Key Points
An optimized protocol for obtaining and preserving RNA from blood cells is provided.
The results serve as guideline for sensitive measurement of mRNA and microRNA expression in clinical material.
Abstract
Assessing messenger RNA (mRNA) and microRNA levels in peripheral blood cells may complement conventional parameters in clinical practice. Working with small, precious samples requires optimal RNA yields and minimal RNA degradation. Several procedures for RNA extraction and complementary DNA (cDNA) synthesis were compared for their efficiency. The effect on RNA quality of freeze-thawing peripheral blood cells and storage in preserving reagents was investigated. In terms of RNA yield and convenience, quality quantitative polymerase chain reaction signals per nanogram of total RNA and using NucleoSpin and mirVana columns is preferable. The SuperScript III protocol results in the highest cDNA yields. During conventional procedures of storing peripheral blood cells at −180°C and thawing them thereafter, RNA integrity is maintained. TRIzol preserves RNA in cells stored at −20°C. Detection of mRNA levels significantly decreases in degraded RNA samples, whereas microRNA molecules remain relatively stable. When standardized to reference targets, mRNA transcripts and microRNAs can be reliably quantified in moderately degraded (quality index 4-7) and severely degraded (quality index <4) RNA samples, respectively. We describe a strategy for obtaining high-quality and quantity RNA from fresh and stored cells from blood. The results serve as a guideline for sensitive mRNA and microRNA expression assessment in clinical material.
Introduction
Data generated with molecular techniques, when combined with clinical parameters and biopsy-derived histopathology data, are instrumental for improved diagnostic and prognostic assessment.
In cancer biology, RNA-derived gene expression signatures of peripheral blood cells or the tumor itself allowed the classification of breast carcinomas,1-3 skin melanomas,4 lung adenocarcinomas,5 and hematologic malignancies.6-8 These signatures were also useful to establish their relationship to prognosis. In solid organ transplantation, gene expression profiling of RNA from biopsied tissues has aided in distinguishing subtypes of acute rejection that respond differently to therapy.9 Using peripheral blood cells as a source of RNA is less invasive for the patient and it enables frequent immunologic monitoring after transplantation.10-13 Messenger RNA (mRNA) biomarkers have been identified in peripheral blood from patients who show operational tolerance toward their graft.14-17
MicroRNAs are small, noncoding RNA molecules that negatively regulate mRNA expression by degradation or translational repression.18,19 In recent years, microRNAs have gained interest for their usefulness in classification of (blood) cancer,20-24 for their role in hematopoiesis and immune cell function,25-28 and for their role in rejection of solid organ transplants.29-31
Reliable quantification of mRNA and microRNA levels for diagnostic and prognostic purposes requires optimal RNA quality and quantity. In the current study, we compared the efficiency of various procedures for obtaining RNA and complementary DNA (cDNA) from peripheral blood cells. We examined the impact of storage of peripheral blood cells on RNA degradation. We also investigated the effect of RNA degradation on mRNA and microRNA signals.
Material and methods
Cells and cell culture
Blood, anticoagulated with heparin, was obtained from anonymous donors (Sanquin Blood Bank, Leiden, the Netherlands) after informed consent, and peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque density gradient centrifugation. Peripheral blood lymphocyte (PBL) blasts were studied because they are a readily available source of RNA for study. Lymphocyte blasts (PBL blasts) were generated by culturing PBMC from 4 donors at 1 × 106/mL for 7 days in RPMI1640 (Gibco, Grand Island, NY), containing 15% human serum and penicillin (100 U/mL)/streptomycin (100 µg/mL) (Gibco, cat. no. 15070-063). The medium was further supplemented with 10 U/mL IL-2 and 2 µg/mL phytohemagglutinin (both from Remel Inc, Lenexa, KS).
Performance of molecular methods was also tested on PBMC without any culturing, for which separate donors were used (n = 3).
Cell storage and thawing
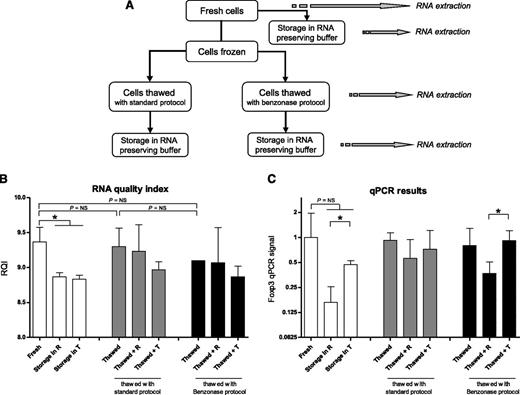
Freshly obtained PBMC were processed for RNA extraction, either directly or after storage for 6 days at −20°C in 25 µL of RNAlater (RNA Stabilization Buffer, Qiagen, Venlo, the Netherlands) or 800 µL of TRIzol (Figure 1). Alternatively, cells were cryopreserved in culture medium (RPMI1640) with 20% fetal calf serum (FCS) and 10% dimethylsulfoxide before RNA extraction. Storage of vials, containing 1 mL cell suspension each, was at −80°C for at least 24 hours and at −180°C for 3 days.
Cell storage and thawing. (A) The scheme shows manipulations of PBMC before RNA extraction. Where appropriate, RNA was preserved in both RNAlater and TRIzol. (B) The effect of freeze-thawing PBMC, combined with the effect of storage in various RNA-preserving reagents, on RNA quality is shown. RNA samples were analyzed on StdSense gels to assess integrity expressed as RNA quality index (RQI). (C) For the same conditions, PCR signals in cDNA that was synthesized from the RNA samples were determined. R, RNAlater; T, TRIzol. *P < .05.
Cell storage and thawing. (A) The scheme shows manipulations of PBMC before RNA extraction. Where appropriate, RNA was preserved in both RNAlater and TRIzol. (B) The effect of freeze-thawing PBMC, combined with the effect of storage in various RNA-preserving reagents, on RNA quality is shown. RNA samples were analyzed on StdSense gels to assess integrity expressed as RNA quality index (RQI). (C) For the same conditions, PCR signals in cDNA that was synthesized from the RNA samples were determined. R, RNAlater; T, TRIzol. *P < .05.
Cells were thawed by two different methods (Figure 1): using a standard protocol or a protocol that includes Benzonase-nuclease (Sigma-Aldrich, Zwijndrecht, the Netherlands, cat. no. E1014).32 In the standard thawing protocol, cells are thawed in a 37°C water bath and are transferred dropwise to 3 mL of FCS at room temperature. After adding 12 mL of culture medium, cells were pelleted and resuspended in 10 mL culture medium with 10% FCS. In the Benzonase thawing protocol, cells are thawed at 37°C and transferred dropwise to 2 mL of a 1:1 FCS/culture medium mixture supplemented with 0.6 µL of Benzonase. Cells were pelleted and resuspended in 5 mL of culture medium with 1 µL Benzonase. After washing steps, cells were resuspended in culture medium. After application of any thawing protocol, 1 × 106 PBMC were processed for RNA extraction either directly or after storage for 6 days at −20°C in 25 µL of RNAlater or 800 µL TRIzol. RNA quality and quantitative polymerase chain reaction (qPCR) signals were determined, as described in “RNA quantity and quality assessment.”
RNA extraction protocols
RNA was extracted using RNeasy spin columns (Qiagen), TRIzol Reagent (Life Technologies), RNA-Bee (Tel-Test, Friendswood, TX), the Ambion mirVana PARIS kit (Life Technologies), and the NucleoSpin miRNA kit (Macherey-Nagel, Düren, Germany). Protocols for the column-based kits (RNeasy, mirVana, NucleoSpin) were followed as described by the manufacturers. With mirVana and NucleoSpin, small and large RNAs were purified in 1 fraction, without separate enrichment of small RNAs. Because the RNeasy procedure does not allow efficient extraction of RNA molecules of less than 200 bases, it was not included in the comparison of microRNA yields. The TRIzol and RNA-Bee reagents were supplemented with 0.2 mL of chloroform per 1 mL of reagent. After rigorous mixing and centrifugation (15 minutes, 12 000g, 4°C), the upper layer was isolated. To pellet the RNA, 0.5 mL of isopropanol (per 1 mL of reagent) was added, followed by 10 minutes of incubation at room temperature and centrifugation (15 minutes, 12 000g, 4°C). The pellet was washed once in 0.5 mL of 75% ethanol.
RNA quantity and quality assessment
RNA quantity was determined on a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific Inc, Asheville, NC). Results are expressed as micrograms RNA per million cells. Signals obtained from qPCR on cDNA that was synthesized (with SuperScript III; see “mRNA”) from a fixed amount of RNA (200 ng) per sample served as an additional criterion of suitability of the RNA extraction methods.
RNA quality was assessed using the StdSense Analysis kit and the Experion RNA analyzer (Bio-Rad, Hercules, CA). The dynamic range of StdSense chips is 5 to 500 ng/µL. The RNA quality index (RQI), also known as RNA integrity number,33 was used as an indicator of RNA integrity, and it ranges from 10 (intact) to 0 (fully degraded). For RQI calculations the 28S ribosomal (r)RNA, 18S rRNA, and pre-18S region (below the 18S rRNA band) on electropherograms are taken into account. A high 28S:18S ratio (ie, around 2:1) and a low number of shadow bands below and above the 18S band are indicative of a high RQI value.
cDNA synthesis protocols
mRNA.
cDNA synthesis from total RNA (extracted with mirVana) with an input of 100 ng, 500 ng, and 1500 ng was carried out with 4 different methods, following the manufacturers’ manuals: AMV-RT (Promega, Madison, WI) (10 U of RT, 0.5 mM dNTP, 20 U of rRNasin); the RevertAid kit (Fermentas, St. Leon-Rot, Germany; 200 U of M-MuLV RT, 1 mM dNTP, 20 U of RiboLock RNAse inhibitor); BioScript RT (Bioline, London, UK; 200 U of MMLV RT, 0.5 mM dNTP); and SuperScript III RT (Invitrogen; 200 U of RT, 0.5 mM dNTP, 40 U of RNAse OUT rRNAse inhibitor, 5 mM DTT). RNA was combined with oligo dT (Invitrogen; 0.25 µg) and random nucleotide hexamers (Invitrogen; 0.25 µg) and incubated at 65°C (70°C for BioScript) for 5 minutes. Subsequently, the remaining constituents supplied by each manufacturer were added on ice, and the reactions were allowed to proceed at 25°C for 5 minutes and at 42°C (50°C for SuperScript III) for 1 hour. Reactions were terminated by increasing the temperature to 70°C for 5 minutes.
microRNA.
cDNA synthesis of microRNAs (100 ng total RNA as input) was carried out with the miRCURY LNA Universal cDNA kit from Exiqon (Vedbaek, Denmark; cat. no. 203300). The reverse transcription reaction was carried out according to the manufacturer’s manual. Reactions were allowed to proceed for 60 minutes at 42°C and were terminated by increasing the temperature to 95°C for 5 minutes.
Quantitative PCR assays
Specific mRNA targets (Table 1) and miRNA targets (Table 2) with various abundance in expression were analyzed. Signals for mRNA transcripts were determined by qPCR on 1:10 diluted cDNA. Optimal primers pairs were selected using Primer 3 (v. 0.4.0), which is an open source project hosted by SourceForge. To prevent amplification of genomic DNA, forward and reverse primers for each transcript were designed to target separate exons, spanning at least one intron with a size of 800 bp or more. To ensure high specificity of the primers without amplification of genomic DNA, all primer sets were tested before use on control cDNA (from Human Reference Total RNA; Clontech, Mountain View, CA) and genomic DNA. All PCR efficiencies were between 90% and 110%. Signals for microRNAs were determined by qPCR on 1:40 diluted cDNA. Primer sequences were obtained from Exiqon (see Table 2 for catalog numbers), and contained built-in locked nucleic acids for enhanced sensitivity and specificity. Further details concerning the qPCR assays have been described elsewhere.34-37
Characteristics of primers for detection of mRNA transcripts
| Transcript . | Forward sequence . | Reverse sequence . | Amplicon size . | Expression level (Cq)* . |
|---|---|---|---|---|
| High expression | ||||
| β-actin | 5′-accacaccttctacaatgag-3′ | 5′-tagcacagcctggatagc-3′ | 161 bp | 13.9 ± 1.4 |
| GAPDH | 5′-acccactcctccacctttgac-3′ | 5′-tccaccaccctgttgctgtag-3′ | 110 bp | 15.1 ± 1.4 |
| Intermediate expression | ||||
| TGF-β1 | 5′-cccagcatctgcaaagctc-3′ | 5′-gtcaatgtacagctgccgca-3′ | 101 bp | 18.6 ± 0.8 |
| Low expression | ||||
| CD25 | 5′-gactgctcacgttcatcatggt-3′ | 5′-aatgtggcgtgtgggatctc-3′ | 82 bp | 20.8 ± 2.8 |
| HPRT-1 | 5′-agatggtcaaggtcgcaagc-3′ | 5′-tcaagggcatatcctacaacaaac-3′ | 165 bp | 21.7 ± 2.6 |
| Foxp3 | 5′-acagcacattcccagagttc-3′ | 5′-caggtggcaggatggtttc-3′ | 190 bp | 21.9 ± 1.4 |
| Transcript . | Forward sequence . | Reverse sequence . | Amplicon size . | Expression level (Cq)* . |
|---|---|---|---|---|
| High expression | ||||
| β-actin | 5′-accacaccttctacaatgag-3′ | 5′-tagcacagcctggatagc-3′ | 161 bp | 13.9 ± 1.4 |
| GAPDH | 5′-acccactcctccacctttgac-3′ | 5′-tccaccaccctgttgctgtag-3′ | 110 bp | 15.1 ± 1.4 |
| Intermediate expression | ||||
| TGF-β1 | 5′-cccagcatctgcaaagctc-3′ | 5′-gtcaatgtacagctgccgca-3′ | 101 bp | 18.6 ± 0.8 |
| Low expression | ||||
| CD25 | 5′-gactgctcacgttcatcatggt-3′ | 5′-aatgtggcgtgtgggatctc-3′ | 82 bp | 20.8 ± 2.8 |
| HPRT-1 | 5′-agatggtcaaggtcgcaagc-3′ | 5′-tcaagggcatatcctacaacaaac-3′ | 165 bp | 21.7 ± 2.6 |
| Foxp3 | 5′-acagcacattcccagagttc-3′ | 5′-caggtggcaggatggtttc-3′ | 190 bp | 21.9 ± 1.4 |
Values represent mean Cq values (with standard deviation) in cells from 4 donors.
Characteristics of primers for detection of microRNAs and small RNAs
| microRNA . | Catalog Number* . | NCBI Accession . | Target Size† . | Expression level (Cq)‡ . |
|---|---|---|---|---|
| High expression (Cq < 20) | ||||
| SNORD49A | 203904 | NR_002744 | 71 bp | 19.0 ± 0.6 |
| SNORD38B | 203901 | NR_001457 | 69 bp | 19.1 ± 1.0 |
| miR-155 | 204308 | HC_040174 | 23 bp | 19.6 ± 0.8 |
| Intermediate expression (Cq 20-25) | ||||
| miR-191 | 204306 | HC_040210 | 23 bp | 21.9 ± 0.9 |
| miR-103 | 204063 | HC_885819 | 23 bp | 22.2 ± 0.9 |
| miR-142-5p | 204722 | HC_040142 | 21 bp | 22.6 ± 1.6 |
| Low expression (Cq > 25) | ||||
| miR-223 | 204256 | HC_040280 | 22 bp | 25.9 ± 1.5 |
| miR-26b | 204172 | HC_885768 | 21 bp | 26.5 ± 2.7 |
| microRNA . | Catalog Number* . | NCBI Accession . | Target Size† . | Expression level (Cq)‡ . |
|---|---|---|---|---|
| High expression (Cq < 20) | ||||
| SNORD49A | 203904 | NR_002744 | 71 bp | 19.0 ± 0.6 |
| SNORD38B | 203901 | NR_001457 | 69 bp | 19.1 ± 1.0 |
| miR-155 | 204308 | HC_040174 | 23 bp | 19.6 ± 0.8 |
| Intermediate expression (Cq 20-25) | ||||
| miR-191 | 204306 | HC_040210 | 23 bp | 21.9 ± 0.9 |
| miR-103 | 204063 | HC_885819 | 23 bp | 22.2 ± 0.9 |
| miR-142-5p | 204722 | HC_040142 | 21 bp | 22.6 ± 1.6 |
| Low expression (Cq > 25) | ||||
| miR-223 | 204256 | HC_040280 | 22 bp | 25.9 ± 1.5 |
| miR-26b | 204172 | HC_885768 | 21 bp | 26.5 ± 2.7 |
NCBI, National Center for Biotechnology Information.
Catalog numbers from Exiqon.
PCR amplicon size for all targets is between 60 and 70 bp on agarose gel.
Values represent mean Cq values (with standard deviation) in cells from 3 donors.
PCR assays were carried out using iQ SYBR Green Supermix and a MyiQ Real-Time PCR detection system (Bio-Rad). The PCR program consisted of 10 minutes of hotstart at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. Upon completion of each run, a melting curve analysis was done to check specificity of the primers.
In real-time PCR, the quantification cycle (Cq) value is a measure of the number of cDNA copies. Cq indicates the number of PCR cycles in which fluorescence increases above the threshold. In experiments for evaluation of the effect of RNA degradation, Cq ratios of individual targets and reference targets (for mRNA: β-actin, GAPDH38 ; for microRNA: miR-103, miR-191, SNORD38B, SNORD49A) were calculated using the 2-ΔΔCq method.39
Effect of RNA degradation
RNA (extracted with mirVana) was diluted to 100 ng/µL and exposed to 90°C. At t=0 and after 15 minutes, 30 minutes, 45 minutes, 1 hour, 1.25 hours, 1.5 hours, 2 hours, 2.5 hours, and 3 hours of heat exposure, RNA quality was determined (StdSense chips). cDNA was synthesized on mRNA (SuperScript III RT) and microRNA (Universal cDNA kit, Exiqon). As read-out for yields, PCR signals of 6 mRNA transcripts (Table 1) and 8 microRNAs (Table 2) were assessed by qPCR. For each marker, the PCR signal per time point was statistically compared with the mean signal of the first 2 time points (t=0, t=15 minutes). In addition, the ratios of individual mRNA transcripts and the mean signal of 2 reference transcripts were calculated. Similarly, ratios for individual microRNAs and the mean signal of 4 reference microRNAs were calculated.
We investigated the effect of RNA degradation on PCR signals for 4 different mRNA transcripts while varying the amplicon size for each transcript. The reverse primer was selected to target a fixed sequence within the coding sequence of GAPDH (5′-TGC TGT AGC CAA ATT CGT TG-3′), transforming growth factor (TGF)-β1 (5′-GTC CTT GCG GAA GTC AAT GT-3′), CD25 (5′-GAC GAG GCA GGA AGT CTC AC-3′), and Foxp3 (5′-CAG GTG GCA GGA TGG TTT C-3′). The location of the forward primer was varied to obtain primer pairs that generated PCR product of around 50 bp, 100 bp, 200 bp, or 400 bp.
Statistics
Experiments were performed with PBL blasts from 3 or more donors and with uncultured PBMC from 3 additional donors. Results are presented as means ± standard deviation, unless stated otherwise. Graphs were made in GraphPad Prism 5 (La Jolla, CA). Differences between groups were tested by independent t tests using SPSS Inc software, version 20 (IBM Corp, Armonk, NY). P < .05 was considered a statistically significant difference.
Results
Cell storage and thawing
The effect on RNA quality of freezing and thawing cells with various protocols, combined with the effect of storage of cells in several RNA-preserving reagents, was investigated. Freshly obtained cells were directly processed for RNA extraction or stored at −20°C in various RNA-preserving reagents (Figure 1A). RNA quality decreased with 0.6 points on the RQI scale (P < .05) after storage in RNAlater (R) or TRIzol (T) (Figure 1B, open bars). Accordingly, qPCR signal with cDNA synthesized from the RNA samples was at least twofold lower after storage in RNAlater or TRIzol, although this difference did not reach statistical significance (Figure 1C).
Freezing the cells in dimethylsulfoxide and thawing them with either a standard protocol (Figure 1B-C, first gray bar) or with the Benzonase protocol (first black bar) did not negatively affect RNA quality compared with freshly processed cells (first open bar; P = NS). Benzonase is reported to prevent cell clumping when PBMC are thawed.32 No adverse effect on RNA quality was observed when Benzonase was included in the thawing procedure (Figure 1B-C: first black bar), compared with the standard thawing protocol (first gray bar; P = NS).
Storage of thawed cells for 6 days in the freezer in TRIzol (T) (Figure 1C: third gray and third black bar) had a slightly higher RNA-preserving effect for downstream qPCR application than storage in RNAlater (R) (second gray and second black bar).
RNA extraction protocols
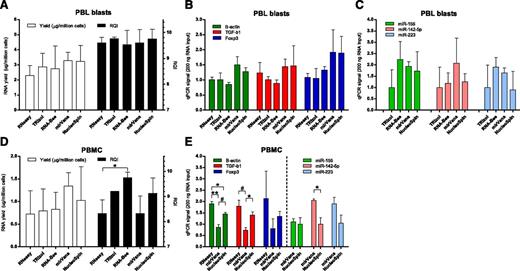
Various methods for extraction of RNA from PBL blasts were compared. The average RNA yield for all 5 methods tested was 2.8 ± 0.8 µg per 1 × 106 PBL blasts, and the average RQI was 9.7 ± 0.3. Yield and quality of RNA did not differ between extraction procedures (Figure 2A). Use of the TRIzol and RNA-Bee procedures in our hands led to suboptimal RNA yields in 50% of cases because of complete or partial loss (0.03 to 0.7 µg per 1 × 106 cells) of the RNA pellet after precipitation. The efficiency of cDNA synthesis may be influenced by the RNA extraction method used. Therefore, as an additional criterion for considering suitability of the procedures, qPCR signals of high-abundance (β-actin, green bars in Figure 2B), medium-abundance (TGF-β1, red bars), and low-abundance transcripts (CD25 and Foxp3, blue bars) were determined in cDNA synthesized from a fixed amount of RNA (200 ng). Signals with NucleoSpin columns were similar to signals obtained with mirVana columns, but were on average 1.5 to 1.7 times higher than those obtained with RNeasy, TRIzol, and RNA-Bee (Figure 2B, P = NS). PCR signals for high-abundance (miR-155, green bars in Figure 2C), medium-abundance (miR-142-5p, red bars), and low-abundance (miR-223, blue bars) microRNAs were analyzed after RNA extraction. On average, mirVana and RNA-Bee gave highest signals per nanogram of RNA, followed closely by NucleoSpin (1.4- to 1.5-fold lower). We found the NucleoSpin columns the most convenient procedure to work with.
RNA extraction protocols. (A) Yield per million PBL blasts (open bars) and integrity (black bars) of RNA obtained with 5 extraction protocols. (B) PCR signals in cDNA synthesized from 200 ng total RNA of PBL blasts on high-abundance (green bars, β-actin), medium-abundance (red bars, TGF-β1), and low-abundance (blue bars, Foxp3) mRNA transcripts, following 5 RNA extraction protocols. (C) PCR signals for high-abundance (green bars, miR-155), medium-abundance (red bars, miR-142-5p), and low-abundance (blue bars, miR-223) microRNAs. Signals for TRIzol were set to 1. (D) Yield per million PBMC (open bars) and integrity (black bars) of RNA obtained with 5 extraction protocols. *P < .05. (E) PCR signals in cDNA synthesized from 200 ng total RNA of PBMC obtained directly from blood. *P < .05. #P < .01. **P < .001.
RNA extraction protocols. (A) Yield per million PBL blasts (open bars) and integrity (black bars) of RNA obtained with 5 extraction protocols. (B) PCR signals in cDNA synthesized from 200 ng total RNA of PBL blasts on high-abundance (green bars, β-actin), medium-abundance (red bars, TGF-β1), and low-abundance (blue bars, Foxp3) mRNA transcripts, following 5 RNA extraction protocols. (C) PCR signals for high-abundance (green bars, miR-155), medium-abundance (red bars, miR-142-5p), and low-abundance (blue bars, miR-223) microRNAs. Signals for TRIzol were set to 1. (D) Yield per million PBMC (open bars) and integrity (black bars) of RNA obtained with 5 extraction protocols. *P < .05. (E) PCR signals in cDNA synthesized from 200 ng total RNA of PBMC obtained directly from blood. *P < .05. #P < .01. **P < .001.
Efficiency of RNA extraction protocols was also tested on mononuclear cells obtained directly from peripheral blood (n = 3), without subsequent culturing. The average RNA yield for the RNA extraction procedures was 0.9 ± 0.4 µg per 1 × 106 cells and the average RQI was 8.8 ± 0.8. Although mirVana and NucleoSpin gave the highest yield of RNA per million PBMC, this was not significantly different from yields obtained with the other extraction procedures (Figure 2D open bars). Although all procedures gave high-quality RNA with an RQI > 8, the use of RNeasy and mirVana gave RNA that on average had the lowest RQI (Figure 2D, black bars). Results of qPCR signals on a fixed amount of RNA (200 ng) are shown for the column-based extraction protocols (RNeasy, mirVana, NucleoSpin) (Figure 2E). Signals obtained for the mRNA transcripts were higher for NucleoSpin compared with mirVana. Signals for microRNAs were highest with mirVana.
cDNA synthesis protocols
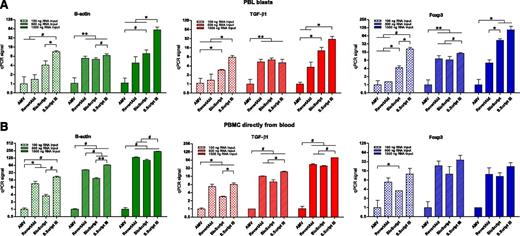
Four protocols for synthesizing cDNA from RNA (extracted with mirVana) were tested. The same mRNA transcripts were analyzed as those in the RNA extraction experiment described previously. With RNA from PBL blasts, the order of cDNA efficiency from highest to lowest was SuperScript III > BioScript > RevertAid > AMV-RT (Figure 3A). This was observed for high-, medium-, and low-abundance transcripts. With both 100 ng and 1500 ng of RNA as input for cDNA synthesis, the use of the SuperScript III protocol led to 2- to 32-fold higher yields of cDNA, as measured by qPCR, compared with the other protocols (Figure 3A).
cDNA synthesis protocols. Relative yields of cDNA, following 4 cDNA synthesis protocols (synthesized from mirVana-extracted RNA), were calculated from PCR signals obtained for high-abundance (green bars, β-actin), medium-abundance (red bars, TGF-β1), and low-abundance (blue bars, Foxp3) mRNA transcripts. This was investigated with RNA from PBL blasts (A) and from PBMC obtained directly from blood (B). Signals for AMV were set to 1. *P < .05. #P < .01. **P < .001.
cDNA synthesis protocols. Relative yields of cDNA, following 4 cDNA synthesis protocols (synthesized from mirVana-extracted RNA), were calculated from PCR signals obtained for high-abundance (green bars, β-actin), medium-abundance (red bars, TGF-β1), and low-abundance (blue bars, Foxp3) mRNA transcripts. This was investigated with RNA from PBL blasts (A) and from PBMC obtained directly from blood (B). Signals for AMV were set to 1. *P < .05. #P < .01. **P < .001.
Experiments were repeated with RNA from PBMC obtained directly from blood (Figure 3B). Results were comparable with the results obtained with PBL blasts. The use of SuperScript III resulted into the highest yields of cDNA, whereas AMV gave the lowest cDNA yields. BioScript and RevertAid did not significantly differ in the amount of cDNA yield that they rendered.
Effect of RNA degradation
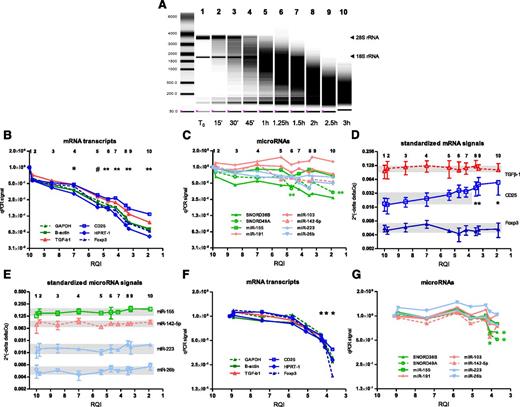
RNA degradation was induced by exposing total RNA from PBL blasts to 90°C for various lengths of time. Over a period of 3 hours, the RQI decreased from 10.0 to 1.9, whereas the ratio of 28S rRNA (upper band) to 18S rRNA (lower band) decreased from 1.8 to 0.18 (Figure 4A). When RNA had reached an RQI of 7.0, the signal for all mRNA transcripts tested, irrespective of transcript abundance, was already at least twofold lower than the mean signal of the first 2 time points (Figure 4B; P < .05). At an RQI of 5.4 or lower, PCR signals for mRNA had three- to fivefold decreased (P < .01). Thus, with decreasing RNA quality the amplification signals declined rapidly. In contrast, all microRNAs tested remained relatively stable (Figure 4C), except small nucleolar RNA SNORD49, which showed a decline of PCR signals at an RQI of 4.7 or lower (two- to threefold, P < .01).
Effect of RNA degradation. RNA was extracted from PBL blasts, and RNA degradation was induced by exposure to 90°C for times indicated. (A) Representative picture of RNA electropherogram showing impact of heat exposure on RNA integrity. (B) RQI, as an indicator of RNA quality, was analyzed (x-axis). The numbers 1 to 10 at the top of the graph indicate time points corresponding with the numbers in the RNA electropherogram. mRNA levels were assessed by qPCR, targeting high-abundance (green), medium-abundance (red), and low-abundance (blue) markers. For each marker, the mean PCR signal at t=0 was set to 1. The graph shows means of 4 experiments. *P < .05; #P < .01; **P < .005 vs mean of the first 2 time points. (C) MicroRNA levels were assessed by qPCR, targeting high-abundance (green), medium-abundance (red), and low-abundance (blue) markers. The graph shows means of 3 experiments. **P < .005 vs mean of first 2 time points for SNORD49A. (D-E) PCR signals for individual mRNA transcripts and microRNAs were corrected for reference markers using the 2-ΔΔCq method. The upper and lower boundaries of the shaded areas indicate maximum and minimum values, respectively. *P < .05 vs first time point. (F-G) Experiments investigating the effect of RNA degradation on PCR signals were repeated with RNA from PBMC obtained directly from blood. mRNA transcripts (F) and microRNAs (G) were analyzed. *P < .05 vs mean of the first 2 time points (F: each mRNA transcript; G: only for SNORD38B and SNORD49A).
Effect of RNA degradation. RNA was extracted from PBL blasts, and RNA degradation was induced by exposure to 90°C for times indicated. (A) Representative picture of RNA electropherogram showing impact of heat exposure on RNA integrity. (B) RQI, as an indicator of RNA quality, was analyzed (x-axis). The numbers 1 to 10 at the top of the graph indicate time points corresponding with the numbers in the RNA electropherogram. mRNA levels were assessed by qPCR, targeting high-abundance (green), medium-abundance (red), and low-abundance (blue) markers. For each marker, the mean PCR signal at t=0 was set to 1. The graph shows means of 4 experiments. *P < .05; #P < .01; **P < .005 vs mean of the first 2 time points. (C) MicroRNA levels were assessed by qPCR, targeting high-abundance (green), medium-abundance (red), and low-abundance (blue) markers. The graph shows means of 3 experiments. **P < .005 vs mean of first 2 time points for SNORD49A. (D-E) PCR signals for individual mRNA transcripts and microRNAs were corrected for reference markers using the 2-ΔΔCq method. The upper and lower boundaries of the shaded areas indicate maximum and minimum values, respectively. *P < .05 vs first time point. (F-G) Experiments investigating the effect of RNA degradation on PCR signals were repeated with RNA from PBMC obtained directly from blood. mRNA transcripts (F) and microRNAs (G) were analyzed. *P < .05 vs mean of the first 2 time points (F: each mRNA transcript; G: only for SNORD38B and SNORD49A).
The ratio of each of 3 mRNA transcripts (TGF-β1, CD25, Foxp3) to reference gene signals was not significantly altered at an RQI as low as 4.2 (Figure 4D). Below this point, CD25 to reference ratios significantly deviated (less than a twofold increase, P < .05) from the ratio observed at the first time point (RQI = 10.0). In contrast, ratio of each of 4 microRNAs tested (miR-155, miR-142-5p, miR-223, miR-26b) and the mean signal of reference microRNAs remained stable even in severely degraded RNA with an RQI as low as 1.9 (Figure 4E).
The effect of RNA degradation on qPCR signals for mRNAs and microRNAs was also tested with mononuclear cells obtained directly from peripheral blood. Similar results were seen as with the RNA from the PBL blasts. Below an RQI of 5, amplification signals for all mRNA transcripts tested had significantly declined (Figure 4F; P < .05). Signals for almost all microRNAs tested, except small nucleolar RNAs 38B and 49A, remained stable at an RQI as low as 3.5 (Figure 4G).
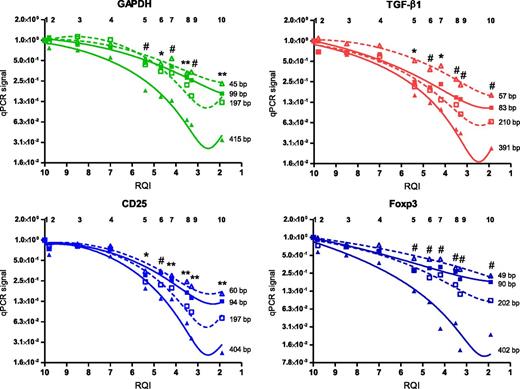
RNA degradation negatively affects PCR signals irrespective of mRNA amplicon size
It is expected that in degraded RNA samples, larger PCR amplicons cease amplification sooner than smaller amplicons. We examined the possibility that amplicon size differences account for the sustained stability of microRNA molecules in relation to increasing degradation of RNA. Amplicon size of each of the miRNA targets tested was between 60 and 70 bp (examined on agarose gel, data not shown). For high-, medium-, and low-abundance mRNA transcripts, we varied the size of the PCR amplicon between 50 and 400 bp and investigated the effect of RNA degradation on PCR signals. For the 4 transcripts tested, larger amplicon sizes resulted in quicker loss of PCR signal upon RNA degradation (Figure 5). However, with an mRNA amplicon size of between 45 and 60 bp (Figure 5, open triangles) the extent of loss of PCR signals remained significantly higher compared with that of the microRNA targets (Figure 4C,G).
RNA degradation negatively affects PCR signals irrespective of mRNA amplicon size. RNA from PBL blasts was degraded by exposure to 90°C. PCR signals for high-abundance (green), medium-abundance (red), and low-abundance (blue) mRNA transcripts were assessed. For each transcript, size of the amplicon was varied (50 bp, 100 bp, 200 bp, or 400 bp). For each condition, the mean PCR signal at t=0 was set to 1. Graph represents means of 4 experiments, with third-order polynomial lines drawn through the data points. *P < .05; #P < .01; **P < .001 vs mean of first two time points for the smallest amplicon.
RNA degradation negatively affects PCR signals irrespective of mRNA amplicon size. RNA from PBL blasts was degraded by exposure to 90°C. PCR signals for high-abundance (green), medium-abundance (red), and low-abundance (blue) mRNA transcripts were assessed. For each transcript, size of the amplicon was varied (50 bp, 100 bp, 200 bp, or 400 bp). For each condition, the mean PCR signal at t=0 was set to 1. Graph represents means of 4 experiments, with third-order polynomial lines drawn through the data points. *P < .05; #P < .01; **P < .001 vs mean of first two time points for the smallest amplicon.
Discussion
Molecular techniques are vital for identification and validation of markers at the RNA level. Novel molecular markers are needed for improving current conventional diagnostic practice. Faced with the challenge of small sizes of clinical samples, researchers optimize their protocols for isolation of mRNA and microRNA. Here we describe an optimized protocol for RNA extraction, cDNA synthesis, and storage of peripheral blood cells for obtaining high yields of high-quality mRNA and microRNA.
RNA integrity was maintained upon freezing and thawing cells, following conventional procedures, when compared with RNA from freshly isolated cells. This finding shows that RNA-preserving agents are not mandatory when workup needs to be postponed and enables retrospective RNA analysis of cells that were cryopreserved for cellular assays. We used the RNA quality index from nanochips to determine RNA quality. This is a suitable means to reliably assess RNA integrity, as was reported by others.33,40 In addition, we compared 2 commonly used protocols for thawing cells, 1 of which incorporates Benzonase-nuclease. Benzonase has no negative effect on the preservation of high-quality RNA and has the additional benefit of preventing clumping of PBMC during thawing.32 In daily laboratory practice, functional cellular tests are often performed with PBMC recovered from cryopreservation. With the additional goal of molecular analyses, one may obviously proceed with RNA extraction from thawed cells immediately. Alternatively, the thawed cells may be stored again for RNA extraction on a later day; for instance, when functional tests need to be performed and interpreted first. In that case, TRIzol (at −20°C) is recommended because its use leads to preservation of RNA integrity, as reflected by RNA quality index and qPCR signal. Because storage time of cell pellets at −20°C in our experiments did not exceed 6 days, a negative impact on RNA quality of extended cell storage cannot be ruled out.
In terms of RNA yield, quality, qPCR signals per nanogram of total RNA, and convenience, the use of NucleoSpin and mirVana columns is preferable. TRIzol and RNA-Bee, which have been used in many studies for RNA extraction, offer rapid procedures but showed a relatively high failure rate in our laboratory because of (partial) loss of the RNA pellet in the precipitation step. Four of the 5 RNA extraction procedures tested were comparable in the amount of PCR signal generated for microRNA targets per nanogram of total RNA. RNeasy spin columns are not suitable for efficient microRNAs extraction (data not shown and Mraz et al.41 ). The miRNeasy kit may better suit this goal, but it was not available at the start of our tests.
Of the cDNA synthesis protocols we tested, SuperScript III resulted in the highest yield of cDNA. This is in line with the recommendation of using a thermostable reverse transcriptase enzyme for cDNA synthesis.42 Of note, SuperScript III was considerably superior to the first generation of SuperScript (data not shown) that we reported on in a previous study.43
Finally, we showed that RNA degradation has a major negative impact on mRNA expression levels, which were determined by qPCR. This finding is in line with the highly negative correlation found between the RNA integrity number (RNA quality index or RQI in our study) and the Cq value from the reverse transcriptase PCR assay in earlier studies.44-46 After correction of individual mRNA transcripts to reference genes, the ratio was not significantly altered with an RQI as low as 4.2. Below this value, the signal to reference ratio was significantly altered for 1 of the 3 transcripts studied, indicating different rates of degradation for different mRNA molecules. We recommend that the RQI of RNA samples should be 4.2 or higher for reliable quantification of standardized transcript levels. In another study, an index of 5 or higher was regarded as suitable for downstream qPCR application.46 In contrast, we observed that individual microRNAs, and especially microRNA signals standardized to reference microRNA signals, remained stable even in severely degraded RNA samples (having an RQI as low as 1.9). In an earlier study, microRNAs did not suffer from degradation during storage of lymphocytes at −80°C,41 but the quality of the RNA was not investigated in that study. Even with mRNA amplicons of approximately 50 bp, we observed significant loss of PCR signals at an RQI of 5.4 or lower. This shows that the increased stability of microRNAs, compared with that of mRNAs, cannot be explained by smaller amplicon sizes. MicroRNAs remain stable in different human bodily fluids.47,48 Our observations and those from others hold promise for reliable quantification of microRNA expression in specimens that are prone to RNA degradation such as blood plasma, serum, and urine.
In summary, we described an optimized protocol for extracting and preserving RNA molecules from blood cells. The results serve to enhance sensitivity of mRNA and microRNA expression assessment for clinical purposes.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr. Arend Mulder for critically reading and editing the manuscript.
Parts of this work were supported by grant C07.2238 from the Dutch Kidney Foundation.
Authorship
Contribution: M.E. analyzed data, interpreted results, and wrote the manuscript; N.V.R. performed experiments and interpreted the results; J.D.H.A. performed experiments and interpreted the results; S.H. performed experiments and interpreted the results; and F.H.J.C. interpreted the data. All authors provided editorial assistance.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael Eikmans, Leiden University Medical Center, Building 1, E3-Q, Department of Immunohematology and Blood Transfusion, Albinusdreef 2, 2333 ZA Leiden, The Netherlands; e-mail: m.eikmans@lumc.nl.