Abstract
Emerging evidence indicates that tumors can follow several evolutionary paths over a patient's disease course. With the use of serial genomic analysis of samples collected at different points during the disease course of 28 patients with multiple myeloma, we found that the genomes of standard-risk patients show few changes over time, whereas those of cytogenetically high-risk patients show significantly more changes over time. The results indicate the existence of 3 temporal tumor types, which can either be genetically stable, linearly evolving, or heterogeneous clonal mixtures with shifting predominant clones. A detailed analysis of one high-risk patient sampled at 7 time points over the entire disease course identified 2 competing subclones that alternate in a back and forth manner for dominance with therapy until one clone underwent a dramatic linear evolution. With the use of the Vk*MYC genetically engineered mouse model of myeloma we modeled this competition between subclones for predominance occurring spontaneously and with therapeutic selection.
Introduction
The contribution of clonal heterogeneity to disease progression and resistance to therapy is increasingly being recognized in cancer. In acute lymphoblastic leukemia, for example, it has been reported that tumors follow 1 of 4 evolutionary pathways: no change over time, linear evolution, evolution from ancestral clones, and genetically distinct relapses supporting a variable branching architecture of tumor evolution.1-3 Interestingly, cytogenetic and early whole genome sequencing studies suggest that not all mutations in a given tumor are conserved over time, again hinting at the presence of multiple clones in addition to probable progression events.4-12 Although these studies suggest that tumor evolution often does not follow the linear models described in textbooks,13 an additional layer of complexity in tumor biology is introduced when one considers that clones do not exist in isolation but as part of a dynamic equilibrium competing for limited resources. Understanding such complex relations between subclones is difficult in humans but can be elegantly modeled in the mouse. For instance, a mouse model of lung cancer was recently used to show that ancestrally related subclones coexist and functionally cooperate in promoting tumor metastasis.14
Multiple myeloma (MM) represents an ideal model system for extending the study of clonal dynamics during disease progression and the effects of drug therapy because it is possible to collect highly purified serial samples over time and because of an often complex disease course characterized by serial cycles of response, remission, and relapse made possible by the availability of several effective therapies. Furthermore, the development of the Vk*MYC genetically engineered mouse model has provided a faithful model of MM in which sporadic MYC activation in germinal center B lymphocytes occurs in a strain of mouse that spontaneously develops monoclonal gammopathy that results in an indolent and low-proliferative MM that remains dependent on the BM microenvironment and displays similar biologic and clinical features to human MM.15,16 This models the critical role that has been postulated for MYC dysregulation in the progression of monoclonal gammopathy of undetermined significance to MM in humans.15,17 As in human patients, MM cells in Vk*MYC mice secrete high level of serum monoclonal immunoglobulins, resulting in an M-spike that is detected by serum protein electrophoresis (SPEP) and represents a clonal marker of tumor burden. Occasionally, as a result of independent MYC activation, Vk*MYC mice develop biclonal or triclonal MM that can be identified and followed longitudinally by the specific SPEP migration pattern of each individual clone.
Taking advantage of all these unique features, we chose to explore the extent of tumor heterogeneity in MM by conducting a survey of genomic changes occurring over time in 28 patients with and without cytogenetically defined high-risk disease [(t(4;14), t(14;16), t(14;20), del(17p)].18 To build on these serial observations we performed a comprehensive analysis of a single patient with high-risk t(4;14) MM from initial diagnosis to secondary plasma cell leukemia with the use of array comparative genomic hybridization (aCGH) and FISH at 7 serially collected time points. To validate the observed findings we modeled our observation in the Vk*MYC mouse to re-create a dynamic picture of clonal competition and tumor evolution.
Methods
Samples
All samples were acquired after patients provided written informed consent in accordance with the Declaration of Helsinki which approved the use of their samples in compliance with Mayo Clinic Institutional Review Board. BM and peripheral blood samples were treated with ACK lysis buffer to remove red cells, and CD138+ cell populations were isolated with anti-CD138 Abs on a StemCell Technologies Robocept. Tumor cell purity was estimated with a slide-based κ/λ assay. Purified tumor cells were stored either lysed in TRIzol (Invitrogen) or as dry pellets at −80°C. Stored tumors not reserved for specific studies were selected from our tumor bank for this study on the basis of the availability of serially acquired tumor samples with confirmed high-purity CD138+ plasma cell enrichments. Nucleic acids were extracted from TRIzol as suggested. RNA was purified with the use of the Purelink Micro-to-Midi kit (Invitrogen), and DNA was purified with a proteinase K digestion and 2 rounds of phenol-chloroform extraction. DNA was isolated from dry pellets with the use of the Puregene kit (QIAGEN) for use in CGH experiments.
Microarrays, FISH, and resequencing
RNA samples were submitted to the Mayo Clinic genomics core for gene expression studies or analyzed as part of the Multiple Myeloma Genomics Initiative.19 Samples were labeled as recommended by the manufacturer with the use of the One-Cycle Labeling Kit (Affymetrix) and hybridized to HG-U133Plus 2.0 gene chips (Affymetrix). Expression estimates were extracted with the MAS5.0 algorithm implemented in Expression Console with the use of default parameters. Gene expression indices were calculated as suggested.20,21 Tumor and control DNA samples were digested with DNAseI, and fragmented DNA was labeled with the BioPrime Array CGH Genomic Labeling kit as previously described.19,22 Labeled DNA was hybridized to either the 44B (4 patients) or 244A Human Genome CGH Microarrays (Agilent) according to the manufacturer's suggestion. Aberration calling was performed in Genomic WorkBench Version 6.5 (Agilent Technologies) with the use of the ADM-2 algorithm and required that aberrations were defined by ≥ 3 consecutive probes and exceeded a 0.2-fold change in log2 space. Regions of copy number change were manually curated to exclude naturally occurring copy number variants from the aberration list. This analysis was limited to copy number changes detected by aCGH, whereas enrichment events were not scored (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We did not score enrichment events because the primary goal was to identify structural changes, and we cannot attribute changes in the percentage of cells showing the same copy number abnormality (CNA) to clonal selection or CD138+ purity differences by aCGH. Appropriate BAC or Fosmid clones residing within the regions of interest defined by the CGH experiments were selected with the University of California Santa Cruz Genome browser (supplemental Table 1). DNA purified from the identified BAC and Fosmid clones were labeled with Spectrum Green, Spectrum Orange, or Aqua and used in cIg-FISH assays as previously described.23 Statistical significance of comparisons was determined by the use of a 2-sided, unpaired t test. Individual exons of TP53 were amplified and sequenced with previously described PCR primers24 and conditions.22
Mouse work
All experiments were performed under The Mayo Foundation Institutional Animal Care and Use Committee approval and conformed to all the regulatory environmental safety standards. The generation of the initial characterization of the Vk*MYC mice has been reported elsewhere.15 Mice were periodically bled by tail grazing, and venous blood was collected into Microtainer tubes (Becton Dickinson), allowed to coagulate at room temperature, and spun for 10 minutes at 2300g. Sera were diluted 1:2 in normal saline buffer and analyzed on a QuickGel Chamber apparatus with the use of precasted QuickGels (Helena Laboratories) according to the manufacturer's instruction.
Mouse transplantations
BM cells were flushed from the tibia of donor Vk*MYC mice with MM, subjected to ACK lysis, suspended in PBS, and immediately injected (∼ 1 ×106 cells/mouse) by intracardiac injection into aged syngeneic Vk*MYC mice with established MM and in parallel into 5- to 12-week-old congenic C57BL/6 wild-type mice. Recipient mice were monitored weekly by SPEP for appearance of the M-spike, indication of tumor engraftment.
Mouse drug treatment
Bortezomib was obtained from our institutional oncology pharmacy. Three Vk*MYC mice were chosen on the basis of the presence of ≥ 2 M-spikes of ≥ 10 g/L and were treated twice a week by intraperitoneal injection of escalating doses of bortezomib, starting at 0.15 mg/kg and increasing to 1 mg/kg, until the emergence of aggressive bortezomib-refractory MM clone detected by SPEP. Carfilzomib, a gift from Onyx Pharmaceutical, was administered at 5 mg/kg in 10% Captisol buffer by intracardiac injection on days 1, 2, 8, and 9. Pomalidomide was purchased from Sequoia Research Products and dosed at 100 mg/kg in 10% Cremaphor by oral gavage at days 1-5 and days 8-12.
Results
More genomic changes over time evident in cytogenetically high-risk MM
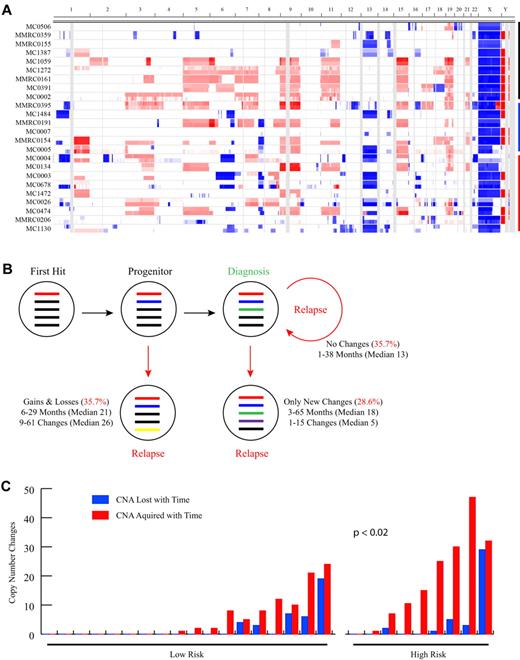
To study the changes in MM genomes over time we analyzed by aCGH paired serial samples from 28 patients (median time between samples, 19.3 months; range, 1.2-65 months) treated with a variety of different regimens and identified a mean of 23 CNAs (median, 19 CNAs; range, 3-77 CNAs) per sample (Figure 1A; Table 1). The number of CNAs detected increased significantly over time (P = .002; Table 1), with a mean of 19.7 CNAs (median, 13.5 CNAs; range, 3-51 CNAs) detected at baseline, increasing to 26.3 CNAs (median, 20 CNAs; range, 10-77 CNAs) at the second time point. Within each serial sample pair, a mean of 16.8 shared CNAs (median, 13 shared CNAs; range, 3-46 shared CNAs) indicated a clonal relation of the temporally distinct tumor populations. Thus, in general the genomic complexity of the MM tumor genome increases over time. Importantly, however, the picture is quite variable for any given patient. Indeed, 3 distinct phenotypic patterns of genomic evolution are observed (Figure 1B). In matching tumors from 35.7% of patients no detectable CNA changes were identified between the initial and subsequent sample. Although the median follow-up time in this subset of 10 patients was the shortest of the 3 groups (median, 13.3 months; range, 1-37 months), there was no statistical difference in the sampling interval between the groups. In 21.4% of patients, instead, only new acquired CNAs were detectable in the matching tumor samples, suggesting these tumors are on a conventional linear evolutionary path. However, a third pattern was observed in 42.9% of cases, characterized by detectable gains but also losses of CNAs, including the reappearance of regions homozygously deleted. This latter finding unequivocally indicates the presence of multiple unique clones at baseline whose relative frequency changes over time. Notably, the presence of cytogenetic marks commonly associated with high-risk disease18 [t(4;14), t(14;16), t(14;20), del(17p13)] was significantly associated (P < .02) with increased copy number changes over time (Figure 1C), suggesting that the poor prognosis of cytogenetically high-risk MM may be related in part to increased clonal heterogeneity and genomic instability. Interestingly among the high-risk patients, those with del(17p13) had significantly more CNAs at diagnosis (P < .02; supplemental Table 2).
Paired sample analysis identifies 3 patterns of clonal evolution. (A) Heat map showing CNAs in the 24 patients with sample pairs analyzed on the Agilent 244k exclusively (4 samples analyzed on Agilent 44k or mixture of 44k and 244k are not shown). The first 2 sequential samples for each patient are stacked, with the first sample shown on the top. Sample pairs are indicated on the y-axis and chromosome location on the x-axis. Blue shading indicates the presence of copy number loss, red indicates copy number gain, and white indicates regions with no CNA. Black, blue, and red bars indicate groups of patients with no copy number changes, with only acquired new changes, or with both losses and gains of CNA over time, respectively. The latter 2 groups are ordered from top to bottom from the least to the most changes. (B) Summarized findings from the 28 patients with a sequential sample pair. Colored bars represent theoretical CNAs perceived to exist at a particular point in the evolution of the tumor. (C) Bar graph showing the relation between copy number changes and standard (n = 18) and high (n = 8) cytogenetic risk status. Samples are ordered left to right, based on the total number of changes. The number of losses and acquired events are shown independently. There is a significant difference in the total number of changes detected between the standard and high-risk cytogenetic groups (P < .02) with a higher number of changes found in the high-risk group.
Paired sample analysis identifies 3 patterns of clonal evolution. (A) Heat map showing CNAs in the 24 patients with sample pairs analyzed on the Agilent 244k exclusively (4 samples analyzed on Agilent 44k or mixture of 44k and 244k are not shown). The first 2 sequential samples for each patient are stacked, with the first sample shown on the top. Sample pairs are indicated on the y-axis and chromosome location on the x-axis. Blue shading indicates the presence of copy number loss, red indicates copy number gain, and white indicates regions with no CNA. Black, blue, and red bars indicate groups of patients with no copy number changes, with only acquired new changes, or with both losses and gains of CNA over time, respectively. The latter 2 groups are ordered from top to bottom from the least to the most changes. (B) Summarized findings from the 28 patients with a sequential sample pair. Colored bars represent theoretical CNAs perceived to exist at a particular point in the evolution of the tumor. (C) Bar graph showing the relation between copy number changes and standard (n = 18) and high (n = 8) cytogenetic risk status. Samples are ordered left to right, based on the total number of changes. The number of losses and acquired events are shown independently. There is a significant difference in the total number of changes detected between the standard and high-risk cytogenetic groups (P < .02) with a higher number of changes found in the high-risk group.
Patients studied serially for CNAs
| Patient . | Cyto risk . | Ploidy . | Sample 1 CNA . | Sample 2 CNA . | Shared CNA . | New CNA . | Lost CNA . | Time, mo . | Treatments . |
|---|---|---|---|---|---|---|---|---|---|
| MC0002 | Low | HD | 13 | 13 | 13 | 0 | 0 | 10 | None |
| MC1059 | Low | HD | 19 | 19 | 19 | 0 | 0 | 12 | MPT |
| MC1272 | Low | HD | 11 | 11 | 11 | 0 | 0 | 38 | Rd + HDT-Tx |
| MMRC0161 | Low | HD | 13 | 13 | 13 | 0 | 0 | 8 | Bort + Temsirolimus |
| MC0391 | Low | HD | 11 | 11 | 11 | 0 | 0 | 19 | Bort + HDT-tx |
| MC0085 | Low | HD | 33 | 33 | 33 | 0 | 0 | 36 | MP |
| MMRC0359 | Low | NHD | 21 | 21 | 21 | 0 | 0 | 1 | Radiation |
| MMRC0155 | Low | NHD | 13 | 13 | 13 | 0 | 0 | 15 | Radiation | TD |
| MC1387 | NA | NHD | 14 | 14 | 14 | 0 | 0 | 5 | AUY922 | Cyclo-Len-Pred |
| MC0506 | High | NHD | 21 | 21 | 21 | 0 | 0 | 24 | MPR | MPV |
| MC1484 | High | NHD | 11 | 12 | 11 | 1 | 0 | 17 | Pd |
| MMRC0395 | Low | HD | 35 | 36 | 35 | 1 | 0 | 3 | DPCE + HDT-Tx |
| MMRC0191 | Low | HD | 31 | 33 | 31 | 2 | 0 | 22 | 5 Regimens |
| MC0453 | Low | HD | 8 | 10 | 8 | 2 | 0 | 43 | TD + HDT-Tx | TD | Bort |
| MC0007 | Low | NHD | 3 | 11 | 3 | 8 | 0 | 20 | Len | Pd |
| MC0518 | High | NHD | 24 | 36 | 24 | 12 | 0 | 6 | Dex |
| MC0134 | Low | HD | 9 | 10 | 5 | 5 | 4 | 12 | None |
| MC0004 | High | HD | 30 | 35 | 28 | 7 | 2 | 21 | Rd |
| MC0496 | Low | HD | 11 | 16 | 8 | 8 | 3 | 23 | Dex + HDT-Tx |
| MMRC0154 | Low | HD | 7 | 19 | 6 | 12 | 1 | 17 | Dex + HDT-Tx |
| MC0005 | Low | HD | 13 | 28 | 12 | 15 | 1 | 65 | Rd |
| MC0003 | Low | NHD | 11 | 14 | 4 | 10 | 7 | 22 | MPR | VD | CyBorD | Rd |
| MC0678 | High | NHD | 36 | 60 | 35 | 25 | 1 | 6 | Dex + HDT-Tx |
| MC1472 | Low | NHD | 10 | 25 | 4 | 21 | 6 | 19 | CyBorD + HDT-Tx |
| MC0026 | High | NHD | 51 | 76 | 46 | 30 | 5 | 10 | VAD + HDT-Tx |
| MC0474 | Low | HD | 26 | 31 | 7 | 24 | 19 | 29 | Dex | Rd + HDT-Tx |
| MMRC0206 | High | NHD | 33 | 77 | 30 | 47 | 3 | 20 | Unknown |
| MC1130 | High | NHD | 34 | 37 | 5 | 32 | 29 | 21 | Rd |
| Patient . | Cyto risk . | Ploidy . | Sample 1 CNA . | Sample 2 CNA . | Shared CNA . | New CNA . | Lost CNA . | Time, mo . | Treatments . |
|---|---|---|---|---|---|---|---|---|---|
| MC0002 | Low | HD | 13 | 13 | 13 | 0 | 0 | 10 | None |
| MC1059 | Low | HD | 19 | 19 | 19 | 0 | 0 | 12 | MPT |
| MC1272 | Low | HD | 11 | 11 | 11 | 0 | 0 | 38 | Rd + HDT-Tx |
| MMRC0161 | Low | HD | 13 | 13 | 13 | 0 | 0 | 8 | Bort + Temsirolimus |
| MC0391 | Low | HD | 11 | 11 | 11 | 0 | 0 | 19 | Bort + HDT-tx |
| MC0085 | Low | HD | 33 | 33 | 33 | 0 | 0 | 36 | MP |
| MMRC0359 | Low | NHD | 21 | 21 | 21 | 0 | 0 | 1 | Radiation |
| MMRC0155 | Low | NHD | 13 | 13 | 13 | 0 | 0 | 15 | Radiation | TD |
| MC1387 | NA | NHD | 14 | 14 | 14 | 0 | 0 | 5 | AUY922 | Cyclo-Len-Pred |
| MC0506 | High | NHD | 21 | 21 | 21 | 0 | 0 | 24 | MPR | MPV |
| MC1484 | High | NHD | 11 | 12 | 11 | 1 | 0 | 17 | Pd |
| MMRC0395 | Low | HD | 35 | 36 | 35 | 1 | 0 | 3 | DPCE + HDT-Tx |
| MMRC0191 | Low | HD | 31 | 33 | 31 | 2 | 0 | 22 | 5 Regimens |
| MC0453 | Low | HD | 8 | 10 | 8 | 2 | 0 | 43 | TD + HDT-Tx | TD | Bort |
| MC0007 | Low | NHD | 3 | 11 | 3 | 8 | 0 | 20 | Len | Pd |
| MC0518 | High | NHD | 24 | 36 | 24 | 12 | 0 | 6 | Dex |
| MC0134 | Low | HD | 9 | 10 | 5 | 5 | 4 | 12 | None |
| MC0004 | High | HD | 30 | 35 | 28 | 7 | 2 | 21 | Rd |
| MC0496 | Low | HD | 11 | 16 | 8 | 8 | 3 | 23 | Dex + HDT-Tx |
| MMRC0154 | Low | HD | 7 | 19 | 6 | 12 | 1 | 17 | Dex + HDT-Tx |
| MC0005 | Low | HD | 13 | 28 | 12 | 15 | 1 | 65 | Rd |
| MC0003 | Low | NHD | 11 | 14 | 4 | 10 | 7 | 22 | MPR | VD | CyBorD | Rd |
| MC0678 | High | NHD | 36 | 60 | 35 | 25 | 1 | 6 | Dex + HDT-Tx |
| MC1472 | Low | NHD | 10 | 25 | 4 | 21 | 6 | 19 | CyBorD + HDT-Tx |
| MC0026 | High | NHD | 51 | 76 | 46 | 30 | 5 | 10 | VAD + HDT-Tx |
| MC0474 | Low | HD | 26 | 31 | 7 | 24 | 19 | 29 | Dex | Rd + HDT-Tx |
| MMRC0206 | High | NHD | 33 | 77 | 30 | 47 | 3 | 20 | Unknown |
| MC1130 | High | NHD | 34 | 37 | 5 | 32 | 29 | 21 | Rd |
Ploidy categories hyperdiploid (HD) and non-hyperdiploid (NHD) are indicated. Different treatment combinations used separately are spaced and separated by a vertical bar (|).
MPT indicates melphalan, prednisone, and thalidomide; Rd, lenalidomide with low-dose dexamethasone; HDT-Tx, high-dose melphalan followed by autologous transplantation; Bort, bortezomib alone; MP, melphalan and prednisone; TD, thalidomide and dexamethasone; MPR, melphalan, prednisone, and lenalidomide; MPV, melphalan, prednisone, and bortezomib; Pd, pomalidomide with low-dose dexamethasone; DPCE, cyclophosphamide, dexamethasone, etoposide, and cisplatin; VD, bortezomib with dexamethasone; CyBorD, cyclophosphamide, bortezomib, and dexamethasone; VAD, vincristine, Adriamycin, and dexamethasone; and NA, not available.
The association between del(17p13) and the number of detected CNAs at diagnosis prompted us to investigate the status of TP53 at each time point in the patients with a del(17p13) detected at any point in their disease course. We were able to determine the TP53 status of 5 of the 6 patients with a detected del(17p13) event (Table 2). Four of the 5 tested patients had lost both TP53 alleles by either 2 independent deletions or a combination of a deletion and mutation. In 2 patients we could show the progression from a monoallelic deletion detected at diagnosis to biallelic inactivation at relapse because of a secondary deletion and a secondary missense mutation in one patient each. In one patient a minor subclone was detected at diagnosis with a del(17p13), which progressed to 2 independent deletions in the terminal phase (supplemental Figure 8). Interestingly, the patient fitting the linear evolution phenotype with the most changes over time had acquired biallelic inactivation of TP53. These results suggest that the poor prognosis associated with del(17p13) at diagnosis is probably associated with an increased risk of biallelic inactivation and a correlation with a higher number of CNAs at diagnosis.
Temporal correlation of 17p13 deletion and TP53 mutation
| Patient . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . |
|---|---|---|---|---|---|
| MC0518 | 1× del not tested (24) | 2× del (36) | |||
| MC0004 | 1× del no mutation (30) | 1× del + N239K (35) | |||
| MC0678 | 1× del no mutation (36) | 1× del no mutation (60) | |||
| MC0026 | 1× del not tested (51) | 1× del + T125-M133del (76) | |||
| MMRC0206* | 2× del (33) | 1× del (77) | |||
| MC1130† | No del WGS (34) | No del WGS (37) | No del WGS (36) | 2× del (63) | 2× del WGS (63) |
| Patient . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . |
|---|---|---|---|---|---|
| MC0518 | 1× del not tested (24) | 2× del (36) | |||
| MC0004 | 1× del no mutation (30) | 1× del + N239K (35) | |||
| MC0678 | 1× del no mutation (36) | 1× del no mutation (60) | |||
| MC0026 | 1× del not tested (51) | 1× del + T125-M133del (76) | |||
| MMRC0206* | 2× del (33) | 1× del (77) | |||
| MC1130† | No del WGS (34) | No del WGS (37) | No del WGS (36) | 2× del (63) | 2× del WGS (63) |
Numbers in parentheses indicate the number of copy number abnormalities at each time point.
These samples, in particular the second sample, are not available for testing and unfortunately were not tested as part of the Multiple Myeloma Research Consortium Genomics Initiative whole genome sequencing project.
MC1130 was sequenced with both SOLiD and HiSeq Whole Genome Sequencing (WGS) at the time points indicated by WGS (Egan et al29 ). No mutations were seen in TP53 at any time.
Variability in the acquisition and loss of CNAs over time in individual patients
For 3 patients, a third sample was available, which extended the follow-up time by 9, 12, and 51 months, respectively (supplemental Table 2). No changes were noted in the first patient during the first 19 months of treatment. However, during a subsequent 9 months of observation 4 new CNAs became apparent. A second patient had no detectable copy number changes throughout the 49-month interval analyzed. The third patient progressed from smoldering to overt MM after 12 months, while gaining 5 and losing 4 CNAs, but in the next 51 months after therapy was initiated the tumor genome of this hyperdiploid patient did not change. These observations indicate that patients are not fixed on a single evolutionary trajectory and can potentially shift between stable and evolving genomes during the course of their disease. Alternatively, a single episode of genomic instability may generate multiple subclones that attain clonal dominance at different rates and under various selective pressures.
Alternating clonal dominance in a patient with high-risk t(4;14) MM
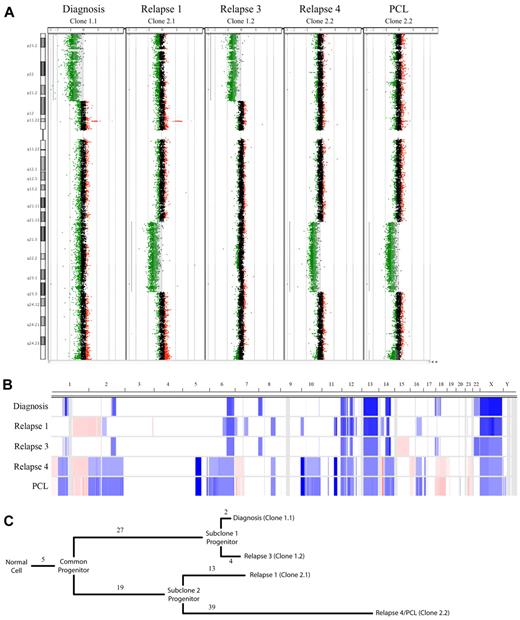
For one patient we collected 7 serial samples covering the entire disease course from diagnosis to terminal plasma cell leukemia (PCL). This patient followed a relatively common clinical disease course, achieving a stable partial response while receiving continuous lenalidomide with low-dose dexamethasone as front-line therapy with no dose reductions or skipped cycles, then progressing while on therapy and subsequently receiving multiple additional therapeutic regimens (Figure 2). The longitudinal aCGH analysis of the MM tumor of this patient exhibited a pattern of 2 major clones with alternating dominance over time by aCGH (Figure 3A-B). We identified 5 CNAs present in all samples studied, indicative of early events occurring in a common progenitor before the 2 major subclone progenitors diverged (Figure 3B-C). There were 27 and 19 unique CNAs that defined the divergence of the 2 subclone progenitors. At each time point studied, unique CNAs were also observed that showed linear evolution from the perceived subclone progenitor. Interestingly, the 2 major subclones diverged at different rates: the offspring of subclone progenitor 1 were differentiated by only 6 CNAs (2 lost and 4 gained), whereas clones related to subclone progenitor 2 were highly divergent with 52 different CNAs (13 lost and 39 gained). In this patient, clonal suppression and recrudescence appear to correspond to drug sensitivity and resistance, suggesting that therapeutic selective pressures are responsible, at least in part.
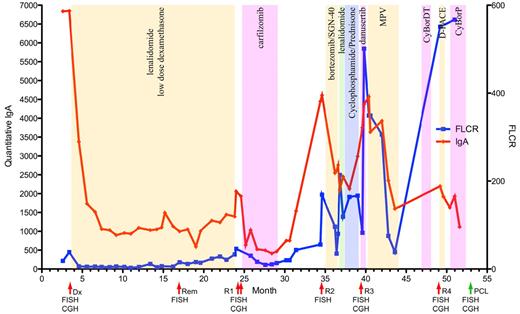
Clinical course of a patient with high-risk MM. The clinical course of a single patient (MC1130) studied throughout the entire disease course is shown. Red line indicates the quantitative IgA level detected and units are shown on the left y-axis. Blue line indicates the free light chain ratio detected with units shown on the right y-axis. Alternating color regions indicate type and durations of treatment received during each interval. Red arrows highlight the time points at which BM aspirates were analyzed, the green arrow indicates the time of collection of the terminal PCL sample from peripheral blood. Dx indicates diagnosis; Rem, remission: R1, relapse 1; R2, relapse 2; R3, relapse 3; and R4, relapse 4. The assays performed at each time point are indicated under their representative arrows.
Clinical course of a patient with high-risk MM. The clinical course of a single patient (MC1130) studied throughout the entire disease course is shown. Red line indicates the quantitative IgA level detected and units are shown on the left y-axis. Blue line indicates the free light chain ratio detected with units shown on the right y-axis. Alternating color regions indicate type and durations of treatment received during each interval. Red arrows highlight the time points at which BM aspirates were analyzed, the green arrow indicates the time of collection of the terminal PCL sample from peripheral blood. Dx indicates diagnosis; Rem, remission: R1, relapse 1; R2, relapse 2; R3, relapse 3; and R4, relapse 4. The assays performed at each time point are indicated under their representative arrows.
Copy number analysis of a patient with high-risk MM. (A) A whole chromosome CGH dot plots for chromosome 8 are shown for each sample assayed. Statistically significant CNAs are indicated by light gray shading, which coincides with regions with extensive green dots (deletions) or red dots (amplifications). The 2 regions used to define subclone progenitors 1 and 2, which ebb and flow with time in this patient, are visible as large deletions on the 8p and 8q, respectively. (B) Heat map showing copy number changes in the 5 samples analyzed by Agilent 244k. Sample types are shown on the y-axis and ordered longitudinally. The chromosome location is on the x-axis. Blue shading indicates the presence of copy number loss; red, copy number gain; and white, no copy number abnormality. (C) Dendrogram showing the relation of the observed subclones. Branch length represents the number of CNAs detected by aCGH and assigned to each evolutionary step. There are 5 CNAs shared in all samples and a clear divergence of 2 subclone progenitors defined by 27 or 19 CNAs, respectively. Clones related to subclone progenitor 1 represent the major tumor population at diagnosis and relapse 3 and are differentiated by 2 and 4 CNAs, respectively. Clones related to subclone progenitor 2 represent the majority of the tumor population at relapse 1 and relapse 4/PCL, which are differentiated by 13 and 39 CNAs, respectively.
Copy number analysis of a patient with high-risk MM. (A) A whole chromosome CGH dot plots for chromosome 8 are shown for each sample assayed. Statistically significant CNAs are indicated by light gray shading, which coincides with regions with extensive green dots (deletions) or red dots (amplifications). The 2 regions used to define subclone progenitors 1 and 2, which ebb and flow with time in this patient, are visible as large deletions on the 8p and 8q, respectively. (B) Heat map showing copy number changes in the 5 samples analyzed by Agilent 244k. Sample types are shown on the y-axis and ordered longitudinally. The chromosome location is on the x-axis. Blue shading indicates the presence of copy number loss; red, copy number gain; and white, no copy number abnormality. (C) Dendrogram showing the relation of the observed subclones. Branch length represents the number of CNAs detected by aCGH and assigned to each evolutionary step. There are 5 CNAs shared in all samples and a clear divergence of 2 subclone progenitors defined by 27 or 19 CNAs, respectively. Clones related to subclone progenitor 1 represent the major tumor population at diagnosis and relapse 3 and are differentiated by 2 and 4 CNAs, respectively. Clones related to subclone progenitor 2 represent the majority of the tumor population at relapse 1 and relapse 4/PCL, which are differentiated by 13 and 39 CNAs, respectively.
Complex clonal dynamics after therapy
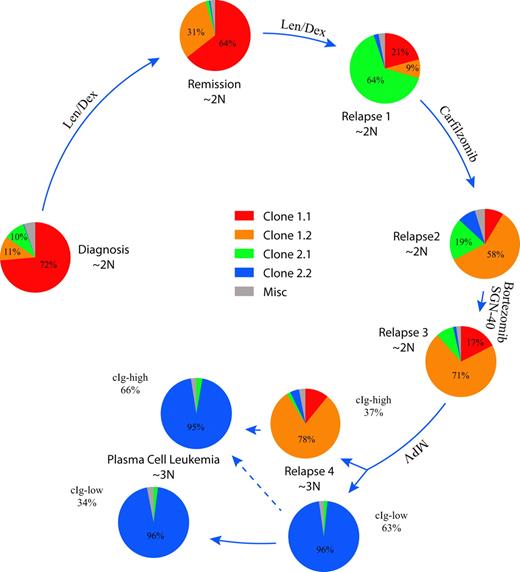
To understand the relative population dynamics associated with the 2 major clonal progenitors and their observed offspring clones 1.1, 1.2, 2.1, and 2.2, we designed a series of 8 custom FISH assays (Figure 4; supplemental Figures 2-9). At diagnosis, the tumor population was primarily composed of cells with a clone 1.1 phenotype, but cells associated with clone 1.2 and 2.1 phenotypes were present as minor populations of 11% and 10%, respectively. Interestingly, after a year of treatment with lenalidomide and dexamethasone, while the patient was in a partial remission, a previously minor clonal population (clone 1.2) had increased in prominence from 11% to 31% of MM cells. Over the next 7 months, the patient progressed while on treatment, and clone 2.1 characterized by a biallelic deletion of BIRC2/3, known to activate the NF-κB pathway,25 became the dominant clone, comprising 64% of the tumor cells. At this time, a sibling clone (clone 2.2) with an independent BIRC2/3 deletion emerged, representing 13% of the tumor population (supplemental Figure 6). At the fourth relapse after a melphalan-containing regimen, the tumor population shifted dramatically, with re-emergence of clone 2.2 now representing 62% of the tumor population. The increased complexity of the clone 2.2 genome (Figure 3C), compared with earlier samples analyzed, raises the possibility of potential harmful genetic effects induced by DNA-damaging agents in clonal populations with inherent genomic instability and defective DNA repair.26 Interestingly, at diagnosis of PCL, the tumor population was almost exclusively triploid clone 2.2 cells, indicating a ploidy shift occurred between the BM localized relapse 4 version of clone 2.2 and the PCL version of clone 2.2. These observations establish that 2 different dominant clones coexisted in this patient and their relative abundance appeared to be modulated by the therapy received.
Clonal dynamics in a patient with high-risk MM. The summarized results of 8 different FISH assays are shown to indicate the relative abundance of each clone defined by aCGH at the 5 time points studied. Pie charts showing the relative proportions of each indicated clone are ordered clockwise starting on the left in longitudinal order. Arrow length is proportional to the time interval. The relative ploidy of the tumor population at each time point is also indicated. The cIg FISH of relapse 4 and the PCL identified clonal cells with low levels of cytoplasmic immunoglobulin (cIg-low) and larger cells with abundant cytoplasmic immunoglobulin (cIg-high) that were scored independently.
Clonal dynamics in a patient with high-risk MM. The summarized results of 8 different FISH assays are shown to indicate the relative abundance of each clone defined by aCGH at the 5 time points studied. Pie charts showing the relative proportions of each indicated clone are ordered clockwise starting on the left in longitudinal order. Arrow length is proportional to the time interval. The relative ploidy of the tumor population at each time point is also indicated. The cIg FISH of relapse 4 and the PCL identified clonal cells with low levels of cytoplasmic immunoglobulin (cIg-low) and larger cells with abundant cytoplasmic immunoglobulin (cIg-high) that were scored independently.
Modeling clonal competition in MM
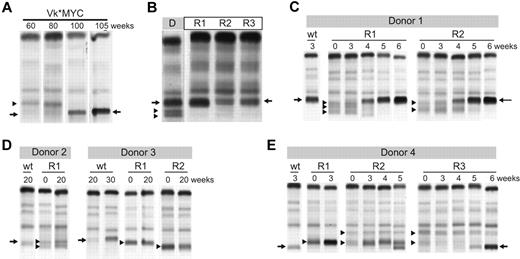
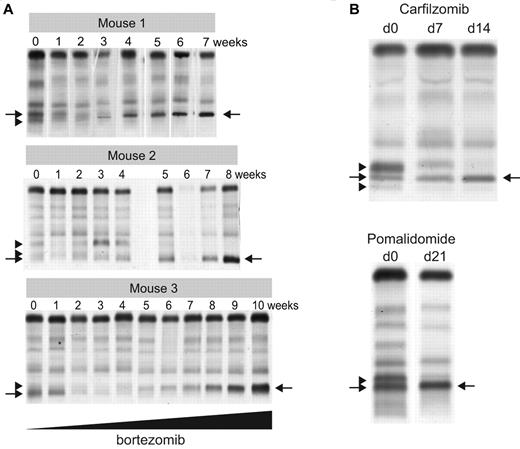
To functionally study clonal dynamics we used the Vk*MYC transgenic mouse model of MM.15 Our ability to identify and follow the behavior of independent clones according to their distinctive M-spikes is a unique feature of this model relevant to understanding the biology of intratumor heterogeneity in human MM. Although these distinctive M-spikes represent independent transformation events, they share the same initiating event, MYC activation, and represent a way to model the effects of secondary events akin to subclonal differences existing in a patient with a presumed initiating IgH translocation. We identified aged Vk*MYC mice that spontaneously developed biclonal or triclonal MM and followed them over time without any intervention. Frequently one M-spike would increase, while the other(s) remained stable (data not shown), but occasionally, however, the other M-spike would disappear (Figure 5A), indicating spontaneous clonal competition in MM in vivo. To model in vivo this spontaneous competition we performed 2 experiments. First, we transplanted BM cells from Vk*MYC mice with biclonal spikes into congenic C57BL/6 wild-type (wt) mice and noted that typically only 1 of the clones would engraft, resulting in a single M-spike in all the recipient mice (Figure 5B). Second, we selected Vk*MYC mice with a monoclonal M-spike produced by either an aggressive or indolent MM and transplanted their BM cells in parallel into wt and Vk*MYC recipient mice with preexisting MM, producing a M-spike of different size. We found that the most aggressive Vk*MYC tumors, which rapidly engrafted (< 3 weeks) into congenic wt recipients, also rapidly engrafted in the tumor-bearing Vk*MYC mice and within 5 weeks after transplantation completely replaced the preexisting MM (Figure 5C). Conversely, Vk*MYC tumors with a more indolent course and longer time to engraftment when transplanted into wt recipients (20 weeks) did not engraft in tumor-bearing Vk*MYC mice (Figure 5D). Occasionally, we found that aggressive MM tumors could either compete out the existing tumor clone (Figure 5E recipient R3), coexist with it (Figure 5E recipient R2), or even stimulate the growth of the existing clone (Figure 5E recipient R1), suggesting a more intricate cross talk between tumor clones. To study the contribution of therapy to clonal dynamics we treated Vk*MYC mice bearing oligoclonal MM with suboptimal doses of bortezomib. The treatment exerted a selective pressure on all of the tumor clones; however, in each case one clone eventually emerged and out-competed the remaining clones (Figure 6A). Similarly, we noted that treatment of Vk*MYC mice with pomalidomide or carfilzomib occasionally produced differential tumor response within the same mouse, with responsive MM clones disappearing and refractory clones progressing (Figure 6B). Taken together these studies graphically show that there is a limiting MM cell microenvironment in which tumor subclones compete for dominance under changing selective pressures of subclonal progression and drug therapy.
Clonal dynamics in a murine model of MM. (A) Serum protein electrophoresis performed at the indicated weeks on a biclonal Vk*MYC mouse shows the emergency at 80 weeks of a secondary clone that becomes dominant by 100 weeks. Arrows highlight the M-spike secreted by the dominant clone. Arrowhead points to the M-spike secreted by the competed out clone. (B) BM cells from a donor mouse (D) with a triclonal myeloma were transplanted into 3 wt congenic mice recipient mice (R). Serum protein electrophoresis shows engraftment of only 1 of the 3 clones in all 3 recipient mice. Arrows highlight the M-spike secreted by the dominant clone. Arrowheads point to the M-spikes secreted by the competed out clones. (C-E) BM cells from Vk*MYC myeloma-bearing mice (Donor) were transplanted into either wt congenic mice (wt) or recipient Vk*MYC mice with preexisting MM (R). Serum protein electrophoresis was performed at the indicated weeks after transplantation. Arrows highlight the M-spikes secreted by the donor mouse MM cells and arrowheads points to the preexisting M-spikes in the recipient mice.
Clonal dynamics in a murine model of MM. (A) Serum protein electrophoresis performed at the indicated weeks on a biclonal Vk*MYC mouse shows the emergency at 80 weeks of a secondary clone that becomes dominant by 100 weeks. Arrows highlight the M-spike secreted by the dominant clone. Arrowhead points to the M-spike secreted by the competed out clone. (B) BM cells from a donor mouse (D) with a triclonal myeloma were transplanted into 3 wt congenic mice recipient mice (R). Serum protein electrophoresis shows engraftment of only 1 of the 3 clones in all 3 recipient mice. Arrows highlight the M-spike secreted by the dominant clone. Arrowheads point to the M-spikes secreted by the competed out clones. (C-E) BM cells from Vk*MYC myeloma-bearing mice (Donor) were transplanted into either wt congenic mice (wt) or recipient Vk*MYC mice with preexisting MM (R). Serum protein electrophoresis was performed at the indicated weeks after transplantation. Arrows highlight the M-spikes secreted by the donor mouse MM cells and arrowheads points to the preexisting M-spikes in the recipient mice.
Therapy induced clonal dynamics in a murine model of MM. (A) Three independent Vk*MYC mice with biclonal or triclonal MM were treated biweekly with escalating doses of bortezomib (0.16-1 mg/kg). SPEP performed at the indicated weeks shows clonal modulation, with arrowheads pointing to the M-spikes secreted by the competed out clones, and arrows indicating the dominant bortezomib-resistant clones. (B) SPEP performed before and after carfilzomib or pomalidomide treatment of Vk*MYC mice with oligoclonal myeloma identifies responsive (arrowheads) and refractory (arrows) clones.
Therapy induced clonal dynamics in a murine model of MM. (A) Three independent Vk*MYC mice with biclonal or triclonal MM were treated biweekly with escalating doses of bortezomib (0.16-1 mg/kg). SPEP performed at the indicated weeks shows clonal modulation, with arrowheads pointing to the M-spikes secreted by the competed out clones, and arrows indicating the dominant bortezomib-resistant clones. (B) SPEP performed before and after carfilzomib or pomalidomide treatment of Vk*MYC mice with oligoclonal myeloma identifies responsive (arrowheads) and refractory (arrows) clones.
Discussion
This comparative study of MM in 28 patients and a faithful mouse model identifies complex clonal dynamics in MM. We found that approximately one-third of patients with MM have stable genomes, particularly those with low-risk hyperdiploid disease, potentially explaining their more favorable clinical outcomes. However, in another one-third of patients, changes over time were evident that are best explained by the existence of clonal heterogeneity at diagnosis (unequivocally so for the cases showing the reappearance of regions previously containing biallelic deletions). In the final one-third of patients, a pattern consistent with linear evolution was the dominant characteristic. The last 2 groups include all but one of the high-risk patients, suggesting high-risk tumors are less stable and more prone to change with time. Given the disseminated nature of MM into multiple discrete foci of lytic bone disease, it is probable that even greater clonal heterogeneity may exist than is evident from a simple serial sampling of iliac crest BM. Although from this analysis it is clear that changes over time were seen in patients receiving different treatment regimens, a larger, comprehensive, and preferably prospective study is required to systematically analyze the contribution of different drugs, depth of response, clinical variables, prognostic significance, and individual high-risk genetic abnormalities. Furthermore, such a study may help identify genetic lesions present in those few patients with cytogenetically low-risk disease that nevertheless displayed marked changes in CNAs over time. Moreover, given currently available technologies, which can detect changes at base pair resolution, future studies will undoubtedly identify changes that are not detectable with the aCGH platforms used in this study.
Importantly, detailed analysis of one patient with t(4;14) MM paints an elaborate picture of how the relative frequency of competing MM subclones can ebb and flow over time and with therapeutic intervention. Although the emergence of an initially minor clone with treatment has been documented through mutational analysis of BCR-ABL in acute lymphoblastic leukemia and chronic myeloid leukemia,27,28 this back and forth competition between subclones has not been documented previously. Furthermore we show in the mouse both a friendly and sinister relation between clones in which the more aggressive clone surprisingly appears to sometimes support the growth and other times lead to the regression of the more indolent clone. We suspect that this is probably a general feature of many tumor types that is simply easier to observe in MM because the M-spike serves as an easily measured clonal marker of tumor burden.
There are important clinical implications of these findings. First, our results indicate that a “partial response” rather than reflecting partial suppression of the entire tumor population could also result from the suppression of a sensitive clone while a refractory clone remains stable and thus becoming proportionally more dominant. Second, combination therapies targeting all coexisting disease subclones will probably be particularly important for cytogenetically high-risk MM, whereby sequential single-agent therapy should be avoided. Third, retreatment of a patient with a regimen on which they have previously progressed is often avoided because of the assumption of continued drug resistance. However, with intervening therapy a sensitive subclone may have re-emerged, and retreatment may be effective. Finally, early suboptimal treatment of tumors may in some cases preferentially eradicate the more indolent clone, making room for the more aggressive one to expand when the microenvironment is limiting. Although these findings require additional exploration in a larger cohort, they highlight that personalized medicine approaches in MM and other cancers will need to be sufficiently nimble to deal with the complex nature of the evolving tumor genome and with shifting clonal tides that can occur throughout a patient's disease course.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kim Henderson, Greg Ahmann, and the associated tissue bank staff at Mayo Clinic Rochester, Mayo Clinic Arizona, and the Multiple Myeloma Research Consortium (MMRC) for their help in processing samples for this project. They also thank the MMRC for making available the serial samples from the MM genomics initiative.19
This work was supported by the National Institutes of Health (grants CA136671 and CA133966, P.L.B.; and CA133115, A.K.S.).
National Institutes of Health
Authorship
Contribution: J.J.K. and M.C. designed research, performed research, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript; V.M.G. and S.E.P. performed mouse experiments; S.V.W. executed FISH experiments; E.B., A.S.B., P.R.B., and J.B.E. helped with execution of experiments and analysis of patient data; A.D., S.K., and S.V.R. provided patient samples and clinical data; J.D.C. and M.B. contributed vital analysis technology and analytical tools; P.R.B. helped retrieve clinical information; A.K.S. and R.F. contributed vital patient samples and provided critical review of the manuscript; P.L.B. conceived the initial hypothesis, designed research, contributed vital patient samples, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The microarray datasets associated with this study have been deposited in GEO as super series under accession GSE36825 the individual platforms are available individually under GSE36822, GSE36823, and GSE36824.
The current affiliation for J.J.K. is Translational Genomics Research Institute, Phoenix, AZ.
Correspondence: P. Leif Bergsagel, Mayo Clinic Collaborative Research Bldg, 13400 E Shea Blvd, Scottsdale, AZ 85259-5494; e-mail: bergsagel.leif@mayo.edu.
References
Author notes
J.J.K. and M.C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal