Abstract
Chronic infantile neurologic cutaneous and articular (CINCA) syndrome is an IL-1–driven autoinflammatory disorder caused mainly by NLRP3 mutations. The pathogenesis of CINCA syndrome patients who carry NLRP3 mutations as somatic mosaicism has not been precisely described because of the difficulty in separating individual cells based on the presence or absence of the mutation. Here we report the generation of NLRP3-mutant and nonmutant-induced pluripotent stem cell (iPSC) lines from 2 CINCA syndrome patients with somatic mosaicism, and describe their differentiation into macrophages (iPS-MPs). We found that mutant cells are predominantly responsible for the pathogenesis in these mosaic patients because only mutant iPS-MPs showed the disease relevant phenotype of abnormal IL-1β secretion. We also confirmed that the existing anti-inflammatory compounds inhibited the abnormal IL-1β secretion, indicating that mutant iPS-MPs are applicable for drug screening for CINCA syndrome and other NLRP3-related inflammatory conditions. Our results illustrate that patient-derived iPSCs are useful for dissecting somatic mosaicism and that NLRP3-mutant iPSCs can provide a valuable platform for drug discovery for multiple NLRP3-related disorders.
Introduction
Chronic infantile neurologic cutaneous and articular syndrome (CINCA syndrome; MIM #607715) is a dominantly inherited autoinflammatory disease characterized by systemic inflammation with an urticaria-like rash, neurologic manifestations, and arthropathy.1 NLRP3 mutation is the first and so far the only identified mutation that is responsible for CINCA syndrome.2,3 NLRP3 is expressed mainly in myelomonocytic lineage cells and chondrocytes3 and acts as an intracellular sensor of danger signals from various cellular insults. In normal macrophages, a first stimulus, such as lipopolysaccharide (LPS), induces the synthesis of NLRP3 and the biologically inactive proIL-1β.4 A second stimulus, such as ATP, enhances the assembly of a protein complex called the NLRP3-inflammasome.5 The inflammasome contains caspase1, which executes the proteolytic maturation and secretion of IL-1β. Although normal monocytes/macrophages show no or limited IL-1β secretion in response to LPS stimulation alone, CINCA patients' cells exhibit robust IL-1β secretion because the mutant NLRP3-inflammasome is autoactivated without the need for any second stimulus.6 It is therefore thought that the manifestations of CINCA syndrome are predominantly caused by the excessive secretion of the proinflammatory cytokine, IL-1β, and this concept is supported by the efficacy of an IL-1 receptor antagonist (IL-1Ra) for decreasing most of the symptoms.7 However, because IL-1Ra treatment does not seem to ameliorate the characteristic arthropathy of cartilage overgrowth and joint contraction,8 a more specific therapeutic approach that directly modulates the NLRP3-inflammasome is desired.
Although approximately half of CINCA patients carry heterozygous gain-of-function mutations of the NLRP3 gene,2,3 30% to 40% of all patients have mutations in NLRP3 in only a small number of somatic cells.9,10 Because the population of mutant cells is relatively small (4.2%-35.8% in blood cells), it remains controversial whether the small fraction of NLRP3-mutated cells actually causes the strong autoinflammation observed in CINCA patients, or whether the NLRP3 mutations found in mosaic patients are just a bystander, with all cells carrying an unknown mutation of another gene that causes the disease.11
Somatic mosaicism refers to the presence of more than 1 genetically distinct cell population in a single person, and has been identified in patients with various diseases.12,13 The relevance of somatic mosaicism to the onset of diseases has been suggested mainly through sequence-based approaches. However, direct evidence that a cell population with a distinct genetic property shows disease-specific characteristics is lacking because it has been impossible to separately extract individual live cells from affected tissues to assess their biologic characteristics. Regarding hematopoietic disorders in which mutant cells show decreased expression of a certain protein, genetic heterogeneity caused by somatic mutations was detected by flow cytometry after intracellular staining,14-16 but sorting out alive mutant and nonmutant cells for evaluating biologic property has been impossible.
Induced pluripotent stem cells (iPSCs) are pluripotent cell lines directly reprogrammed from somatic cells.17 Patient-derived iPSCs can provide somatic cells, which cannot be directly obtained from patients, and this discovery has led to the development of a new field of disease modeling (reviewed by Grskovic et al18 ). In addition, iPSC technology has another interesting characteristic that each iPSC clone originates from a single cell,19 which may make it possible to obtain genetically different iPSC clones from a person.
In this study, we established mutant and nonmutant iPSC lines from the same patients by deriving iPSCs from patients carrying a mutation of an autosomal gene as somatic mosaicism. By analyzing the disease-relevant characteristic of IL-1β secretion, we demonstrated that mutant macrophages are mainly responsible for the disease phenotype in the mosaic patients. Moreover, using a robust differentiation protocol to generate macrophages and purifying them by their surface marker expression, we showed that drug candidates inhibit the IL-1β secretion from mutant macrophages. Our data prove the usefulness of iPSC technology both for dissecting somatic mosaicism and as a platform for drug discovery of multiple NLRP3-related inflammatory diseases.
Methods
Human iPSC generation
We obtained skin biopsy specimens from 2 independent patients (patient 1, CIRA188Ai; and patient 2, CIRA086Ai). This study was approved by Ethics Committee of Kyoto University, and informed consent was obtained from both the patients and their guardians in accordance with the Declaration of Helsinki. We expanded the fibroblasts in DMEM (Nacalai Tesque) containing 10% FBS (Invitrogen) and 0.5% penicillin and streptomycin (Invitrogen). Generation of iPS cells was performed as described previously.17 In brief, we introduced OCT3/4, SOX2, KLF4, and c-MYC using ecotropic retroviral transduction into fibroblasts expressing the mouse Slc7a1 gene. Six days after transduction, the cells were harvested and replated onto mitotically inactivated SNL feeder cells. The next day, we replaced the medium with Primate ES cell medium (ReproCELL) supplemented with 4 ng/mL bFGF (Wako). Three weeks after this period, individual colonies were isolated and expanded. Cell culture was performed under 37°C, with 5% CO2 and 21% O2 unless otherwise stated. Cells were examined using Olympus CKX41 inverted microscope equipped with Nikon Digital Sight DS-L2 camera. A UPlan FLN 4×/0.13 objective (Nikon) was used for image acquisition.
Genetic analysis
Genomic DNA from either fibroblasts or iPSCs was isolated. The PCR product of exon 3 of NLRP3 was sequenced directly or after subcloning with a TOPO TA cloning kit (Invitrogen), using an ABI 3100 sequencer (Applied Biosystems). For pyrosequencing, the PCR product of exon 3 of NLRP3 was analyzed by PyroMarkQ96ID (QIAGEN).
RNA isolation and quantitative PCR for NANOG and the transgene
Total RNA was purified with the Trizol reagent (Invitrogen) and treated with a Turbo DNA-free kit (Ambion) to remove genomic DNA contamination. A total of 1 μg of total RNA was used for a reverse transcription reaction with ReverTraAce-α (Toyobo) and the dT20 primer, according to the manufacturer's instructions. Quantitative PCR was performed on the 7900HT Fast Real-Time PCR System (Applied Biosystems) with SYBR Premix ExTaqII (Takara). The primer sequences are described in supplemental Table 4 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Southern blotting
Genomic DNA (5 μg) was digested with BglII and ScaI overnight. The digested DNA fragments were separated on 1% agarose gels and were transferred to a nylon membrane (GE Healthcare). The membrane was incubated with a digoxigenin (DIG)–labeled human cMYC DNA probe in DIG Easy Hyb buffer (Roche Diagnostics) at 42°C overnight with constant agitation. After washing, an alkaline phosphatase-conjugated anti-DIG antibody (1:10 000; Roche Diagnostics) was added to a membrane. Signals were obtained using CDP-star (Roche Diagnostics) and detected by an LAS4000 imaging system.
Teratoma formation
Approximately 2 × 106 cells were injected subcutaneously into the dorsal flank of immunocompromised NOD/scid/γcnull mice (Central Institute for Experimental Animals). Masses were excised 8 to 10 weeks after injection and fixed with PBS containing 4% paraformaldehyde. Paraffin-embedded tissues were sliced and stained with hematoxylin and eosin. Slides were examined using BIOREVO BZ-9000 (KEYENCE). A PlanApo 20×/0.75 objective (Nikon) and BZ-II Viewer software (KEYENCE) were used for image acquisition.
In vitro differentiation into macrophages
Undifferentiated human embryonic stem cell (ESC) and iPSC lines were cultured on mitotically inactivated SNL feeder cells with Primate ES cell medium supplemented with 4 ng/mL bFGF. During the differentiation of the cells into macrophages, cells were cultured under 37°C, with 5% CO2 and 5% O2. On day 0, the iPSCs were plated at a ratio of 1:15 onto a mitotically inactivated OP9 feeder layer on 100-mm cell culture plates in α-MEM (Invitrogen) containing 10% FBS and 1% Antibiotic-Antimycotic (Invitrogen) supplemented with 50 ng/mL VEGFα (R&D Systems). On day 5, the medium was changed. On day 10, the differentiating iPSCs were collected by trypsinization, and Tra-1-85+ CD34+ and KDR+ hematopoietic progenitors were sorted on a FACSAria II instrument (BD Biosciences). The progenitors were plated at 2 × 104 cells on another mitotically inactivated OP9 feeder layer on 100-mm cell culture plates or at 3 × 103 cells/well in 6-well cell culture plates in α-MEM containing 10% FBS and 1% Antibiotic-Antimycotic supplemented with 50 ng/mL IL-3, 50 ng/mL stem cell factor, 10 ng/mL thrombopoietin, 50 ng/mL Flt-3 ligand, and 50 ng/mL M-CSF (all R&D Systems). On day 18, the medium was changed. On day 26, differentiating cells were collected with Accumax (Innovative Cell Technologies), and CD14+ iPSC-derived macrophages were purified on an autoMACSpro instrument (Miltenyi Biotec).
Peripheral blood mononuclear cells (PBs) were obtained from healthy volunteers, and CD14+ monocytes were purified on the autoMACSpro instrument. For macrophage differentiation, 5 × 105 monocytes were plated in 6-well cell culture plates in RPMI 1640 (Sigma-Aldrich) containing 10% FBS and 1% Antibiotic-Antimycotic supplemented with 50 ng/mL M-CSF. On day 5, the adherent cells were collected with Accumax, and CD14+ blood-derived macrophages (B-MPs) were purified on the autoMACSpro instrument. May-Giemsa–stained slides were examined using BIOREVO BZ-9000. A PlanApo 40×/0.95 objective (Nikon) and BZ-II Viewer software were used for image acquisition.
FACS analysis
Hematopoietic marker expression was evaluated on a MACSQuant Analyzer (Miltenyi Biotec). Primary antibodies Tra-1-85-FITC (R&D Systems), CD34-PE (Beckman Coulter), KDR–AlexaFluor-647 (BioLegend), CD45-PE (BD Biosciences PharMingen), and CD14-APC (Beckman Coulter) were used.
Immunocytochemistry
For immunocytochemistry, cells were fixed with PBS containing 4% paraformaldehyde for 5 minutes, permeabilized in PBS containing 0.1% Tween 20 for 5 minutes, and blocked in PBS containing 3% BSA for 10 minutes, all at room temperature. The primary antibody was for CD68 (1:50; Santa Cruz Biotechnology), and the secondary antibody was Cy3-conjugated AffiniPure Donkey Anti–Mouse IgG (1:100; Jackson ImmunoResearch Laboratories). Nuclei were stained with 1 μg/mL Hoechst 33342 (Invitrogen). Cells were examined using BIOREVO BZ-9000. A Plan Fluor DL 10×/0.30 Ph1 objective (Nikon) and BZ-II Viewer software were used for image acquisition.
Electron microscopy
The 5 × 104 macrophages in 20 μL suspension were placed on the poly-L-lysine treated, carbon-coated sapphire disks (3 mm in diameter) and incubated for 30 minutes at 37°C with 5% CO2. The cell-adsorbed disk was then subjected to chemical fixation with 2.5% glutaraldehyde in NaHCa buffer (100mM NaCl, 30mM HEPES, 2mM CaCl2, adjusted at pH 7.4 with NaOH). These specimens were postfixed with 1% osmium and 1.5% K4Fe(CN)6 in 0.1M PBS buffer, washed, dehydrated with a series of ethanol, and embedded in Epoxy resin (TAAB EPON812). After the polymerization at 70°C, the ultra-sections (70 nm) obtained by Ultramicrotome (Leica FC6) were mounted in EM grids, stained with uranyl acetate/lead citrate, and then observed by conventional TEM (JEOL JEM1400).
PCR and microarray analysis of macrophages
Total RNA was column-purified with the RNeasy kit (QIAGEN) and treated with RNase-free DNase (QIAGEN). A total of 20 ng of total RNA was reverse transcribed into cDNA using random primers and the Sensiscript RT Kit (QIAGEN). Quantitative PCR was performed on a StepOne Plus Real-Time PCR System (Applied Biosystems) with TaqMan Gene Expression Master Mix (Applied Biosystems). The primer sequences are described in supplemental Table 4. For the microarray analysis, RNA probes were hybridized to SurePrint G3 Human GE 8 × 60K Microarrays (Agilent Technologies) according to the manufacturer's protocols. Microarrays were scanned, and the data were analyzed using the GeneSpring GX Version 11 software program (Agilent Technologies). The complete dataset from this analysis is available at the NCBI Gene Expression Omnibus using accession no. GSE38626.
LM infection
Listeria monocytogenes EGD (LM) were grown in brain heart infusion broth (Eieken Chemical), washed, suspended in PBS supplemented with 10% glycerol, and stored in aliquots at −80°C. Macrophages were seeded into an 8-well chamber slide at 2 × 105 cells/well in RPMI containing 10% FBS and then infected with bacteria at a multiplicity of infection of 10 for 60 minutes at 37°C. Cells were cultured for further 1 or 5 hours in the presence of 5 μg/mL gentamicin. The cells were fixed in 4% paraformaldehyde and incubated with PBS containing 10% Blocking One (Nacalai Tesque) and 0.1% saponin. F-actin and nuclei were visualized by staining with Alexa-488-phalloidin (Invitrogen) and 4′,6-diamidino-2-phenylindole (Dojindo), respectively. The bacteria were stained by treatment with a goat anti-Listeria polyclonal antibody (Kirkegaard & Perry Laboratories) and then with the Alexa 546 anti–goat IgG antibody (Invitrogen). Slides were examined using BIOREVO BZ-9000. A PlanApo_VC 100×H/1.40 objective (Nikon) and BZ-II Viewer software were used for image acquisition, and BZ-II Analyzer (KEYENCE) was used for image processing. Immunofluorescence was evaluated with the IN Cell Analyzer 2000, and samples were analyzed with the IN Cell Developer Toolbox Version 1.8 software program (GE Healthcare).
Cytokine secretion from macrophages
Purified iPS-MPs or B-MPs were seeded at the indicated counts per well or 5 × 104 cells/well unless otherwise stated in 96-well cell culture plates in RPMI 1640 containing 10% FBS and 1% Antibiotic-Antimycotic. Cells were cultured for 2 hours in the presence or absence of inhibitors. The plates were centrifuged at 300g for 10 minutes; then the medium was changed. Cells were cultured for 4 hours in the presence of LPS or recombinant human IL-1β. LPS concentration was 1 μg/mL unless otherwise stated. After the 30 minute or 1-hour culture after the addition of 1mM ATP (Sigma-Aldrich), we collected the supernatants and cell lysates. As second signal stimulants, we also used 500 μg/mL silica crystals (U.S. silica) for 1 hour, or 100 μg/mL monosodium urate crystals (Sigma-Aldrich) for 3 hours. For the supernatant transfer experiments, we harvested the supernatant from the wells of mutant or wild-type iPS-MPs, which were stimulated with LPS for 4 hours. After centrifugation, we transferred the supernatants to the wells of other iPS-MPs and cultured them for another 4 hours. The cytokine concentration of the supernatants was determined using a Th1/Th2 11plex FlowCytomix Kit (Bender MedSystems) following the manufacturer's instructions. Reagents were purchased as follows: CA074Me (Calbiochem), IL-1Ra (R&D Systems), oxidized ATP (oATP; Sigma-Aldrich), pyridoxalphosphate-6-azophenyl-2′,4′-disulphonic acid (PPADS; Sigma-Aldrich), cycloheximide (Sigma-Aldrich), MG132 (Calbiochem), Bay11-7082 (Sigma-Aldrich), and Ac-YVAD-CHO (Calbiochem).
LDH secretion assay
The lactate dehydrogenase (LDH) concentration of the supernatants of iPS-MPs after a 4-hour culture with LPS was determined with an LDH Cytotoxicity Detection kit (Takara) following the manufacturer's instructions.
Statistical analysis
The data were processed using the SPSS Statistics Version 18 software package. The values are reported as the mean ± SEM. Comparisons between groups were performed using the unpaired Student t test. P < .05 was considered statistically significant.
Results
Establishment and characterization of iPSCs
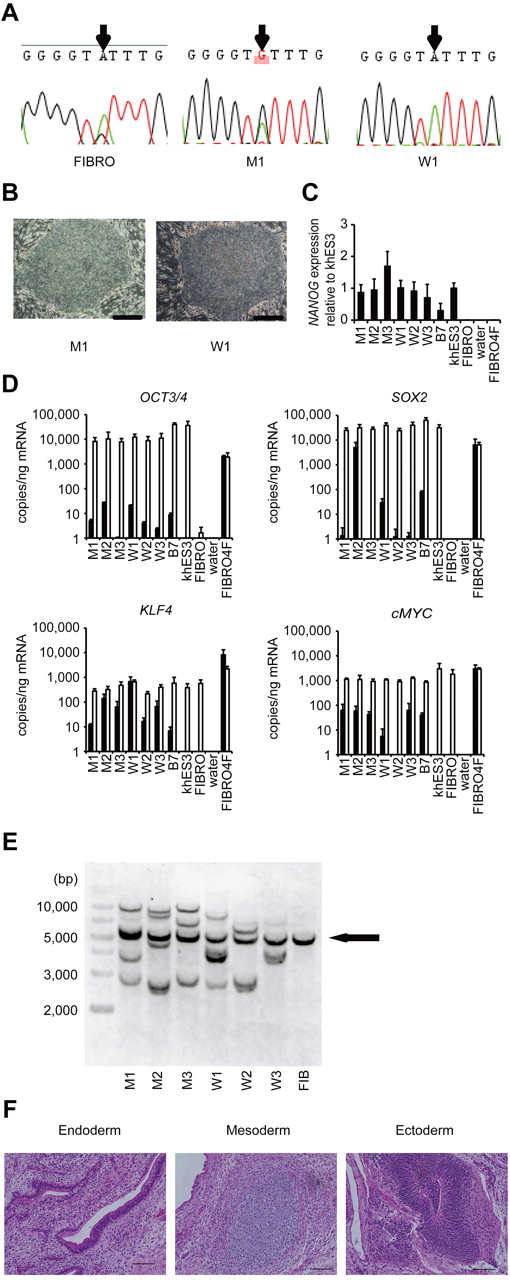
Dermal fibroblasts were obtained from 2 male CINCA patients who had mutations of NLRP3 as somatic mosaicism. Both patients had nonsynonymous point mutations in the NLRP3 coding region. The fibroblasts from patients 1 and 2 contained 34% and 9.8% mutant cells, respectively (Figure 1A; supplemental Figure 1A). These fibroblasts were reprogrammed to iPSCs after transduction with retroviral vectors encoding OCT3/4, SOX2, KLF4, and cMYC.17 Twelve of the 28 isolated clones from patient 1, and 3 of 30 clones from patient 2 had a heterozygous mutation of the NLRP3 gene, whereas the rest of the clones were wild-type (Figure 1A; supplemental Figure 1B-C). The frequency of mutants was comparable among blood cells,9,20 fibroblasts, and iPSCs (Table 1). We randomly selected 3 mutant (M1-M3) and 3 wild-type clones (W1-W3) from patient 1 and 3 mutant (m1-m3) and 3 wild-type clones (w1-w3) from patient 2 for the propagation and subsequent analyses.
Establishment and characterization of iPSCs. (A) Sequencing of the NLRP3 1709 A > G mutation (Y570C) in fibroblasts (FIBRO), mutant iPSCs (M1), and wild-type iPSCs (W1) in patient 1. (B) The morphology of the mutant and wild-type iPSCs. (C) NANOG expression in CINCA iPSCs, control iPSCs (B7), control ESCs (khES3), fibroblasts (FIBRO), and fibroblasts transduced with 4 factors (FIBRO4F) normalized to GAPDH. n = 3. (D) A quantitative RT-PCR assay for the expression of OCT3/4, SOX2, KLF4, and cMYC in iPSCs. One primer set detects only the transgene (in black), and the other primer set detects both the transgene and endogenous gene (in white). n = 3. (E) Retroviral transgene integration analyses. Southern blot analyses were performed with DIG-labeled DNA probes against c-MYC. The parental fibroblasts carried a band in common with all of the iPSC lines (arrow). (F) A teratoma derived from a mutant iPSC clone, M1. Scale bars represent 100 μm. Data are mean ± SEM.
Establishment and characterization of iPSCs. (A) Sequencing of the NLRP3 1709 A > G mutation (Y570C) in fibroblasts (FIBRO), mutant iPSCs (M1), and wild-type iPSCs (W1) in patient 1. (B) The morphology of the mutant and wild-type iPSCs. (C) NANOG expression in CINCA iPSCs, control iPSCs (B7), control ESCs (khES3), fibroblasts (FIBRO), and fibroblasts transduced with 4 factors (FIBRO4F) normalized to GAPDH. n = 3. (D) A quantitative RT-PCR assay for the expression of OCT3/4, SOX2, KLF4, and cMYC in iPSCs. One primer set detects only the transgene (in black), and the other primer set detects both the transgene and endogenous gene (in white). n = 3. (E) Retroviral transgene integration analyses. Southern blot analyses were performed with DIG-labeled DNA probes against c-MYC. The parental fibroblasts carried a band in common with all of the iPSC lines (arrow). (F) A teratoma derived from a mutant iPSC clone, M1. Scale bars represent 100 μm. Data are mean ± SEM.
Mutation frequency among different cell types
| Patient no. . | Site of mutation . | Frequency (%) of mutant cells . | ||
|---|---|---|---|---|
| Whole blood* . | Fibroblasts . | iPSCs . | ||
| 1 | 1709A > G (Y570C) | 33.3 | 34.3 | 42.9 |
| 2 | 919G > A (G307S) | 8.5 | 9.8 | 10.0 |
| Patient no. . | Site of mutation . | Frequency (%) of mutant cells . | ||
|---|---|---|---|---|
| Whole blood* . | Fibroblasts . | iPSCs . | ||
| 1 | 1709A > G (Y570C) | 33.3 | 34.3 | 42.9 |
| 2 | 919G > A (G307S) | 8.5 | 9.8 | 10.0 |
All iPSC clones showed a characteristic human ESC-like morphology (Figure 1B), the reactivation of endogenous pluripotency genes (OCT3/4, SOX2, NANOG; Figure 1C-D; supplemental Figure 1D) and the demethylation of the OCT3/4 promoter regions (supplemental Figure 1E). Transgene expression was rarely detected (Figure 1D; supplemental Figure 1D), and the retroviral integration patterns were confirmed by a Southern blot analysis (Figure 1E; supplemental Figure 1F). All of the iPSC clones maintained a normal karyotype (data not shown). There were neither proviral integration nor copy number changes observed in any of the genes that might affect the function of the NLRP3 inflammasome (supplemental Tables 1 and 2). Genetic identity was proven by a short tandem repeat analysis (supplemental Table 3), and the pluripotency of the iPSC clones was confirmed by the presence of cell derivatives of all 3 germ layers by teratoma formation after injection of undifferentiated iPSCs into immunocompromised NOD/scid/γcnull mice (Figure 1F; supplemental Figure 1G).
Differentiation and characterization of iPSC-derived macrophages
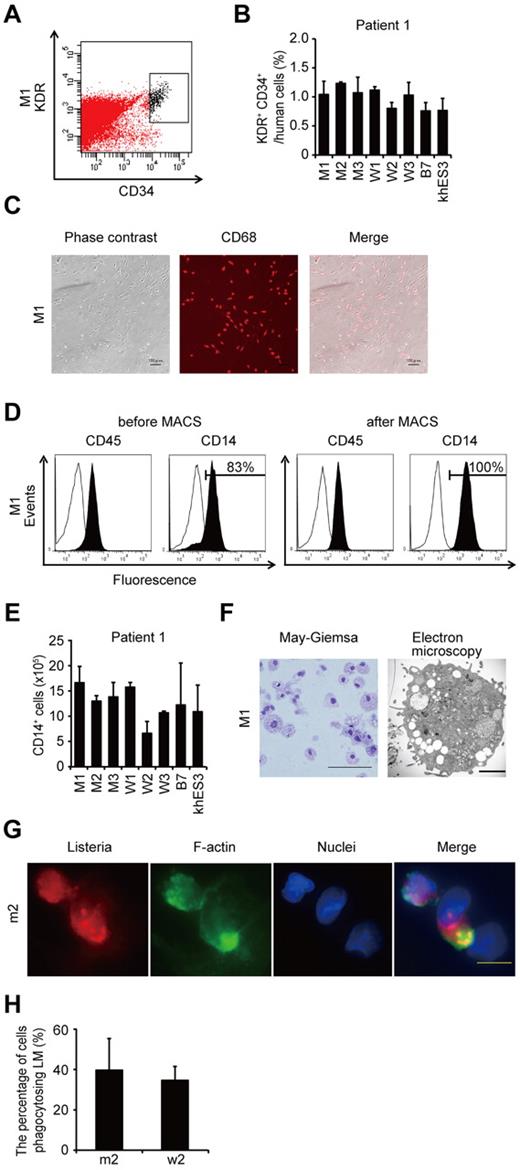
To compare the most prominent features of the disease, we differentiated the patient-derived iPSCs into the monocyte/macrophage lineage using a murine stromal cell line, OP9.21 After culturing the iPSCs on an OP9 feeder layer for 10 days, we collected KDR+ CD34+ hemangioblasts (Figure 2A). All of the iPSC clones, whether they carried an NLRP3 mutation or not, differentiated into KDR+ CD34+ progenitors as efficiently as the control ESC or iPSC clones (Figure 2B; supplemental Figure 2A). Adherent CD68+ macrophages emerged after culturing the KDR+ CD34+ cells on another OP9 feeder layer for 16 days (Figure 2C; supplemental Figure 2B). Approximately 80% of the differentiated cells expressed CD14, and magnetic-activated cell sorting increased the purity to almost 100% (Figure 2D). All of the clones we used efficiently produced comparable amounts of iPSC-derived macrophages (iPS-MPs; Figure 2E; supplemental Figure 2C). The iPS-MPs visualized by light and electron microscopy showed a typical morphology, with a high cytoplasm-to-nucleus ratio and cytoplasmic vacuoles (Figure 2F; supplemental Figure 2D). The iPS-MPs showed a global gene expression pattern closer to that of blood-derived macrophages than to the parental iPSC clone (supplemental Figure 2E-F). Both mutant and wild-type iPS-MPs phagocytosed bacteria to the same extent when we infected the cells with Gram-positive LM, an intracellular bacterium that escapes into the cytosol (Figure 2G-H). These data indicate that both the mutant and wild-type iPS-MPs derived from mosaic CINCA patients are indistinguishable based on their gene expression and their phagocytic function.
Differentiation and characterization of iPSCs-derived macrophages. (A) KDR+ CD34+ hematopoietic progenitors purified 10 days after differentiation. (B) The percentage of KDR+ CD34+ cells in Tra-1-85+ human cells. n = 3. (C) CD68 immunostaining of macrophages. Scale bars represent 100 μm. (D) The histograms show antibody staining (in black) relative to the isotype-matched controls (in white) for a blood cell marker (CD45), and a macrophage marker (CD14), in cells before (left 2 panels) or after (right 2 panels) magnetic-activated cell sorting purification. (E) CD14+ cell counts obtained from iPSCs plated on an OP9 feeder layer on one 100-mm dish. n = 3. (F) Representative morphology of iPS-MPs evaluated by May-Giemsa staining or transmission electron microscopy. Scale bars represent 100 μm and 2 μm, respectively. (G) The phagocytosis by iPS-MPs after LM infection. The cells were treated with anti-LM antibody, phalloidin, and 4′,6-diamidino-2-phenylindole. Scale bar represents 20 μm. (H) The percentage of iPS-MPs phagocytosing LM was calculated as the average of 9 fields of vision. Data are mean ± SEM.
Differentiation and characterization of iPSCs-derived macrophages. (A) KDR+ CD34+ hematopoietic progenitors purified 10 days after differentiation. (B) The percentage of KDR+ CD34+ cells in Tra-1-85+ human cells. n = 3. (C) CD68 immunostaining of macrophages. Scale bars represent 100 μm. (D) The histograms show antibody staining (in black) relative to the isotype-matched controls (in white) for a blood cell marker (CD45), and a macrophage marker (CD14), in cells before (left 2 panels) or after (right 2 panels) magnetic-activated cell sorting purification. (E) CD14+ cell counts obtained from iPSCs plated on an OP9 feeder layer on one 100-mm dish. n = 3. (F) Representative morphology of iPS-MPs evaluated by May-Giemsa staining or transmission electron microscopy. Scale bars represent 100 μm and 2 μm, respectively. (G) The phagocytosis by iPS-MPs after LM infection. The cells were treated with anti-LM antibody, phalloidin, and 4′,6-diamidino-2-phenylindole. Scale bar represents 20 μm. (H) The percentage of iPS-MPs phagocytosing LM was calculated as the average of 9 fields of vision. Data are mean ± SEM.
Elucidation of the pathogenesis of somatic mosaic CINCA syndrome
Monocytes derived from CINCA syndrome patients usually do not spontaneously secrete IL-1β and become active after LPS stimulation.6 Monocytes or mononuclear cells from untreated CINCA syndrome patients, however, sometimes show an increased synthesis of proIL-1β2 and secretion of mature IL-1β,7 even in the absence of LPS stimulation, because they can be activated by persistent inflammation or by the purification procedure. As spontaneous activation complicates the functional analysis, we herein evaluated the IL-1β activation status both before and after the stimulation. We observed that the mRNA expression of IL1B was low in unstimulated iPS-MPs and increased to comparable levels in mutant and wild-type iPS-MPs in response to LPS stimulation (supplemental Figure 3A). Similarly, the mRNA level of NLRP3 was relatively low before LPS stimulation (supplemental Figure 3A). Mature IL-1β was not detectable in the supernatant of the cell culture medium (data not shown). Collectively, these data indicate that the unstimulated iPS-MPs were in an “inactive” state before stimulation.
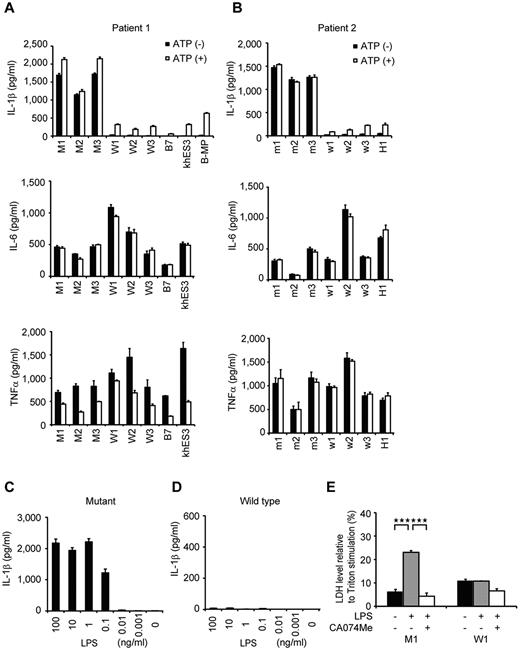
To identify which iPS-MP clones showed the specific features compatible to patients' monocytes, we evaluated their IL-1β secretion. Although LPS stimulation alone led to IL-1β secretion from the mutant iPS-MPs, the addition of ATP was necessary to induce IL-1β secretion from wild-type iPS-MPs, as it was from either ESC-derived or blood-derived macrophages (Figure 3A). The IL-1β level from mutant iPS-MPs was significantly higher than that from wild-type macrophages, even in the presence of LPS plus ATP. Both groups of macrophages showed similar kinetics in their secretion of other cytokines, such as IL-6 or TNFα (Figure 3A). The results were similar in the iPS-MPs from patient 2 (Figure 3B). Although iPS-MPs showed a similar response at lower LPS concentrations (Figure 3C-D; supplemental Figure 3B-C), no IL-1β secretion was detectable from mutant iPSCs, wild-type iPSCs, or parental fibroblasts in response to stimulation with 1 μg/mL LPS (data not shown). These data demonstrate that the abnormal function of the iPS-MPs is predominantly determined by the NLRP3 mutation, and not by some unknown genetic alteration(s) prevalent in all cells. We next investigated whether iPS-MPs show pyronecrosis: a pathogen-induced, cathepsin B-dependent, necrosis-like programmed cell death that is characteristically observed in NLRP3-mutant monocytes/macrophages.22,23 When we compared LDH secretion as a marker of membrane rupture, we found that LPS stimulation evoked a significantly higher LDH secretion only from the mutant iPS-MPs, which was inhibited by the cathepsin B inhibitor, CA074Me (Figure 3E).
Elucidation of the pathogenesis of somatic mosaic CINCA syndrome. (A) Cytokine secretion from iPS-MPs derived from patient 1. After stimulating iPS-MPs by LPS with or without ATP, we determined the IL-1β (top panel), IL-6 (middle panel), or TNFα (bottom panel) level of the supernatant. n = 3. (B) Cytokine secretion from iPS-MPs derived from patient 2, determined as in panel A. (C) IL-1β secretion from mutant iPS-MPs in the presence of 10-fold dilutions of LPS from 100 ng/mL. n = 3. (D) IL-1β secretion from wild-type iPS-MPs, determined as in panel C. (E) LDH secretion from iPS-MPs stimulated with LPS in the presence or absence of the cathepsin B inhibitor, CA074Me. n = 3. Data are mean ± SEM. ***P < .001 (Student t test).
Elucidation of the pathogenesis of somatic mosaic CINCA syndrome. (A) Cytokine secretion from iPS-MPs derived from patient 1. After stimulating iPS-MPs by LPS with or without ATP, we determined the IL-1β (top panel), IL-6 (middle panel), or TNFα (bottom panel) level of the supernatant. n = 3. (B) Cytokine secretion from iPS-MPs derived from patient 2, determined as in panel A. (C) IL-1β secretion from mutant iPS-MPs in the presence of 10-fold dilutions of LPS from 100 ng/mL. n = 3. (D) IL-1β secretion from wild-type iPS-MPs, determined as in panel C. (E) LDH secretion from iPS-MPs stimulated with LPS in the presence or absence of the cathepsin B inhibitor, CA074Me. n = 3. Data are mean ± SEM. ***P < .001 (Student t test).
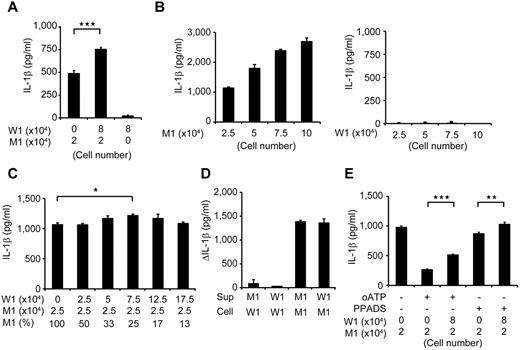
Despite the low percentage of mutant cells, the clinical manifestation of mosaic CINCA patients is similar to that of patients with a heterozygous mutation.9,10 We hypothesized that an interaction between the mutant and wild-type macrophages leads to exacerbation of the inflammation. To test this hypothesis, we modeled a mosaic condition by coculturing mutant and wild-type cells. After stimulating mutant iPS-MPs with LPS in separate cultures or in cocultures with wild-type counterparts, we determined the IL-1β level in the supernatant. We found that the IL-1β secretion significantly increased after coculture (Figure 4A; supplemental Figure 4A). Although increasing the cell concentration raised the total amount of the IL-1β secretion from mutants, it did not accelerate the IL-1β secretion per cell from mutant iPS-MPs or enhance the secretion from wild-type macrophages (Figure 4B). To determine the ratio of mutant/wild-type cells at which the additional IL-1β secretion is most enhanced, we changed the ratio using a fixed number of mutant iPS-MPs and increasing the number of wild-type iPS-MPs. We observed a significant increase only at a percentage of 25% mutant macrophages (Figure 4C). Thus, we capitulated, at least in part, the patient's mosaic condition in vitro.
Remodeling mosaicism by coculturing mutant and wild-type iPS-MPs. (A) IL-1β secretion from cocultured iPS-MPs. We used 2 × 104 mutant iPS-MPs (M1) and 8 × 104 wild-type iPS-MPs (W1) as indicated. n = 6. (B) IL-1β secretion from various numbers of mutant (left panel) or wild-type (right panel) iPS-MPs. The iPS-MPs were seeded at the indicated numbers. n = 3. (C) IL-1β secretion from iPS-MPs that were cocultured at various ratios. The wild-type or mutant iPS-MPs were seeded at the numbers indicated in the first and second rows, respectively. The percentage of mutants is indicated in the third row; n = 3. (D) Increase of IL-1β levels during stimulation by the supernatant. The supernatant was harvested from the wells of the indicated iPS-MPs (Sup) and transferred to the wells of other iPS-MPs (Cell); n = 3. (E) IL-1β secretion from cocultured iPS-MPs in the presence of the ATP receptor antagonist, oATP (300μM) or PPADS (300μM). We used 2 × 104 mutant iPS-MPs (M1) and 8 × 104 wild-type iPS-MPs (W1) as indicated. n = 6. Data are mean ± SEM. ***P < .001 (Student t test). **P < .01 (Student t test). *P < .05 (Student t test).
Remodeling mosaicism by coculturing mutant and wild-type iPS-MPs. (A) IL-1β secretion from cocultured iPS-MPs. We used 2 × 104 mutant iPS-MPs (M1) and 8 × 104 wild-type iPS-MPs (W1) as indicated. n = 6. (B) IL-1β secretion from various numbers of mutant (left panel) or wild-type (right panel) iPS-MPs. The iPS-MPs were seeded at the indicated numbers. n = 3. (C) IL-1β secretion from iPS-MPs that were cocultured at various ratios. The wild-type or mutant iPS-MPs were seeded at the numbers indicated in the first and second rows, respectively. The percentage of mutants is indicated in the third row; n = 3. (D) Increase of IL-1β levels during stimulation by the supernatant. The supernatant was harvested from the wells of the indicated iPS-MPs (Sup) and transferred to the wells of other iPS-MPs (Cell); n = 3. (E) IL-1β secretion from cocultured iPS-MPs in the presence of the ATP receptor antagonist, oATP (300μM) or PPADS (300μM). We used 2 × 104 mutant iPS-MPs (M1) and 8 × 104 wild-type iPS-MPs (W1) as indicated. n = 6. Data are mean ± SEM. ***P < .001 (Student t test). **P < .01 (Student t test). *P < .05 (Student t test).
Next, we tried to elucidate whether the interaction is mediated by some humoral factor(s), but supernatant transfer did not facilitate the IL-1β secretion (Figure 4D). As a candidate that may mediate this interaction, we selected ATP because necrotic cells trigger NLRP3-inflammasome activation in part through ATP release.24 We therefore investigated whether the necrosis-induced ATP secretion activates the wild-type iPS-MPs using ATP receptor antagonists, oxidized ATP (oATP) and PPADS. Although both antagonists markedly inhibited the IL-1β secretion after LPS plus ATP stimulation (supplemental Figure 4B), neither of them abrogated the additional IL-1β secretion in the mixed culture (Figure 4E; compare column 2 with column 3, and column 4 with column 5). The IL-1β secretion from mutant iPS-MPs may have decreased because of off-target effects of oATP.25 Overall, although it remains to be elucidated how this effect is mediated, these results suggest that the interaction between mutant and wild-type macrophages may enhance IL-1β secretion in mosaic patients.
Validation for future applications for drug screening
An NLRP3-targeted therapeutic approach would be attractive because (1) the progressive arthropathy despite anti–IL-1 therapy indicates that the presence of additional proteins processed by the inflammasome is also involved in the pathogenesis of CINCA syndrome; (2) specific inhibition of the NLRP3-inflammasome can avoid unfavorable suppression of other IL-1β–processing pathways in response to various triggers; and (3) these drugs may be also effective for various other NLRP3-related chronic inflammatory conditions, such as Alzheimer disease, diabetes, severe gout, and atherosclerosis.26-30 Because drug screening using NLRP3 autoactivated cells has not been described previously, we examined whether the iPS-MPs from CINCA patients can serve as a prototype for seeking drug candidates that directly modulate NLRP3-inflammasome activation.
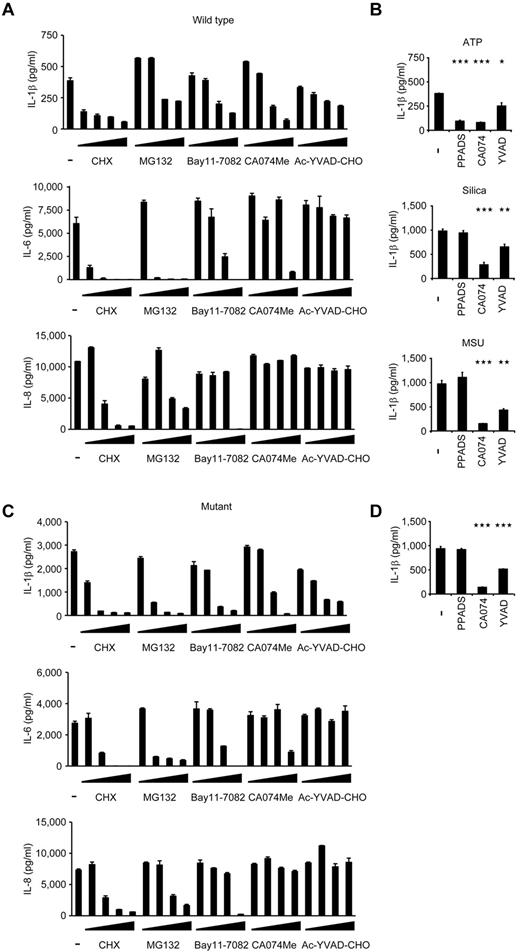
When wild-type iPS-MPs were stimulated with LPS and ATP in the presence of various inhibitors, inhibitors known to modulate molecules upstream of the NLRP3-inflammasome (a protein synthesis inhibitor, cycloheximide, and an NF-κB inhibitor, MG132), downstream of the inflammasome (a caspase-1 inhibitor, Ac-YVAD-CHO), and both upstream of and the inflammasome itself31 (Bay11-7082) successfully inhibited IL-1β secretion (Figure 5A). Although the precise mechanism is unknown, a cathepsin B inhibitor, CA074Me, also efficiently inhibited IL-1β secretion. As expected, upstream inhibitors inhibited the secretion of other cytokines, such as IL-6 and IL-8, but a downstream inhibitor, Ac-YVAD-CHO, specifically affected IL-1β secretion (Figure 5A). Although CA074Me and Ac-YVAD-CHO inhibited IL-1β secretion regardless of the second signals that were present, PPADS, an inhibitor of extracellular ATP signaling, failed to inhibit IL-1β secretion by following exposure to other second signals, such as monosodium urate and silica crystals (Figure 5B), proving that wild-type iPS-MPs can be activated in a second signal-dependent manner. Therefore, the results of the wild-type iPS-MP-based compound screening depended on the choice of second signals, and such a screening makes it possible to extract candidate compounds that modulate specific second signaling pathways.
Validation of the cells for future applications for drug screening. (A) Inhibition of IL-1β (top panel), IL-6 (middle panel), or IL-8 (bottom panel) secretion from wild-type iPS-MPs by various inhibitors. The iPS-MPs were cultured for 2 hours in the presence of 100μM cycloheximide (CHX), 100μM MG132, 10μM Bay11-7082, 25μM CA074Me, 50μM Ac-YVAD-CHO, as well as 10-fold dilutions of each inhibitor, except CA074Me (which was diluted 5-fold), followed by LPS treatment plus ATP stimulation. n = 3. (B) The differential inhibition of IL-1β secretion from wild-type iPS-MPs by various inhibitors. In the presence of inhibitors, such as PPADS (300μM), CA074Me (25μM), or Ac-YVAD-CHO (50μM), LPS-primed wild-type iPS-MPs were stimulated with second signal triggers, such as ATP for 1 hour (top panel), silica crystals for 1 hour (middle panel), or monosodium urate crystals for 3 hours (bottom panel). n = 3. (C) Inhibition of IL-1β (top panel), IL-6 (middle panel), or IL-8 (bottom panel) secretion from mutant iPS-MPs by various inhibitors was evaluated as in panel A; n = 3. (D) Inhibition of IL-1β secretion from mutant iPS-MPs by various inhibitors. In the presence of inhibitors, such as PPADS (300μM), CA074Me (25μM), or Ac-YVAD-CHO (50μM), mutant iPS-MPs were stimulated with LPS for 4 hours. n = 3. Data are mean ± SEM. ***P < .001 (Student t test). **P < .01 (Student t test). *P < .05 (Student t test).
Validation of the cells for future applications for drug screening. (A) Inhibition of IL-1β (top panel), IL-6 (middle panel), or IL-8 (bottom panel) secretion from wild-type iPS-MPs by various inhibitors. The iPS-MPs were cultured for 2 hours in the presence of 100μM cycloheximide (CHX), 100μM MG132, 10μM Bay11-7082, 25μM CA074Me, 50μM Ac-YVAD-CHO, as well as 10-fold dilutions of each inhibitor, except CA074Me (which was diluted 5-fold), followed by LPS treatment plus ATP stimulation. n = 3. (B) The differential inhibition of IL-1β secretion from wild-type iPS-MPs by various inhibitors. In the presence of inhibitors, such as PPADS (300μM), CA074Me (25μM), or Ac-YVAD-CHO (50μM), LPS-primed wild-type iPS-MPs were stimulated with second signal triggers, such as ATP for 1 hour (top panel), silica crystals for 1 hour (middle panel), or monosodium urate crystals for 3 hours (bottom panel). n = 3. (C) Inhibition of IL-1β (top panel), IL-6 (middle panel), or IL-8 (bottom panel) secretion from mutant iPS-MPs by various inhibitors was evaluated as in panel A; n = 3. (D) Inhibition of IL-1β secretion from mutant iPS-MPs by various inhibitors. In the presence of inhibitors, such as PPADS (300μM), CA074Me (25μM), or Ac-YVAD-CHO (50μM), mutant iPS-MPs were stimulated with LPS for 4 hours. n = 3. Data are mean ± SEM. ***P < .001 (Student t test). **P < .01 (Student t test). *P < .05 (Student t test).
Next, we examined the response of mutant iPS-MPs to the inhibitors. In the absence of inhibitors, mutant iPS-MPs secreted a higher level of IL-1β, but treatment with inhibitors dose-dependently decreased IL-1β secretion to the comparable level produced by WT iPS-MPs (Figure 5C). We thus demonstrated the efficacy of these chemical compounds, even for excessive IL-1β production by constitutively hyperactivated inflammasomes. As expected, the mutant iPS-MPs did not respond to PPADS, confirming their autoactivation in a second signal-independent manner (Figure 5D). Therefore, because they can be activated independently from the type of second signals, mutant iPS-MP–based screening would enable the exclusion of compounds that inhibit IL-1β secretion depending on a specific type of second signal transduction. Overall, through using the IL-1β inhibition as the initial criteria and weeding out upstream inhibitors by measuring the levels of other cytokines, we can use NLRP3-mutant iPS-MPs to screen for drugs for CINCA syndrome and possibly for other NLRP3-related chronic inflammatory conditions.
Discussion
Since the first identification of a CINCA syndrome patient carrying NLRP3 mutation as somatic mosaicism,20 it has been controversial whether the small fraction of NLRP3-mutated cells actually causes the strong autoinflammation. It remained unanswered because of the difficulty to separately obtain live mutant and nonmutant blood cells. In this study, we reprogrammed fibroblasts from mosaic patients and obtained macrophages with different genotypes. By showing that only NLRP3-mutant iPS-MPs exhibit the distinct proinflammatory phenotype, we demonstrated that the NLRP3-mutant macrophages are mainly responsible for the pathogenesis of mosaic CINCA syndrome.
In this study, we established both NLRP3-mutant and nonmutant iPSC clones from the same person. One of the potential limitations of studies with patient-derived iPSCs is the difficulty in obtaining isogenic control counterparts, which do not carry the responsible mutations. One possible strategy to solve this problem is to correct the affected gene locus of patient-derived iPSC clones using novel techniques that facilitate homologous recombination.32,33 As another solution, both affected and control iPSC clones can be obtained from patients of some X-linked hereditary diseases because each iPSC clone originated from somatic cells carrying either a mutated or nonmutated allele as an active X chromosome.34-36 In the present study, we have retrieved both mutant and wild-type iPSC clones from patients with somatic autosomal mutations. These clones theoretically have the same genetic backgrounds, except for the NLRP3 gene, and should serve as an ideal pair of mutant and control clones for disease research.
In addition to obtaining isogenic controls, iPSCs from patients with somatic autosomal mutations enable dissection and modeling of somatic mosaicism. Despite the fact that each person contains various minor somatic mutations,37 the effects of mosaicism can often be overlooked because of the difficulty in assessing the possible biologic effects caused by the small cell populations carrying the genetic alterations. Here we dissected somatic mosaicism by obtaining the component cells with heterogeneous genetic identity separately and established an in vitro model to evaluate the interaction between these cells, although precise mechanism of interaction remains to be elucidated. As an approach to determining the disease-causing potential of a specific somatic mutation found in a person, iPSC technology provides advantages compared with ordinary methods, such as the use of transgenic cell lines. First, iPSCs can be differentiated into the affected cell types or tissues, allowing direct functional assays to be performed that are associated with the pathology. Second, because the disease-causing potential of some mutations is dependent on the genetic backgrounds of the patients,38 it may be better to obtain both mutant and wild-type clones from a single mosaic patient to more accurately assess the impact of the mutation(s).
Considering that a mutation of NLRP3 in 10% of the cells is sufficient to cause a distinct disease phenotype, somatic mutations of various genes at an even rarer frequency may also affect the biologic characteristics of a person. Because the presence of the NLRP3 mutation did not affect the efficacy of reprogramming to the iPSCs, we may be able to obtain both mutant and wild-type iPSC clones from CINCA syndrome patients who carry NLRP3 mutant cells at a lower percentage. In some diseases, such as Fanconi anemia, however, mutant cells may be resistant to reprogramming.39,40 Even though there are some possible limitations, establishing both mutant and wild-type iPSC clones is a promising approach to dissect the extent and role of somatic mosaicism.
We demonstrated that several inhibitors that are considered to be effective against CINCA syndrome actually attenuated the disease-relevant phenotype of iPSC-derived macrophages. Before a successful drug screening using iPSC-derived somatic cells can be developed, several limitations need to be overcome, such as the heterogeneity of differentiation and difficulties associated with purification.18 In this report, we used an efficient and robust differentiation protocol and obtained plenty of macrophages free from the clonal variations.
In conclusion, we elucidated the pathologic roles of both mutant and wild-type cells in mosaic CINCA syndrome patients. After obtaining iPSC-derived macrophages in large quantity and with high purity, we showed they are applicable for drug screening. The iPSC-based approach may help to illuminate the pathogenesis of various diseases that are caused by somatic mosaicism, and facilitate drug discovery for the treatment of NLRP3-related inflammatory diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the CINCA syndrome patients who participated in this study; Y. Sasaki, Y. Jindai, A. Okada, M. Narita, A. Nagahashi, T. Ohkame, S. Nishimoto, Y. Inoue, and S. Arai for technical assistance; I. Kato for help with animal experiments; M. Nakagawa, K. Okita, Y. Yoshida, T. Aoi, and M. Yanagimachi for scientific comments; and R. Kato, E. Nishikawa, S. Takeshima, Y. Otsu, H. Hasaba, H. Watanabe, T. Ishii, H. Kurokawa, N. Takasu, and Y. Takao for administrative assistance.
This work was supported by the Ministry of Health, Labor and Welfare (N.M. and T.N.), the Ministry of Education, Culture, Sports, Science and Technology (MEXT; N.M. and T.N.), the Leading Project of MEXT (S.Y. and T.N.), the Promotion of Fundamental Studies in Health Sciences of National Institute of Biomedical Innovation (S.Y.), the Funding Program for World-Leading Innovative Research and Development on Science and Technology (FIRST Program) of Japan Society for the Promotion of Science (JSPS; T.N., and S.Y.), JSPS and MEXT (Grants-in-Aid for Scientific Research; S.Y.), JSPS (T.N., T.T., and M.K.S.), the Takeda Science Foundation, SENSHIN Medical Research Foundation, and Suzuken Memorial Foundation to (M.K.S.).
Authorship
Contribution: T.T. planned the project, established iPSCs, performed experimental work, analyzed data, and prepared the manuscript; K.T. planned the project, established iPSCs, and analyzed data; M.Y., S.T., and S.N. performed experimental work; K.O., A.N., and T.H. analyzed data; R.N. and N.K. planned the project; H.H. and M.M. performed L monocytogenes infection; N.M. and J.E.H. performed electron microscopy; T.Y. identified retroviral integration sites; A.W. performed bisulfite sequencing; A.S.-O. and S.O. analyzed CNV; I.A. established iPSCs; S.Y. and T.N. planned the project and analyzed data; M.K.S. planned the project, analyzed data, and prepared the manuscript; and all authors read and approved the manuscript.
Conflict-of-interest disclosure: S.Y. is a member without salary of the scientific advisory boards of iPierian, iPS Academia Japan, and Megakaryon Corporation. The remaining authors declare no competing financial interests.
Correspondence: Megumu K. Saito, Center for iPS Cell Research and Application, Kyoto University, Kyoto 606-8507, Japan; e-mail: msaito@cira.kyoto-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal