell identity7 and a major negative regulator of plasma cell differentiation,8 stood out as one of the most significant direct target genes upregulated by SOX11 in MCL cells.
Consistently, SOX11 knockdown cells showed transcriptional and immunophenotypic changes consistent with repression of the B-cell program and upregulated PRDM1/BLIMP1, a transcription factor tumor suppressor gene involved in the termination of the B-cell program during plasma cell differentiation.9 Notably, PRDM1/BLIMP1 is a known direct target gene repressed by PAX5.8 Moreover, xenograft studies demonstrated a marked loss of tumorigenic potential in SOX11 knockdown cells.
The relevance of these findings was then elegantly highlighted by integrative analyses of SOX11 small hairpin RNA knockdown induced signatures with those derived from a panel of well-characterized MCL clinical samples. These studies showed significant enrichment of SOX11 upregulated genes in SOX11-positive tumors and SOX11 downregulated transcripts in SOX11-negative lymphomas, respectively. Consistently, SOX11-positive MCL signatures were enriched in B-cell vs plasmablast and PAX5 activated gene sets, whereas the SOX11-negative MCL program was characteristically enriched in plasmablast-associated transcripts. Moreover, histologic and flow cytometry examination showed signs of focal plasmacytic differentiation and downregulation of B-cell marker expression in SOX11-negative tumors.
Overall, these results demonstrate an essential role for SOX11 in the growth of MCLs in vivo and support a role for the SOX11-PAX5-PRDM1/BLIMP1 regulatory axis in the maintenance of B-cell features and suppression of plasma cell differentiation programs in MCL. Still, several questions remain to be elucidated: What drives the aberrant expression of SOX11 in MCL? What are the specific mechanisms mediating the antilymphoma effects of SOX11 inactivation observed in mouse tumor xenografts? Is there a physiologic counterpart of the MCL-associated SOX11-PAX5-PRDM1/BLIMP1 regulatory axis in normal B cells? If so, what are the SOX factors upstream of PAX5-PRDM1/BLIMP1 in B-cell development?
The studies by Vegliante et al highlight the power of integrative analyses coupling carefully crafted mechanistic studies, genomic analyses, and expert histopathologic examination of clinical samples to uncover basic mechanisms underlying the biology of MCL.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal