Abstract
The developmental progression of immature thymocytes requires cooperative input from several pathways, with Notch signals playing an indispensable role at the T-cell receptor (TCR)–β selection checkpoint. Notch signals affect the activation of the PI3K/Akt pathway, which is required for pTα/TCRβ (pre-TCR)–induced survival, differentiation, and proliferation of developing αβ-lineage thymocytes. However, the molecular players responsible for the interaction between the Notch and PI3K pathways at this critical developmental stage are unknown. Here, we show that Notch induction of Hes1 is necessary to repress the PI3K/Akt pathway inhibitor, PTEN (phosphatase and tensin homolog), which in turn facilitates pre-TCR–induced differentiation. In support of this mechanism, deletion or down-regulation of Pten overcomes the Notch signaling requirement for survival and differentiation during β-selection. In addition, c-Myc is a critical target of Notch at this stage, as c-Myc expression overcomes the Notch signaling requirement for proliferation during β-selection. Collectively, our results point to HES1, via repression of PTEN, and c-Myc as critical mediators of Notch function at the β-selection checkpoint.
Introduction
In the thymus, incoming lymphocyte progenitors encounter an inductive environment known to support intrathymic T-cell development, which includes the Notch ligand Delta-like 4 (Dll4),1,2 the cytokine interleukin (IL)–7 3,4 and the chemokine CXCL12.5,6 However, how signals derived from these factors are integrated by a developing thymocyte to realize the T-cell differentiation program remains to be elucidated.
T-cell development is a highly ordered process typically characterized by the surface expression of CD4 and CD8, with the earliest T-cell subset contained among CD4− CD8−, double-negative (DN), cells,7 which can be further defined based on the expression of CD44, CD117, and CD25. The most primitive CD44+CD117+CD25− DN1 cell-subset contains multipotent progenitors8,9 and expression of CD25 marks entry into the T-lineage specified DN2 stage.7 Here, expression of recombination-activating gene-1 (Rag1) and Rag2 induces T-cell receptor (TCR)β, TCRγ, and TCRδ gene loci to rearrange V(D)J gene segments, which continues into the subsequent CD44−CD117−CD25+ DN3 stage, wherein thymocytes irreversibly commit to the T-lineage and are subjected to their first developmental checkpoint, β-selection.7,10 DN3 cells expressing a productively rearranged TCRβ chain with its partner pTα and CD3 form the pre-TCR complex that mediates passage across β-selection, resulting in rescue from apoptosis, cellular proliferation, TCRβ gene allelic exclusion, and differentiation of DN3 cells to the subsequent CD4+CD8+, double-positive (DP), stage.10,11
Intrathymic Notch signaling is initiated when the Notch receptor (Notch1) engages its ligand (Dll4), which leads to the transcriptional activation of Notch target genes.12,13 Notch signals induce adoption of the T-cell fate in progenitors that enter the thymus,14 and are essential for the survival, proliferation, and differentiation of DN thymocytes along the αβ-lineage, to the DP stage.7,14 Previously, our findings revealed that Notch receptor-ligand interactions are crucial for maintaining cell size, glucose metabolism, and survival of DN3 cells before the initiation of β-selection.15 This was because of Notch signals supporting the activation of the phosphatidylinositol-3-kinase (PI3K) pathway, leading to Akt/PKB phosphorylation. In support of this notion, pre-T cells deficient in phosphoinositide-dependent kinase 1 (PDK1), an enzyme which phosphorylates and activates AGC serine kinases, including Akt,16 were found to be unresponsive to trophic effects of Notch signaling. Despite these studies establishing the critical role for Notch in activating PI3K signaling in developing T cells, the identity of relevant targets downstream of Notch responsible for bridging the 2 pathways remained unclear. In addition, other signaling pathways mediated by IL-7R and CXCR4, known to promote PI3K/Akt activation were shown to act along with the pre-TCR during β-selection.5,6,17
Recent studies examining the role of Notch in T-cell acute lymphoblastic leukemia (T-ALL) have implicated HES1 and c-Myc as critical targets of Notch signaling in leukemic cells.18,19 Furthermore, PTEN (phosphatase and tensin homolog), an inhibitor of the PI3K pathway, was found to be an indirect target of activated Notch1 in T-ALL cells, via an HES1-mediated repression of the Pten promoter.20 Together, these results suggested a potential mechanism for developing thymocytes by which Notch signaling supported the activation of the PI3K pathway, involving HES1 and PTEN as probable candidate genes.
Here, we investigate the role of HES1, PTEN and c-Myc downstream of Notch signaling in DN3 thymocytes. Using the OP9-DL1 T-cell differentiation system,21,22 we show that loss of Notch-ligand interactions in DN3 cells led to the down-regulation of Hes1 with a concomitant rise in Pten mRNA expression. DN3 cells with reduced HES1 function exhibited a phenotype similar to loss of Notch signaling, including elevated levels of PTEN expression even in the presence of Notch signaling, supporting the previous report identifying HES1 as a transcriptional repressor of the Pten promoter.20 This was accompanied with impaired proliferation and differentiation along the αβ-cell lineage to the DP stage. Thus, HES1 plays an important role in mediating PI3K regulation and trophic effects by Notch at the β-selection checkpoint. In support of this connection, restoration of PI3K signaling in pre-T cells, through the loss or down-regulation of PTEN, was sufficient to mediate β-selection in the absence of Notch signaling. However, without Notch signals, ectopic expression of c-Myc was critical to also ensure cellular proliferation. Taken together, these findings suggest that Notch signals at β-selection serve to promote PI3K-mediated survival and differentiation through HES1 repression of PTEN, as well as induce c-Myc expression to drive proliferation of thymocytes as they reach the CD4+ CD8+ stage of T-cell development, at which point Notch signaling ceases, which serves to avoid an otherwise inevitable path to leukemic transformation.
Methods
Mice
Rag2-deficient mice were bred and maintained in the animal facility of Sunnybrook Research Institute in specific pathogen-free conditions. Ptenf/f Lck-cre mice23 were obtained from the University Health Network. Rag2−/−Ptenf/f;Lck−cre+ mice were generated by crossing Rag2−/− mice with Ptenf/f;Lck−cre+ mice. C57Bl/6 and CD1 timed-pregnant mice were obtained from Charles River laboratories. All animal procedures were approved by the Sunnybrook Health Science Center Animal Care Committee.
Generation of HES1 and PTEN shRNA constructs and viral-producing cells
shRNA sequences were ordered from Sigma-Aldrich, or obtained from the RNAi consortium and oligonucleotides were ordered from Invitrogen. shRNA sequences were subsequently digested with XhoI and EcoRI and ligated with the Rapid DNA Ligation Kit (Roche) into the MSCV-LTR miR30-PIG (LMP) vector digested with the same enzymes. Bacterial clones were screened for insert by polymerase chain reaction (PCR). GP+E.86 cells transfected for each of these shRNA constructs were generated.
OP9 coculture and retroviral transduction
HES1 and dominant-negative HES1 constructs were kindly provided by Dr R. Kageyama (Kyoto University, Japan) and Dr A. Strom (Karolinska Institute, Sweden). Retroviral constructs were generated by subcloning cDNAs into MigR1, and stable retroviral-producing GP+E.86 packaging lines were generated for each construct. OP9-DL1, OP9-DL4, and OP9-Ctrl cells were produced and maintained as previously described,24 and cultures were supplemented with 1 ng/mL mouse IL-7 and 5 ng/mL human recombinant Flt-3L (Peprotech). Fetal liver was obtained from timed-pregnant Rag2–/– or CD1 female mice on day 14 of gestation, and bone marrow was harvested from 6- to 8-week-old Rag2−/−Ptenf/f;Lck−cre+ or Rag2−/−Pten+/+;Lck−cre+ mice. Single-cell suspensions were generated by disruption through a 40-mm nylon mesh screen using a syringe plunger. Bone marrow cell suspensions were purified for Lin− CD117+ Sca1+ hematopoietic progenitor cells (HPCs) by cell sorting before culture with OP9-DL1 cells. For retroviral transduction of HPCs, CD24lo/− CD1 fetal liver (FL) cells were enriched for HPCs by complement-mediated lysis with CD24 antibody, and transduced by overnight culture with stable retrovirus-producing GP+E.86 packaging cells. Transduced (GFP+ or GFP+YFP+) or nontransduced Lin−CD117+ Sca1+ HPCs were purified by cell sorting cultures as previously described25 and returned to culture with OP9-DL4 cells for T-lineage differentiation. For retroviral transduction of DN3 cells, day 7 HPC OP9-DL1 cocultures were passaged onto an overnight culture with stable retrovirus-producing GP+E.86 packaging cells. Transduced (GFP+ or GFP+YFP+) or nontransduced CD44− CD25+ DN3 cells were purified by cell sorting from day 8 cultures as previously described.25
Flow cytometry and cell sorting
All single-cell suspensions were stained with commercially available antibodies (BD Pharmigen and e-biosciences) and analyzed with a BD-LSRII flow cytometer, using FlowJo Version 6.4.7 software (TreeStar). Dead cells were excluded from the analyses using 4,6-diamidion-2-phenylindole (DAPI) gating.
Quantitative real-time PCR
Thymocyte populations were purified by flow cytometry or selection using magnetic anti-CD45 beads (Miltenyi Biotec). Total RNA was extracted using TRIzol (Invitrogen) and converted to cDNA using Quantitect reverse transcription kit (QIAGEN). Expression of the indicated genes was measured by quantitative real-time PCR using SYBR GreenER (Invitrogen). Primer sequences will be supplied on request. β-actin was used to normalize cycle thresholds.
Immunoblots
Whole cells lysates were prepared by resuspending cell pellets in radioimmunoprecipitation assay (RIPA) lysis buffer with protease inhibitors. Protein concentrations were determined by Bradford Assay. Equal amounts of protein from each sample were loaded and resolved using 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE), and transferred onto polyvinyldifluoride membranes (Amersham Biosciences). PTEN (Cell Signaling Technology), phospho-GSK3β (Ser9; Cell Signaling Technology) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Millipore) specific antibodies were used to probe the immunoblots.
PTEN-luciferase reporter assays
293T cells were transfected with a PTEN-luciferase reporter construct (pGL3 PTEN HindIII-NotI)20 along with plasmids encoding HES1 (pcDNA3-HES1, generously provided by Dr A. Strom) and/or dnHES1 (pcDNA3-dnHES1, generously provided by Dr A. Strom) and/or shHES1. PTEN reporter activity was normalized to pRL-CMV Renilla-luciferase expression plasmid. PTEN reporter activity and renilla luciferase levels (normalization control) were measured 48 hours after transfection with the Dual-Luciferase Reporter Assay kit (Promega).
Results
Loss of Notch signaling in DN3 cells leads to cellular atrophy but not decreased levels of CD127 and CXCR4 expression
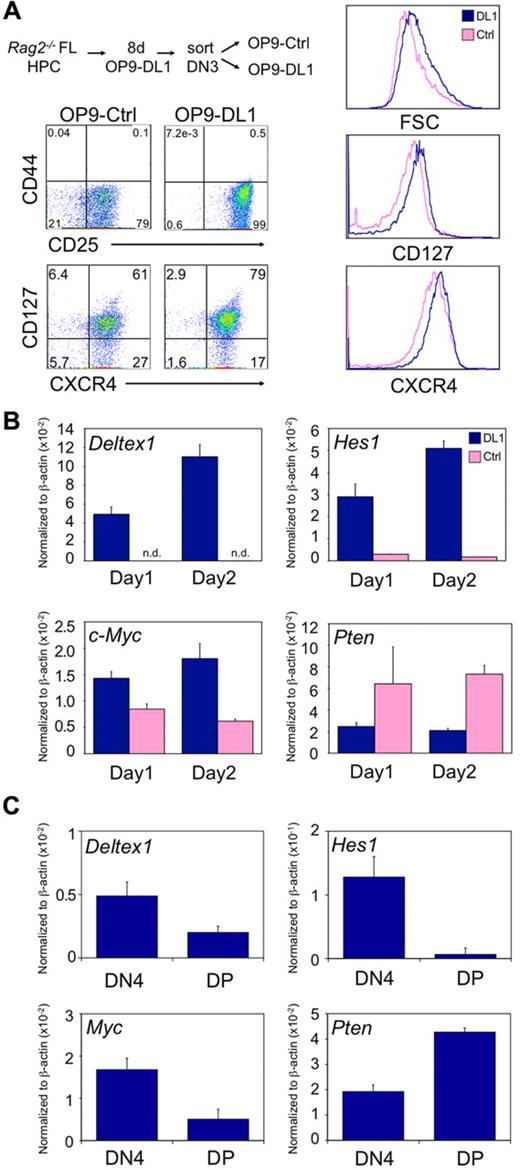
As cytokine and chemokine-driven responses in developing T cells are often regulated at the level of receptor expression,17,26 we sought to address whether Notch signals target the PI3K pathway directly, or indirectly by affecting the expression of these receptor-mediated pathways. In particular, we compared the expression levels of CD127 (IL7Rα) and CXCR4 in Rag2−/− DN3 cells cultured in the presence or absence of Notch signaling. To this end, HPCs from Rag2−/− FLs (E14) were cultured for 8 days on OP9-DL1 cells to allow for T-cell lineage commitment and differentiation to the DN3 stage.25 Cocultures were subsequently sorted for DN3 cells, returned to OP9-DL1 or OP9-Ctrl cells, and analyzed 2 days later (Figure 1A). As IL-7 is supplemented at 1 ng/mL, and stromal derived factor 1 (SDF-1)α is endogenously expressed in OP9 cells,6 IL-7 and SDF-1 (CXCL12) levels are equivalent in OP9-DL1/Ctrl cocultures. Importantly, Rag2−/− cells were used to circumvent the confounding effects of pre-TCR signaling on survival, proliferation, and differentiation. As expected, Rag2−/− DN3 cells remained CD44−CD25+ on both OP9-DL1 and OP9-Ctrl cells, albeit CD25 expression appeared to be reduced in the absence of Notch signals, consistent with the report of Notch signaling targeting CD25 expression.27 In addition, Rag2−/− DN3 cells experienced accelerated cell death and atrophy in the absence of Notch ligand, demonstrated by decreased cell numbers (data not shown) and cell size (forward side scatter, FSC), respectively (Figure 1A), consistent with our previous findings.15 This was accompanied by a slight decrease in surface expression of CD127 and CXCR4. However, these changes in surface expression were probably because of the observed cellular atrophy.
Notch signaling in developing thymocytes inversely affects the expression of HES1 and PTEN. (A) Rag2−/− E14 FL-derived HPCs cultured with OP9-DL1 cells for 8 days are used to give rise to CD44− CD25+ DN3 cells, which are then sorted and returned to either OP9-DL1 or OP9-Ctrl cells for 2 days before analysis. Flow cytometric analysis of CD44, CD25, CD127, and CXCR4 expression is shown for Rag2−/− DN3 cells cultured for 2 days in the absence (Ctrl) or presence (DL1) of Notch signaling. Overlay histograms showing cell size (FSC), and surface expression of CD127 and CXCR4 of DN3 cells, cultured as indicated, are shown on the right. Data are representative of at least 3 independent experiments. (B) qRT-PCR analysis of mRNA expression, normalized to β-actin, of Notch downstream target genes (Deltex1, Hes1, and c-Myc) and Pten is shown for Rag2−/− DN3 cells cultured for 1 or 2 days as indicated. Data are representative of at least 3 independent experiments, with standard deviation of the mean shown as error bars. (C) qRT-PCR analysis of mRNA expression (as in panel B) is shown for C57BL/6 ex vivo–isolated DN4 and DP thymocyte subsets. Data are representative of at least 3 independent experiments.
Notch signaling in developing thymocytes inversely affects the expression of HES1 and PTEN. (A) Rag2−/− E14 FL-derived HPCs cultured with OP9-DL1 cells for 8 days are used to give rise to CD44− CD25+ DN3 cells, which are then sorted and returned to either OP9-DL1 or OP9-Ctrl cells for 2 days before analysis. Flow cytometric analysis of CD44, CD25, CD127, and CXCR4 expression is shown for Rag2−/− DN3 cells cultured for 2 days in the absence (Ctrl) or presence (DL1) of Notch signaling. Overlay histograms showing cell size (FSC), and surface expression of CD127 and CXCR4 of DN3 cells, cultured as indicated, are shown on the right. Data are representative of at least 3 independent experiments. (B) qRT-PCR analysis of mRNA expression, normalized to β-actin, of Notch downstream target genes (Deltex1, Hes1, and c-Myc) and Pten is shown for Rag2−/− DN3 cells cultured for 1 or 2 days as indicated. Data are representative of at least 3 independent experiments, with standard deviation of the mean shown as error bars. (C) qRT-PCR analysis of mRNA expression (as in panel B) is shown for C57BL/6 ex vivo–isolated DN4 and DP thymocyte subsets. Data are representative of at least 3 independent experiments.
Transcriptional changes in DN3 cells on loss of Notch signaling
As loss of PI3K signaling in Rag2−/− DN3 cells cultured in the absence of Notch signals was probably not because of alterations in CD127 and CXCR4 expression, we sought to identify relevant downstream Notch targets responsible for Notch interaction with PI3K at the β-selection checkpoint. To this end, we identified and measured by qRT-PCR the changes in transcript levels that occur on Notch signaling withdrawal, using mRNA from Rag2−/− DN3 cells cultured for 24 and 48 hours on OP9-DL1 or OP9-Ctrl cells. As expected, loss of Notch signaling in Rag2−/− DN3 cells was accompanied by decreased transcript levels for known Notch target genes Deltex1, c-Myc, and Hes1 (Figure 1B). Recently, HES1 was found to bind and repress the promoter of Pten, an inhibitor of the PI3K pathway.20 A gene expression reporter assay confirmed that HES1 expression decreased Pten promoter activity (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Furthermore, Notch signaling withdrawal in Rag2−/− DN3 cells resulted in decreased Hes1 mRNA, coupled with increased Pten transcripts (Figure 1B), making HES1 and PTEN likely candidates for bridging upstream Notch signals to downstream effects on the PI3K pathway. In support of this, analyses of ex vivo thymocytes revealed that, on loss of Notch signaling associated with traversing β-selection,7 similar changes in transcript levels are observed. Specifically, on the down-regulation of Notch signaling from the DN4 to the DP stage of development, transcript levels of Notch target genes Deltex1, c-Myc, and Hes1 are decreased, concomitantly with increased Pten transcripts (Figure 1C). Considering these data together, particularly the inverse relationship between HES1 and PTEN levels observed with loss of Notch signaling, we sought to determine whether HES1 played a critical role in the Notch-mediated activation of the PI3K pathway in DN3 cells at the β-selection checkpoint.
Transcriptional changes, decreased cellularity and DP development after inhibition of HES1 function
HES1 is critically required for proliferation in early T-cell progenitors.28-30 Here, we retrovirally cotransduced DN3 cells to express 2 key Notch target genes, Hes1 and c-Myc, and assessed whether these cells could traverse the β-selection checkpoint in the absence of Notch signaling (OP9-Ctrl cultures). Flow cytometric analysis revealed that HES1 and c-Myc overexpression in DN3 cells did not overcome the need for Notch-mediated signals at this checkpoint, as seen by the failure to give rise to DP cells (supplemental Figure 2). Interestingly, HES1/c-Myc–transduced DN3 cells were smaller in size, compared with c-Myc only (MigR1/c-Myc) transduced cells, but expressed higher levels of CD71, an indicator of increased PI3K/Akt pathway activity.16
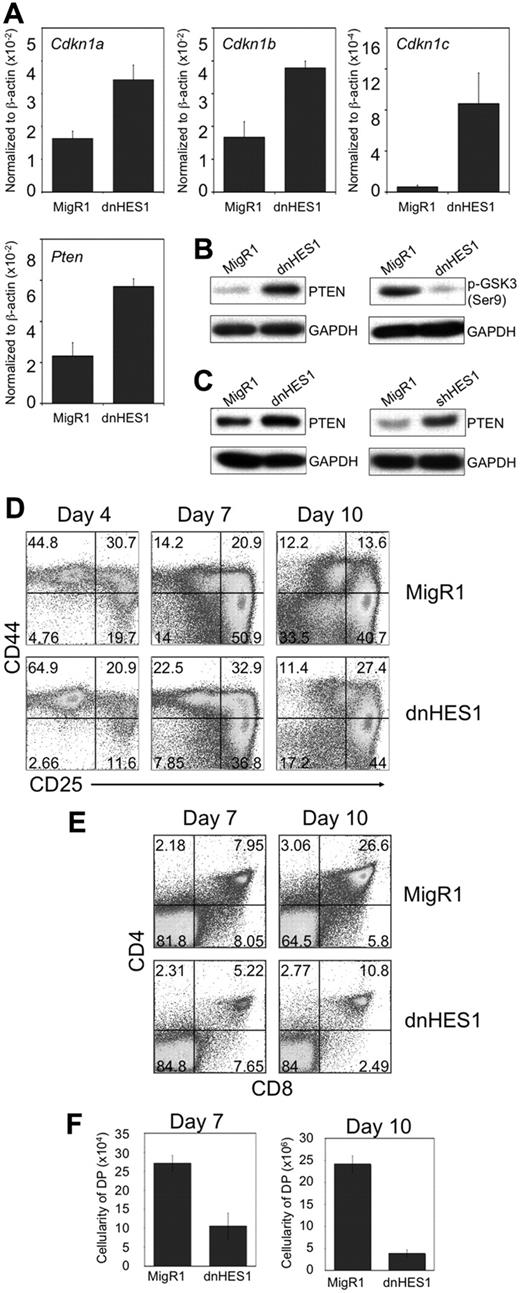
To more clearly evaluate the role of HES1 in early T-cell development, we expressed a dominant-negative version of HES1 (dnHES1)31 in DN3 cells. Importantly, a gene expression reporter assay confirmed that dnHES1 expression functionally repressed HES1 activity (supplemental Figure 1A). HES1 has been proposed to influence proliferation through inhibition of cyclin-dependent kinase inhibitors.32,33 In agreement with this notion, expression of dnHES1 in Rag2−/− DN3 cells cultured on OP9-DL1 cells increased Cdkn1a, Cdkn1b, and Cdkn1c transcript levels (Figure 2A). GSK-3β is an important downstream target of PI3K/Akt signaling,34 and its phosphorylation at Ser9 is mediated by Akt. In support of our hypothesis that decreased HES1 function decreases PI3K/Akt pathway activity, dnHES1-expressing cells show increased PTEN mRNA and protein levels (Figure 2A-C), corresponding to a decrease in phosphorylated GSK3β protein levels (Figure 2B). Together, these results further confirm HES1's role as a repressor of PTEN expression in DN3 cells, and further support a mechanism by which Notch signaling influences PI3K pathway activation in thymocytes undergoing β-selection.
Expression of a dominant negative form of HES1 (dnHES1) leads to up-regulation of Cdkn1 and Pten expression, and impaired T-cell development. (A) QRT-PCR analysis of mRNA expression of HES1 target genes (Cdkn1a,b,c, and Pten) in Rag2−/− DN3 cells retrovirally transduced to express dnHES1 and/or GFP (MigR1), and then cultured with OP9-DL1 cells for 2 days before analysis. qRT-PCR results are normalized to β-actin expression levels, and the data are representative of 3 independent experiments. (B-C) Analysis of PTEN and phosphorylated GSK3β (Ser9) protein expression in (B) BWZ.36 cells or (C) DN3 cells retrovirally transduced to express dnHES1, shHES1, and/or GFP (MigR1) is shown as immunoblots of whole cell lysates probed with antibodies specific for PTEN, GSK3β (Ser9), or GAPDH. Data are representative of 3 independent experiments. (D-F) Developmental progression of FL-derived HPCs transduced to express dnHES1 and/or GFP (MigR1) and subsequently cultured for 10 days with OP9-DL4 cells. Flow cytometric analysis of (D) CD44 and CD25, and (E) CD4 and CD8, surface expression is shown for GFP+ gated cells on days 4, 7, and 10 of coculture as indicated, whereas panel F shows the corresponding cellularity of DP cells present in the cultures, as indicated. DP cellularity was obtained by multiplying the total cellularity by the percentage of DP cells present in the cultures of each independent experiment. Data are representative of 3 independent experiments.
Expression of a dominant negative form of HES1 (dnHES1) leads to up-regulation of Cdkn1 and Pten expression, and impaired T-cell development. (A) QRT-PCR analysis of mRNA expression of HES1 target genes (Cdkn1a,b,c, and Pten) in Rag2−/− DN3 cells retrovirally transduced to express dnHES1 and/or GFP (MigR1), and then cultured with OP9-DL1 cells for 2 days before analysis. qRT-PCR results are normalized to β-actin expression levels, and the data are representative of 3 independent experiments. (B-C) Analysis of PTEN and phosphorylated GSK3β (Ser9) protein expression in (B) BWZ.36 cells or (C) DN3 cells retrovirally transduced to express dnHES1, shHES1, and/or GFP (MigR1) is shown as immunoblots of whole cell lysates probed with antibodies specific for PTEN, GSK3β (Ser9), or GAPDH. Data are representative of 3 independent experiments. (D-F) Developmental progression of FL-derived HPCs transduced to express dnHES1 and/or GFP (MigR1) and subsequently cultured for 10 days with OP9-DL4 cells. Flow cytometric analysis of (D) CD44 and CD25, and (E) CD4 and CD8, surface expression is shown for GFP+ gated cells on days 4, 7, and 10 of coculture as indicated, whereas panel F shows the corresponding cellularity of DP cells present in the cultures, as indicated. DP cellularity was obtained by multiplying the total cellularity by the percentage of DP cells present in the cultures of each independent experiment. Data are representative of 3 independent experiments.
We addressed the role of HES1 during T-lineage differentiation by retrovirally transducing FL-derived HPCs to express dnHES1 and/or GFP (MigR1), and using flow cytometry to assess their ability to respond to Notch signals, in coculture with OP9-DL4 cells.24 Figure 2D-F shows that dnHES1-transduced HPCs displayed a reduced efficiency in T-cell differentiation that is particularly noticeable by day 10 of culture, when more than 6-times more control (MigR1)–transduced HPCs differentiated to the DP stage than dnHES1-transduced cells (Figure 2F). To circumvent the possibility of nonspecific interference by dnHES1 on other bHLH or HES-family transcription factors, shRNA targeting Hes1 was used.35 A gene expression reporter assay confirmed that shHES1 expression functionally repressed HES1 activity (supplemental Figure 1B). Consistent with results from dnHES1-transduced cells, shHES1-transduced DN3 cells showed increased PTEN protein levels (Figure 2C), and shHES1-transduced HPCs showed reduced efficiency in differentiation along the T-cell lineage (supplemental Figure 3A-C). Together, these findings are consistent with previous reports showing that Hes1-deficiency in hematopoietic precursors arrests T-cell development.28,29
Inhibition of HES1 function allows for non–T-lineage differentiation in the presence of Notch signals
The apparent delay and impairment of T-cell development in dnHES1-transduced HPCs is evident before and during β-selection. Previous studies using human hematopoietic cells showed that, although overexpression of HES1 could induce a partial block on B-cell development, but could not impose T-cell differentiation.36 Here, we find that loss of HES1 function with dnHES1-expression did not promote B-cell development of HPCs in OP9-DL1 cultures, but instead increased myeloid-lineage (CD11b+) cell potential (supplemental Figure 4), based on absolute numbers (data not shown) and percentages. Although HES1 does not appear to play a critical role in Notch-mediated T versus B lineage bifurcation, it is involved downstream of Notch signaling in the divergence away from myeloid lineage outcomes.
HES1 function is required for efficient β-selection
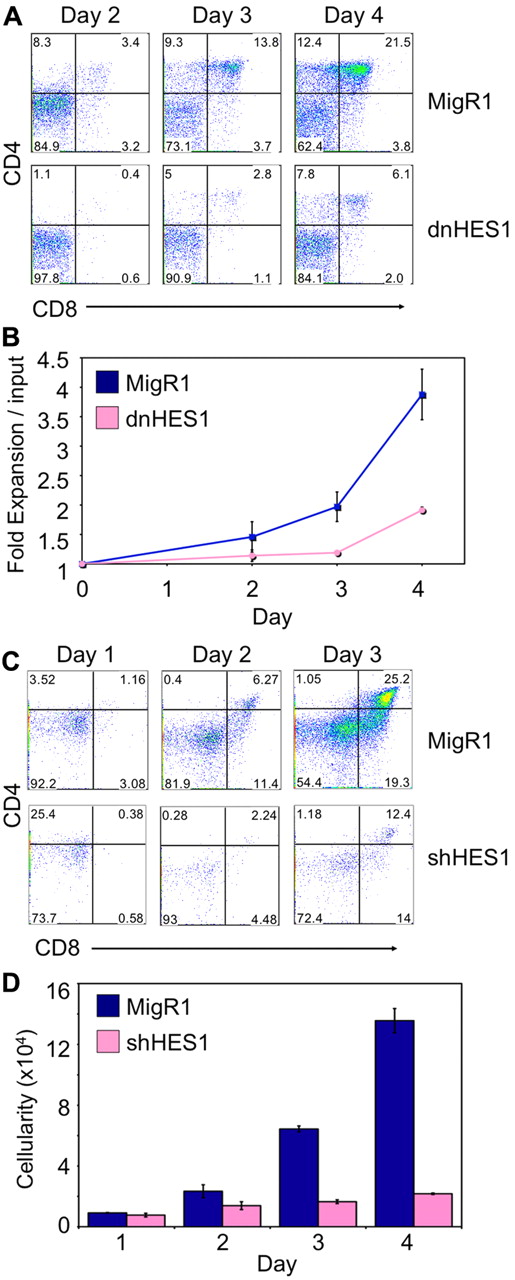
To examine the specific role of HES1 in T-cell differentiation at the β-selection checkpoint separately of its role in early proliferation and differentiation, HES1 function was manipulated at the later DN3 stage of development. To this end, DN3 cells were isolated and transduced to express dnHES1 and/or GFP (MigR1), and cultured on OP9-DL4 cells. Expression of dnHES1 resulted in a marked reduction in the number of cells reaching the DP stage of differentiation (Figure 3A-B). In addition, DN3 cells transduced to express an shRNA targeting Hes1 when cultured on OP9-DL4 cells showed a similar decrease in their ability to develop to the DP stage, compared with GFP (MigR1)–only transduced DN3 cells (Figure 3C-D). Furthermore, to more precisely examine the effect of interfering with HES1 function at the β-selection checkpoint, Rag2−/− DN3 cells were cotransduced to express dnHES1 and/or a rearranged TCRβ chain, and pre-TCR–induced differentiation was analyzed. As expected, in the absence of a TCRβ chain, after 6 days of culture with OP9-DL4 cells, MigR1/MIY- and dnHES1/MIY-transduced Rag2−/− DN3 cells remained at the DN stage, whereas MigR1/TCRβ-expressing cells underwent differentiation from the DN to DP stage (supplemental Figure 5A). In contrast, dnHES1/TCRβ-expressing Rag2−/− DN3 cells showed a marked decrease in differentiation to the DP stage and failed to expand, whereas MigR1/TCRβ-transduced cells proliferated extensively in response to pre-TCR–derived signals (supplemental Figure 5B). In addition, dnHES1/TCRβ-expressing Rag2−/− DN3 cells showed lower levels of CD71 surface expression, but differences in cell size were not observed (supplemental Figure 5C). In keeping with the results from HPCs or DN3 cells transduced to express dnHES1, Rag2−/− DN3 cells cotransduced with dnHES1 and TCRβ developed beyond the DN3 stage but exhibited defects in differentiation and decreased cell numbers compared with controls, suggesting that the Notch-dependent differentiation and survival of DN3 cells at the β-selection checkpoint is at least partially mediated by HES1 and probably because of its ability to repress the PI3K inhibitor, PTEN.
Expression of a dominant negative form of HES1 (dnHES1) or shRNA targeting HES1 (shHES1) in DN3a cells impairs T-cell differentiation across the β-selection checkpoint. (A) Developmental progression of DN3a cells transduced to express dnHES1 and/or GFP (MigR1) and subsequently cultured with OP9-DL4 cells for 2 to 4 days. Flow cytometric analysis of CD4 and CD8 surface expression is shown for GFP+ gated cells on days 2, 3, and 4 of coculture as indicated, with (B) the cellular fold expansion (total cellularity at each time point divided by the number of cells used at the start of the culture, input) observed in the cultures shown for the indicated times and conditions. (C) Developmental progression of DN3a cells transduced to express shHES1 and/or GFP (MigR1) and subsequently cultured with OP9-DL4 cells for 1 to 3 days. Flow cytometric analysis of CD4 and CD8 surface expression is shown for GFP+ gated cells on days 1, 2, and 3 of coculture as indicated with (D) the total cellularity observed in the cultures shown for the indicated times and conditions. Data are representative of 3 independent experiments, with standard deviation of the mean shown as error bars.
Expression of a dominant negative form of HES1 (dnHES1) or shRNA targeting HES1 (shHES1) in DN3a cells impairs T-cell differentiation across the β-selection checkpoint. (A) Developmental progression of DN3a cells transduced to express dnHES1 and/or GFP (MigR1) and subsequently cultured with OP9-DL4 cells for 2 to 4 days. Flow cytometric analysis of CD4 and CD8 surface expression is shown for GFP+ gated cells on days 2, 3, and 4 of coculture as indicated, with (B) the cellular fold expansion (total cellularity at each time point divided by the number of cells used at the start of the culture, input) observed in the cultures shown for the indicated times and conditions. (C) Developmental progression of DN3a cells transduced to express shHES1 and/or GFP (MigR1) and subsequently cultured with OP9-DL4 cells for 1 to 3 days. Flow cytometric analysis of CD4 and CD8 surface expression is shown for GFP+ gated cells on days 1, 2, and 3 of coculture as indicated with (D) the total cellularity observed in the cultures shown for the indicated times and conditions. Data are representative of 3 independent experiments, with standard deviation of the mean shown as error bars.
PTEN enforces the Notch-dependent survival and differentiation of DN3 cells across the β-selection checkpoint
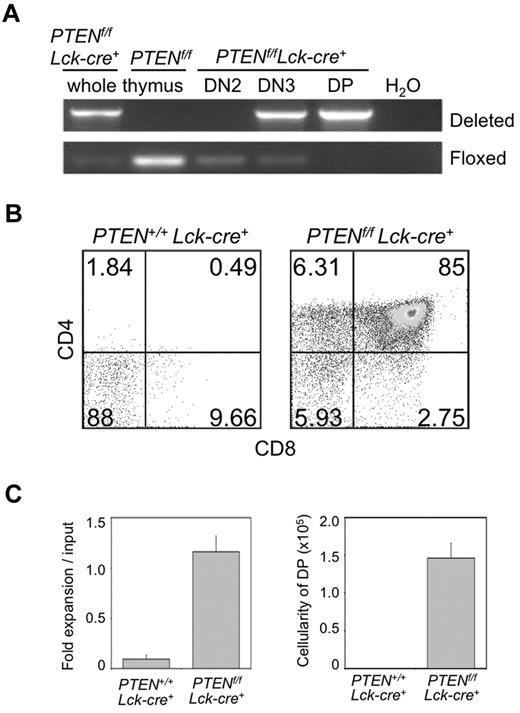
PTEN dephosphorylates phosphatidylinositol-3,4,5-triphosphate (PIP3), thus opposing the activity of PI3K. Deletion of PTEN in pre-T cells was found to substitute for IL-7 and pre-TCR signals, both of which lead to downstream activation of the PI3K pathway, and mediate survival and differentiation to the DP stage.37 As Notch signals also provide trophic effects at the β-selection checkpoint via PI3K pathway activation, and considering previous20 and our present findings (Figures 1–2 and supplemental Figure 1) showing that HES1 represses PTEN expression, we sought to define the relationship between PTEN and Notch in early thymocytes. To this end, we made use of Ptenflox/flox (Lck-cre+) mice, in which deletion of Pten in Ptenf/f;Lck−cre+ T cells begins at the DN3 stage and is complete by the DP stage (Figure 4A). To test whether the absence of PTEN allows DN3 cells to survive and differentiate across the β-selection checkpoint without Notch signals, bone marrow–derived HPCs from of Ptenf/f;Lck−cre+ and Pten+/+;Lck−cre+ mice were cultured with OP9-DL1 cells for 14 days. From these cultures, DN3a (before β-selection)38 cells were sort-purified, placed back in culture in the presence or absence of Notch-ligand interactions, and analyzed 5 days later (Figure 4B, supplemental Figure 6). Consistent with our hypothesis, DN3a cells from Ptenf/f;Lck−cre mice were able to differentiate into DP cells, whereas Pten+/+;Lck−cre+ DN3a cells failed to survive and differentiate in the absence of Notch signals (Figure 4B-C).
Conditional Pten deletion in DN3 cells allows for T-cell differentiation across the β-selection checkpoint in the absence of Notch signals. (A) Deletion of exons 4 and 5 of the Pten allele in PTENf/f;Lck−cre+ mice is initiated at the DN3 stage of development and completed by the DP stage. DNA from whole thymus of PTENf/f;Lck−cre+ or PTENf/f mice, and from sorted DN2, DN3, DN4, and DP thymocyte subsets of PTENf/f;Lck−cre+ mice was extracted and amplified by PCR. Agarose gels with the PCR products corresponding to the deleted and floxed alleles are shown, as indicated. Data are representative of at least 3 independent experiments. (B) Developmental progression of culture-derived PTENf/f;Lck−cre+ DN3 cells cultured for 6 days with OP9-Ctrl cells. Flow cytometric analysis of CD4 and CD8 cell surface expression is shown for CD45+ gated cells, whereas panel C shows the corresponding fold expansion and DP cellularity, as indicated. Lin− c-Kit+ Sca-1+ cells sorted from bone marrow of PTENf/f;Lck−cre+ or PTEN+/+;Lck−cre+ mice were cultured with OP9-DL1 cells for 14 days, sorted for DN3a cells, and returned to OP9-Ctrl cells for 6 days. Fold expansion was obtained from the total cellularity divided by the number of cells used at the start of the culture (input), and DP cellularity by multiplication of the total cellularity by the percentage of DP cells present in the cultures. Results are representative of 3 independent experiments.
Conditional Pten deletion in DN3 cells allows for T-cell differentiation across the β-selection checkpoint in the absence of Notch signals. (A) Deletion of exons 4 and 5 of the Pten allele in PTENf/f;Lck−cre+ mice is initiated at the DN3 stage of development and completed by the DP stage. DNA from whole thymus of PTENf/f;Lck−cre+ or PTENf/f mice, and from sorted DN2, DN3, DN4, and DP thymocyte subsets of PTENf/f;Lck−cre+ mice was extracted and amplified by PCR. Agarose gels with the PCR products corresponding to the deleted and floxed alleles are shown, as indicated. Data are representative of at least 3 independent experiments. (B) Developmental progression of culture-derived PTENf/f;Lck−cre+ DN3 cells cultured for 6 days with OP9-Ctrl cells. Flow cytometric analysis of CD4 and CD8 cell surface expression is shown for CD45+ gated cells, whereas panel C shows the corresponding fold expansion and DP cellularity, as indicated. Lin− c-Kit+ Sca-1+ cells sorted from bone marrow of PTENf/f;Lck−cre+ or PTEN+/+;Lck−cre+ mice were cultured with OP9-DL1 cells for 14 days, sorted for DN3a cells, and returned to OP9-Ctrl cells for 6 days. Fold expansion was obtained from the total cellularity divided by the number of cells used at the start of the culture (input), and DP cellularity by multiplication of the total cellularity by the percentage of DP cells present in the cultures. Results are representative of 3 independent experiments.
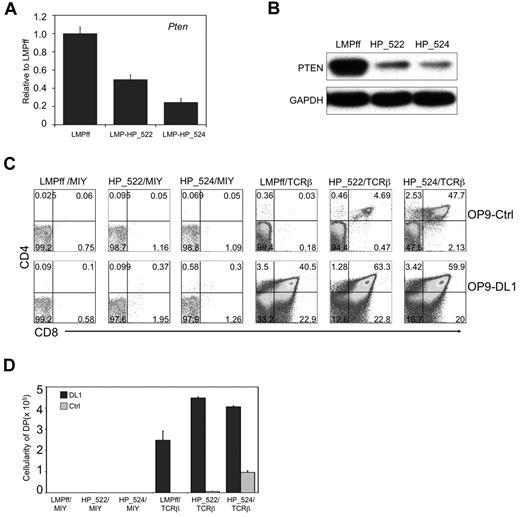
DN3a cells cultured on OP9-Ctrl cells simultaneously experience the absence of 2 critical signals: Notch and pre-TCR. To circumvent this issue, and to more precisely time the induction of a concomitant gain of pre-TCR signals and a reduction of PTEN expression, we used Rag2−/− DN3 cells cotransduced to express TCRβ and PTEN-shRNAs, respectively. This approach not only coordinates the timing of pre-TCR expression, but ensures that any death of DN3 cells is not because of a lack of pre-TCR signals.25 Two PTEN-shRNA lentiviral constructs (LMP-HP_522, LMP-HP_524) were tested in Rag2−/− DN3 cells, with HP_522 and and HP_524 shRNA constructs knocking down Pten mRNA by 50% and 75%, respectively (Figure 5A). A similar decrease was seen in PTEN protein expression (Figure 5B). Although PTEN-shRNA transduced TCRβ+Rag2−/− DN3 cells bypassed the requirement for Notch signals at the β-selection checkpoint, as measured by the presence of DP cells on OP9-Ctrl cells (Figure 5C-D), control firefly luciferase-shRNA (LMPff)–transduced TCRβ-expressing cells failed to traverse the β-selection checkpoint in the absence of Notch signals. The survival and differentiation of PTEN-shRNA–transduced TCRβ+Rag2−/− DN3 cells was PTEN-dose dependent, as greater knockdown of PTEN with HP_524 shRNA led to greater differentiation capacity in the absence of Notch signals. Similar to what was seen with Ptenf/f;Lck−cre+ cells, PTEN-knockdown in TCRβ+Rag2−/− DN3 cells cultured without Notch signals had dramatically reduced proliferative potential compared with those receiving Notch signals.
Knockdown of Pten expression in Rag2−/− DN3 cells with TCRβ allows for T-cell differentiation across the β-selection checkpoint in the absence of Notch signals. (A) qRT-PCR analysis (normalized to β-actin) of Pten mRNA expression in Rag2−/− DN3 cells transduced to express PTEN shRNA (LMP-HP_522 or LMP-HP_524) or firefly luciferase shRNA (LMPff). Data are representative of 3 independent experiments. (B) Analysis of PTEN protein expression in NIH3T3 cells retrovirally transduced to express the indicated shRNAs is shown as immunoblots of whole cell lysates probed with antibodies specific for PTEN or GAPDH. Data are representative of 3 independent experiments. (C-D) Developmental progression of culture-derived Rag2−/− DN3a cells transduced to express shRNAs as indicated and subsequently cultured for 6 days with OP9-DL1 or OP9-Ctrl cells. (C) Flow cytometric analysis of CD4 and CD8 surface expression is shown for GFP+ (shRNAs) and YFP+ (TCRβ or MIY) gated cells, whereas panel D shows the corresponding cellularity of DP cells present in the cultures, as indicated. DP cellularity was obtained by multiplying the total cellularity by the percentage of DP cells present in the cultures of each independent experiment. Data are representative of 3 independent experiments.
Knockdown of Pten expression in Rag2−/− DN3 cells with TCRβ allows for T-cell differentiation across the β-selection checkpoint in the absence of Notch signals. (A) qRT-PCR analysis (normalized to β-actin) of Pten mRNA expression in Rag2−/− DN3 cells transduced to express PTEN shRNA (LMP-HP_522 or LMP-HP_524) or firefly luciferase shRNA (LMPff). Data are representative of 3 independent experiments. (B) Analysis of PTEN protein expression in NIH3T3 cells retrovirally transduced to express the indicated shRNAs is shown as immunoblots of whole cell lysates probed with antibodies specific for PTEN or GAPDH. Data are representative of 3 independent experiments. (C-D) Developmental progression of culture-derived Rag2−/− DN3a cells transduced to express shRNAs as indicated and subsequently cultured for 6 days with OP9-DL1 or OP9-Ctrl cells. (C) Flow cytometric analysis of CD4 and CD8 surface expression is shown for GFP+ (shRNAs) and YFP+ (TCRβ or MIY) gated cells, whereas panel D shows the corresponding cellularity of DP cells present in the cultures, as indicated. DP cellularity was obtained by multiplying the total cellularity by the percentage of DP cells present in the cultures of each independent experiment. Data are representative of 3 independent experiments.
c-Myc induction is required for Notch-mediated cellular proliferation
Although conditional Pten-deleted DN3a cells survive and differentiate to DP cells in the absence of Notch signaling, they fail to undergo cellular proliferation. This may be because of the loss of additional Notch-mediated proliferation mechanisms that cannot be compensated by the absence of PTEN. To address this, we generated Rag2−/−Ptenf/f;Lck−cre+ mice, which allowed for a more precise definition of the interactions between pre-TCR, Notch, and PTEN at the β-selection checkpoint, as the timing of pre-TCR expression, loss of PTEN, and Notch availability can be manipulated by TCRβ-transduction, Lck-mediated Pten deletion and Delta-like availability in OP9 cell cultures. In vitro–derived DN3 cells from Rag2−/−Ptenf/f;Lck−cre+ and Rag2−/−Pten+/+;Lck−cre+ mice were transduced to express TCRβ and YFP (MIY) and cultured in the absence or presence of Notch signals (Figure 6). As seen before, proliferation of TCRβ-transduced Rag2−/−Ptenf/f;Lck−cre+ DN3 cells receiving Notch signals was dramatically higher than that of the other groups (Figure 6C). Also in agreement with the previous experiments, TCRβ-transduced Rag2−/−Pten+/+;Lck−cre+ DN3 cells could not survive or differentiate in the absence of Notch signals, whereas Pten-deleted cells overcame this block (Figure 6), albeit with much reduced cellularity compared with cells receiving Notch signals.
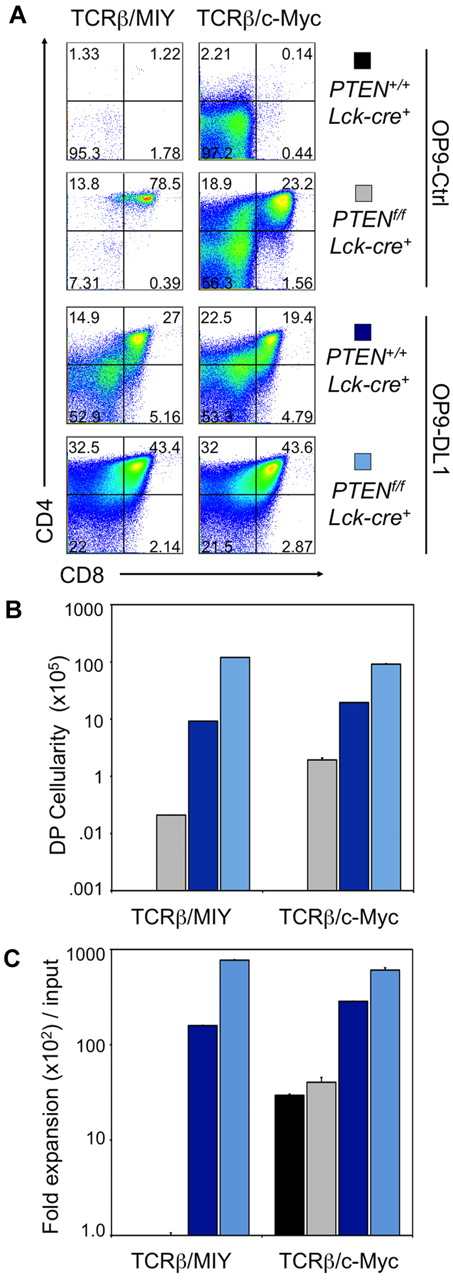
Conditional Pten deletion and ectopic expression of c-Myc allows for survival, differentiation, and proliferation of DN3 cells across the β-selection checkpoint in the absence of Notch signals. Developmental progression of culture-derived Rag2−/−PTEN+/+;Lck−cre+ or Rag2−/−PTENf/f;Lck−cre+ DN3 cells retrovirally cotransduced to express TCRβ (GFP+) and MIY or c-Myc (YFP+) and cultured with OP9-Ctrl or OP9-DL1 cells for 6 days. (A) Flow cytometric analysis of CD4 and CD8 cell surface expression is shown for GFP+, YFP+, CD45+ gated cells, whereas panels B and C show the corresponding DP cellularity and fold expansion, respectively, as indicated. Lin− c-Kit+ Sca-1+ cells sorted from bone marrow of PTENf/f;Lck−cre+ or PTEN+/+;Lck−cre+Rag2−/− mice were cultured with OP9-DL1 cells for 14 days, retrovirally transduced, then sorted for YFP+ and GFP+ DN3 cells, and cultured as indicated. Fold expansion was obtained from the total cellularity divided by the number of cells used at the start of the culture (input), and DP cellularity by multiplication of the total cellularity by the percentage of DP cells present in the cultures. Results are representative of 3 independent experiments.
Conditional Pten deletion and ectopic expression of c-Myc allows for survival, differentiation, and proliferation of DN3 cells across the β-selection checkpoint in the absence of Notch signals. Developmental progression of culture-derived Rag2−/−PTEN+/+;Lck−cre+ or Rag2−/−PTENf/f;Lck−cre+ DN3 cells retrovirally cotransduced to express TCRβ (GFP+) and MIY or c-Myc (YFP+) and cultured with OP9-Ctrl or OP9-DL1 cells for 6 days. (A) Flow cytometric analysis of CD4 and CD8 cell surface expression is shown for GFP+, YFP+, CD45+ gated cells, whereas panels B and C show the corresponding DP cellularity and fold expansion, respectively, as indicated. Lin− c-Kit+ Sca-1+ cells sorted from bone marrow of PTENf/f;Lck−cre+ or PTEN+/+;Lck−cre+Rag2−/− mice were cultured with OP9-DL1 cells for 14 days, retrovirally transduced, then sorted for YFP+ and GFP+ DN3 cells, and cultured as indicated. Fold expansion was obtained from the total cellularity divided by the number of cells used at the start of the culture (input), and DP cellularity by multiplication of the total cellularity by the percentage of DP cells present in the cultures. Results are representative of 3 independent experiments.
Considering c-Myc transcript levels are reduced in the absence of Notch signals (Figure 1B), we examined whether restorating c-Myc expression in DN3 cells could allow for cellular proliferation in the absence of Notch signals. To this end, we cotransduced Pten+/+Lck−cre+or Ptenf/f;Lck−cre+ Rag2−/− DN3 cells to express c-Myc and TCRβ. In the absence of Notch signaling, only TCRβ/c-Myc-expressing Ptenf/f;Lck−cre+, but not Pten+/+Lck−cre+, DN3 cells traversed the β-selection checkpoint and differentiated into DP cells (Figure 6A-B). Furthermore, Rag2−/−Ptenf/f;Lck−cre+ DN3 cells proliferated appreciably, generating DP cells at percentages comparable with TCRβ+Rag2−/−Pten+/+Lck−cre+ cells cultured on OP9-DL1 (Figure 6). In the presence of Notch signaling, TCRβ/c-Myc cotransduced DN3 cells from mice of both genotypes differentiated across β-selection and proliferated extensively.
Together, these results indicate that loss of PTEN partly substitutes for the required Notch receptor-ligand interactions, with an added requirement for ectopic c-Myc expression to restore the Notch-induced proliferative burst associated with cells traversing the β-selection checkpoint.
Discussion
In this study, we addressed the mechanism by which Notch signals mediate trophic effects at the β-selection checkpoint. Our laboratory recently showed that Notch-ligand interactions were crucial for maintaining PI3K/Akt pathway activity, leading to survival and glucose metabolism in DN3 cells. Despite these studies demonstrating a relationship between Notch signaling and the PI3K pathway, the precise mechanism for this interaction was unknown. Here, we find HES1, PTEN, and c-Myc as key molecular players downstream of Notch for the regulation of survival, differentiation, and proliferation at the β-selection checkpoint (supplemental Figure 7).
PI3K signaling and downstream Akt/PKB activation is essential for the survival and metabolism of proliferating pre-T cells.15,39 In the thymus, known upstream inducers of this pathway include IL-7R (CD127)34,37 and CXCR4.5,6 However, whether these signaling pathways or their molecular intermediates are the targets of Notch, and the means by which Notch regulates activation of the PI3K pathway remained unclear. Although cytokine and chemokine-based responsiveness of a developing T cells is often regulated at receptor expression levels,26,40 no changes in CD127 and CXCR4 expression were found with the loss of Notch signaling, suggesting that decreased PI3K signals are not because of receptor down-regulation. In addition, we previously showed that IL-7R function, as measured by signal transducer and activator of transcription (STAT)5 phosphorylation, is retained after the loss of Notch signals.15 Recently, IGF1R expression was reported to be regulated by Notch signals in T-ALL,41 providing a potential link to PI3K pathway activation in leukemic cells. However, a gene expression microarray analysis from Rag2−/− DN3 cells cultured on OP9-DL1 or OP9-Ctrl cells failed to show changes in Igf1r expression on loss of Notch signals (M. Ciofani and J.C.Z.-P., unpublished results, January 2008). These results suggest that receptors that can activate PI3K are operationally available for DN3 cells, but that concomitant Notch signals are required to ensure that this signaling pathway becomes active and responsive to external cues.
The importance for Notch signaling at the DN3 stage and its ability to support PI3K activation is additionally critical when considering the dual signaling properties of CXCR4,42 namely its ability to promote either cell survival or apoptosis via PI3K/Akt or p38/MAPK, respectively. Under circumstances where Notch signaling is discontinued and CXCR4 continues to operate with a diminished capacity to induce PI3K signals, the observed loss of DN3 cells could be because of CXCR4-induced p38 signaling, which has been shown to disrupt early thymocyte differentiation.43 In this regard, we previously showed that inhibition of p38 signaling leads to increased survival and proliferation of β-selected DN3 cells cultured on OP9-DL1 cells.25
A key question that arises is how a signaling pathway such as Notch, which directly regulates gene transcription within the nucleus, can affect the activity of receptor proximal signals that engage the PI3K pathway. Here, we establish a mechanism of interaction between Notch and the PI3K pathway, whereby Notch-dependent transcriptional activation of HES1 mediates the down-regulation of PI3K pathway inhibitor, PTEN.20 This interplay explains how Notch is able to influence the signaling outcomes from cell surface receptors that use the PI3K pathway (supplemental Figure 7). The functional interaction among these players was also observed in T-ALL,20 highlighting the need for tight regulation of these interactions during normal T-cell development. This is achieved by the temporal regulation of Notch receptor expression after the β-selection checkpoint,44 as well as the auto-inhibitory loop of HES1, which limits the heightened level of PI3K responsiveness in DN cells.
Our findings point to HES1 as the key executor of Notch signals that ultimately affects PI3K activity. HES1 appears to not only repress the expression of various cyclin-dependent kinase inhibitors,32,33 but also represses Pten promoter activity, ensuring that β-selected cells can maximize their ability to proliferate and respond to the growth-promoting cues provided by the pre-TCR and the thymus microenvironment. Experimentally, interfering with HES1 function in DN3 cells increases PTEN expression, resulting in decreased ability to differentiate and progress across β-selection. However, a complete blockage in T-cell development is not seen, and this is probably because of incomplete inhibition of HES1 function by dnHES1 or shHES1, or compensation by Hes1 related genes, such as Hey genes.29,36 However, HES5 and HES6 did not appear to play compensatory roles in dnHES1-expressing cells (data not shown). Although HES1 was previously observed to promote proliferation of thymocytes,28,30,33 HES1 overexpression, even with ectopic c-Myc expression, was unable to compensate for Notch signal withdrawal at the β-selection checkpoint. This is probably because of the strong transcriptional repressor activity of HES1,28 which becomes unchecked wtih a retroviral expression system. Nonetheless, HES1 overexpression in DN3 cells led to increased CD71 expression, an indicator of PI3K/Akt pathway activity in thymocytes,16 supporting the proposed regulatory gene network.
A recent report using Hes1f/f mice revealed a critical role for HES1 in early T-lineage commitment, proliferation, and differentiation, although it appeared to be dispensable through and beyond the β-selection checkpoint.29 Our findings that interfering with HES1 function affected early T-cell development and expansion, and led to an increase in non–T-lineage cells are consistent with that report. Similarly, the lower cell yields observed in the thymus of Hes1-deleted mice are in agreement with the effects of decreased cellular proliferation in cells expressing dnHES1 or shHES1. However, our results point to a mechanistically important role for HES1 at the β-selection checkpoint, which was not seen in the Hes1f/f mice. Several potential explanations exist for the apparent discrepancies. Key differences exist between the approaches used, including the stage of T-cell development at which HES1 function is manipulated, and the timing for when the developmental effects are interrogated. For example, we used of Rag2−/− DN3 cells to more precisely pinpoint the developmental stage at which the requirement for HES1 function was examined, either before or after DN3 cells are induced to receive signals from the pre-TCR or Notch receptors. With this approach, we were able to discern a clear role for HES1 in β-selecting DN cells to down-regulate PTEN expression. However Wendorff et al did not offer a potential mechanism of action for HES1 in T-cell development, and did not see a role for HES1 in regulating PTEN expression.29 This last conclusion was based on an analysis of DP leukemic cells, and not normal DN cells, and as such, it is probable that DP cells, which typically do not express HES1, would not show a change in PTEN expression when the Hes1 gene is deleted. In addition, we found that Pten deletion efficiently restored DN3 survival and differentiation to the DP stage on Notch signal withdrawal.
Although conditional loss of PTEN in DN3a cells allows for their survival and differentiation to the DP stage in the absence of Notch signals, their proliferative capacity is greatly diminished, indicating that other pathways downstream of Notch are responsible for this outcome. Several studies of T-ALL have implicated c-Myc as a direct downstream target of Notch signaling, and as a critical component in transformation and cell growth.18,19 In T-cell development, c-Myc is reported as a mediator of proliferation, but not developmental progression.45,46 In agreement with these reports, we find that Notch-induced c-Myc expression at this stage of development is responsible for promoting cellular expansion, leading to a large DP cell pool, and proliferation within the DN cell subset. Although we showed that loss of HES1 function leads to increased expression of cell cycle inhibitors, ectopic expression of c-Myc in the absence of Notch signals would probably counter the loss of HES1-mediated Cdkn gene repression,47 thus enabling cell cycle progression and bypassing the need for Notch signaling to support the proliferative burst typically associated with pre-TCR signaling. Collectively, our findings show that more than one downstream effector is responsible for the trophic effects of Notch signaling at the β-selection checkpoint.
Notch signaling is required by pre-T cells to traverse the β-selection checkpoint. Here, we identify key signaling intermediates downstream of Notch that are responsible for T-lineage differentiation, proliferation, survival, and cellular metabolism. HES1 and PTEN are largely responsible for coordinating differentiation, survival and metabolism of pre-T cells at the critical β-selection checkpoint by bridging Notch signals to the activation of the PI3K/Akt pathway, whereas Notch induction of c-Myc expression drives the proliferation of β-selected cells that reach the DP stage of T-cell development, at which point Notch signaling ceases to avoid an otherwise inevitable path to leukemic transformation.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Dr Maria Ciofani for her initial insights and advice. They highly appreciate the expert support provided by the Sunnybrook Research Institute's Comparative Research Facility.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) MOP42387, the Terry Fox Foundation, and the Krembil Foundation. G.W.W. was supported by a CIHR Studentship Award. A.A.F. is supported by the Leukemia & Lymphoma Society Scholar Award, and National Institutes of Health grants R01CA120196 and R01CA129382. J.C.Z.-P. is supported by a Canadian Research Chair in Developmental Immunology.
National Institutes of Health
Authorship
Contribution: G.W.W. designed and performed experiments; G.C.K. performed flow cytometric cell sorting and data analysis; T.W.M. and A.A.F. provided critical reagents and experimental expertise and advice; J.C.Z.-P. designed experiments and supervised the study; and G.W.W. and J.C.Z.-P. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Juan Carlos Zúñiga-Pflücker, Sunnybrook Research Institute, Dept of Immunology, University of Toronto, 2075 Bayview Ave, A3-31, Toronto, ON M4N 3M5 Canada; e-mail: jczp@sri.utoronto.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal