Abstract
Additional chromosomal abnormalities (ACAs) in Philadelphia-positive cells have been reported in ∼ 5% of patients with newly diagnosed chronic myeloid leukemia (CML) in chronic phase (CP). Few studies addressing the prognostic significance of baseline ACAs in patients treated with imatinib have been published previously. The European LeukemiaNet recommendations suggest that the presence of ACAs at diagnosis is a “warning” for patients in early CP, but there is not much information about their outcome after therapy with tyrosine kinase inhibitors. To investigate the role of ACAs in early CP CML patients treated with imatinib mesylate, we performed an analysis in a large series of 559 patients enrolled in 3 prospective trials of the Gruppo Italiano Malattie Ematologiche dell'Adulto Working Party on CML: 378 patients were evaluable and ACAs occurred in 21 patients (5.6%). The overall cytogenetic and molecular response rates were significantly lower and the time to response was significantly longer in patients with ACAs. The long-term outcome of patients with ACAs was inferior, but the differences were not significant. The prognostic significance of each specific cytogenetic abnormality was not assessable. Therefore, we confirm that ACAs constitute an adverse prognostic factor in CML patients treated with imatinib as frontline therapy. This study was registered with clinicaltrials.gov as NCT00514488 and NCT00510926.
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder characterized by the presence of the Philadelphia (Ph) chromosome, produced by the reciprocal translocation t(9;22)(q34;q11).1,2 This translocation leads to the generation of a chimeric gene that results from the fusion of the ABL gene on chromosome 9 with the BCR gene on chromosome 22. The new leukemia-specific fusion gene encodes a constitutionally activated protein tyrosine kinases (PTKs) of different molecular weights (p185/190, p210, and p230). The oncogenic PTK, located in the cytoplasm, is responsible of the leukemic phenotype, through the constitutive activation of multiple signaling pathways.3 At diagnosis, in most of cases the classic t(9;22)(q34;q11) or its variant occur as the sole abnormality; additional chromosomal abnormalities in Ph+ cells (ACAs) may appear in ∼ 5% of cases, according to several series.4–6 The clinical impact of these changes may be different depending on the treatment.
Almost 10 years ago, the CML therapy was radically changed by the introduction of imatinib mesylate, a break point cluster-Abelson (BCR-ABL)–targeting tyrosine kinase inhibitor (TKI).7–10 Despite the efficacy of imatinib in chronic phase (CP), treatment failure or suboptimal response have been reported.11,12
The appearance of ACAs during treatment is commonly known as clonal evolution (CE) and seems to play an important role in imatinib mesylate resistance8 ; the emergence of ACAs during the treatment is considered a poor prognostic feature. The World Health Organization classification suggests that those patients showing ACAs emerging during treatment should be considered in accelerated phase (AP).13 The European LeukemiaNet recommendations9,14 suggest that the presence of ACAs at diagnosis may represent a “warning” feature, requiring careful monitoring of the patient; however, the level of evidence at that time was low because few studies addressing the prognostic meaning of ACAs at diagnosis, before starting imatinib treatment, have been published. Conversely, ACAs emerging during the course of treatment are considered a failure.
We report an analysis of Gruppo Italiano Malattie Ematologiche dell'Adulto (GIMEMA) CML Working Party (WP), aiming to clarify the impact of baseline ACAs on the response to the therapy and on the outcome in newly diagnosed CML patients in early CP, treated with imatinib as first-line therapy.
Methods
Patients
Patients (559) with previously untreated Ph and BCR-ABL–positive CML in early CP were enrolled in 3 concurrent studies, promoted by GIMEMA CML WP and opened to enrollment in 2004: CML/021 (NCT00514488), a phase 2 trial exploring imatinib 800 mg in intermediate Sokal risk CP CML; CML/022 (NCT00510926), a phase 3 trial comparing imatinib 400 versus 800 mg in high Sokal risk CP CML; and CML/023, an observational trial of imatinib 400 mg in CP CML. The studies were approved by the Internal Review Board of all participating institutions and performed according to Good Clinical Practices and the Declaration of Helsinki.
Treatment monitoring and definition of response
Blood count and serum chemistry were performed at enrollment, monthly until the 12th month of treatment and every 3 months thereafter. A complete hematologic response (CHR) was defined as a white blood cell count of < 10 × 109/L, a platelet count of < 450 × 109/L, no immature cells (blasts, promyelocytes, and myelocytes) in the peripheral blood, and the disappearance of all signs and symptoms related to leukemia (including palpable splenomegaly).
All these study protocols required chromosome banding analysis and fluorescence in situ hybridization (FISH) performed on bone marrow cells at baseline, after 6 and 12 months of treatment, and every 6 months thereafter or in case of failure or disease progression. Cytogenetic response was established on at least 20 evaluable metaphases15 ; if < 20 metaphases were scored, FISH analysis was considered to confirm a complete cytogenetic response (CCgR) when the rate of BCR-ABL–positive nuclei was < 1%.16
Real-time quantitative PCR was performed on peripheral blood and bone marrow samples at baseline and after 3, 6, and 12 months and on peripheral blood every 6 months thereafter. The molecular monitoring was based on peripheral blood samples, and the molecular response was defined as major (MMolR) if the BCR-ABL/ABL ratio was < 0.10% on the International Scale.17–19
Cytogenetics: chromosome banding analysis and FISH analysis
The GIMEMA CML WP has established a network of cytogenetic laboratories covering all the country. Fourteen of these laboratories performed cytogenetic studies for more than 1 clinical center (reference laboratories), and 24 laboratories performed cytogenetic studies for their respective clinical center (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Chromosome banding analysis was performed on bone marrow cells after short-term culture (24 hours, 48 hours, or both) according to the International System for Human Cytogenetic Nomenclature.20 A central review was not performed. At least 20 metaphases were required.
The most frequent chromosomal abnormalities are defined as “major route” abnormalities, and they include trisomy 8, duplication of Ph chromosome, isochromosome 17q, and trisomy 19.21
FISH was performed on bone marrow cells prepared according to cytogenetic techniques and by using DNA probes that hybridize at the BCR and ABL regions.16–22 FISH analysis was performed on at least 200 cells.
Molecular studies
All samples and tests were centralized in Bologna, Italy. RNA extraction, RT-PCR, and real-time quantitative PCR were performed according to European recommendations, as described previously.23–25 Real-time quantitative PCR was performed on an ABI PRISM 7900 sequence detector (Applied Biosystems).19 ABL was used as the housekeeping gene to correct differences in RNA quality, RT efficacy, or both. The results are expressed as ratio between BCR-ABL and ABL copies' percentages. The BCR-ABL/ABL ratios percentages were further multiplied by the conversion factor of the Bologna laboratory to set the results on an international scale.17–19
Definition of progression, failure, and events
The progression to advanced phases was identified by at least 1 of the following criteria: peripheral blood myeloblast ≥ 10%; peripheral blood myeloblasts and promyelocytes ≥ 30%; and any extramedullary blast involvement, excluding spleen and liver. All the cases that did not meet any of the above-mentioned criteria were defined as CP.
According to the international recommendations, failures were defined as follows: no CHR at 3 months, no cytogenetic response (CgR) at 6 months, no partial CgR at 12 months, no CCgR at 18 months, loss of CHR or CCgR previously achieved, new BCR-ABL mutation insensitive to imatinib, and emergence of new ACAs or progression to the AP/blast phase (BP).14,26
Events were defined as treatment failure or permanent discontinuation of imatinib for any reason, including toxicity, patient refusal, or loss to follow-up.
Statistical analysis
All comparisons between the 2 different groups of patients were assessed with the Student t test and with Fisher exact test for categorical variables, as appropriate. Times to CCgR and MMolR were calculated from the date of start of treatment until the achievement of the first response. Overall survival (OS), progression-free survival (PFS), failure-free survival (FFS), and event-free survival (EFS) were calculated from the date of the first imatinib dose until death (OS), until progression to AP or BP or death (PFS), until failure or death (FFS), and until any event (EFS). Probabilities of CCgR, MMolR, OS, PFS, FFS, and EFS were estimated using the Kaplan-Meier method.27 Time to response and survival times were compared with the log-rank test.28 All analysis were performed according to the intention-to-treat principle. Multivariate logistic regression analysis was used to assess the relationship between various predictors of interest and response.
Results
Of the 559 patients enrolled into the GIMEMA CML WP studies CML/021, CML/022, and CML/023 (see details in “Patients”), 378 (68%) had at least 20 evaluable metaphases at diagnosis and were included in the present analysis and 181 had < 20 evaluable metaphases and were excluded from this study. In this way, we have avoided the inclusion of eventual cases with a small clone with ACAs in the group of cases without ACAs. However, no excluded case showed ACAs.
In addition, 357 patients showed the presence of t(9;22)(q34;q11) without additional abnormalities, and 21 patients (5.6%) showed clonal ACAs.
Detailed baseline characteristics of the 2 groups are presented in Table 1. The 2 groups, with and without ACAs, were similar for demographic and hematologic characteristics except for sex and percentage of peripheral blasts: the rate of male patients was 86% and 59% (P = .02) in patients with and without ACAs, respectively; the median percentage of peripheral blasts was 2.5% and 1% (P = .03) in patients with and without ACAs, respectively. The Sokal, Hasford, and European Treatment and Outcome Study risk score distributions were comparable. The proportion of patients treated with high-dose imatinib was similar in the 2 groups. No association of ACAs with variant Ph translocations and with der(9) deletions was observed.
Comparison of patients' characteristics at diagnosis
| Characteristic . | Patients with ACAs (N = 21) . | Patients without ACAs (N = 357) . | P . |
|---|---|---|---|
| Median age, y (range) | 45 (24-74) | 50 (18-84) | .18 |
| Sex, male/female, N (%) | 18/3 (86/14) | 210/147 (59/41) | .02 |
| Median spleen, cm (range) | 4 (0-21) | 2 (0-23) | .10 |
| Median Hb level, g/dL (range) | 11.8 (8.2-16.0) | 12.0 (6.4-17.5) | .77 |
| Median PLT count, 109/L (range) | 406 (143-979) | 346 (107-4920) | .79 |
| Median peripheral blasts, % (range) | 2.5 (1-8) | 1 (0-10) | .03 |
| Median eosinophils, % (range) | 3 (0-6) | 2 (0-15) | .53 |
| Medin basophils, % (range) | 2 (0-10) | 2 (0-19) | .82 |
| Sokal score, N (%) | .66 | ||
| Low | 7 (33) | 140 (39) | |
| Intermediate | 6 (29) | 134 (38) | |
| High | 8 (38) | 83 (23) | |
| Hasford score, N (%) | .35 | ||
| Low | 6 (29) | 159 (45) | |
| Intermediate | 13 (62) | 173 (48) | |
| High | 2 (10) | 25 (7) | |
| EUTOS score, N (%) | .20 | ||
| Low | 18 (86) | 338 (95) | |
| High | 3 (14) | 19 (5) | |
| Variant Ph translocations, N (%) | 1 (5) | 24 (7) | 1.00 |
| Deletions der(9), N (%) | 4 (19) | 36 (10) | .26 |
| Imatinib dose (mg), N (%) | .81 | ||
| 400 | 16 (76) | 258 (72) | |
| 800 | 5 (24) | 99 (28) |
| Characteristic . | Patients with ACAs (N = 21) . | Patients without ACAs (N = 357) . | P . |
|---|---|---|---|
| Median age, y (range) | 45 (24-74) | 50 (18-84) | .18 |
| Sex, male/female, N (%) | 18/3 (86/14) | 210/147 (59/41) | .02 |
| Median spleen, cm (range) | 4 (0-21) | 2 (0-23) | .10 |
| Median Hb level, g/dL (range) | 11.8 (8.2-16.0) | 12.0 (6.4-17.5) | .77 |
| Median PLT count, 109/L (range) | 406 (143-979) | 346 (107-4920) | .79 |
| Median peripheral blasts, % (range) | 2.5 (1-8) | 1 (0-10) | .03 |
| Median eosinophils, % (range) | 3 (0-6) | 2 (0-15) | .53 |
| Medin basophils, % (range) | 2 (0-10) | 2 (0-19) | .82 |
| Sokal score, N (%) | .66 | ||
| Low | 7 (33) | 140 (39) | |
| Intermediate | 6 (29) | 134 (38) | |
| High | 8 (38) | 83 (23) | |
| Hasford score, N (%) | .35 | ||
| Low | 6 (29) | 159 (45) | |
| Intermediate | 13 (62) | 173 (48) | |
| High | 2 (10) | 25 (7) | |
| EUTOS score, N (%) | .20 | ||
| Low | 18 (86) | 338 (95) | |
| High | 3 (14) | 19 (5) | |
| Variant Ph translocations, N (%) | 1 (5) | 24 (7) | 1.00 |
| Deletions der(9), N (%) | 4 (19) | 36 (10) | .26 |
| Imatinib dose (mg), N (%) | .81 | ||
| 400 | 16 (76) | 258 (72) | |
| 800 | 5 (24) | 99 (28) |
Hb indicates hemoglobin; PLTs, platelets; and EUTOS, European Treatment and Outcome Study.
Cytogenetic analysis
In total, 21 CML cases with clonal ACAs were identified, and they are described in Table 2. We observed loss of Y chromosome in 9 patients (43%; no. 1-9), trisomy 8 in 3 patients (14%; no. 10-12), trisomy 19 in 2 patients (10%; no. 13 and 14), other different single abnormalities in 6 patients (28%; no. 15-20), and complex karyotype with double ACAs in only 1 patient (5%; no.21).
Cytogenetic details and treatment response of the 21 patients with clonal ACAs
| Patient . | Sex . | Karyotype at diagnosis (% metaphases with ACAs) . | Der(9) deletion . | CgR 12m . | Outcome . |
|---|---|---|---|---|---|
| 1 | Male | 45,X,-Y,t(9;22)(q34;q11) (65) | Yes | CCgR | CCgR |
| 2 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 3 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 4 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | NoCgR | Death |
| 5 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 6 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | NE | Failure |
| 7 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | Yes | PCgR | CCgR |
| 8 | Male | 45,X,-Y,t(9;22)(q34;q11) (33) | No | CCgR | CCgR |
| 9 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | PCgR | CCgR |
| 10 | Male | 47,XY,+8,t(9;22)(q34;q11) (14) | No | mCgR | Death |
| 11 | Male | 47,XY,+8,t(9;22)(q34;q11) (100) | No | PCgR | Failure |
| 12 | Male | 47,XY,+8,t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 13 | Male | 47,XY,t(9;22)(q34;q11),+19 (100) | No | PCgR | Drop-out |
| 14 | Male | 47,XY,t(9;22)(q34;q11),+19 (100) | No | CCgR | Failure |
| 15 | Male | 46,XY,t(9;22)(q34;q11),del(20)(q11q13) (100) | Yes | CCgR | CCgR |
| 16 | Male | 46,XY,del(7)(p21),t(9;22)(q34;q11) (100) | No | NE | Failure |
| 17 | Female | 46,XX,t(1;7)(p36;q31),t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 18 | Male | 46,XY,t(6;11)(q13;q13)t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 19 | Female | 46,XX,t(7;19)(q21;p13),t(9;22;22)(q34;q11;q11) (100) | No | CCgR | CCgR |
| 20 | Male | 46,XY,t(9;22)(q34;q11),t(X;13)(q13;q32) (100) | Yes | CCgR | Failure |
| 21 | Female | 46,XX,t(2;14)(p13;q32),der(14)t(2;14)(p13;q32), t(9;22)(q34;q11) (100) | No | NE | Failure |
| Patient . | Sex . | Karyotype at diagnosis (% metaphases with ACAs) . | Der(9) deletion . | CgR 12m . | Outcome . |
|---|---|---|---|---|---|
| 1 | Male | 45,X,-Y,t(9;22)(q34;q11) (65) | Yes | CCgR | CCgR |
| 2 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 3 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 4 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | NoCgR | Death |
| 5 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 6 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | NE | Failure |
| 7 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | Yes | PCgR | CCgR |
| 8 | Male | 45,X,-Y,t(9;22)(q34;q11) (33) | No | CCgR | CCgR |
| 9 | Male | 45,X,-Y,t(9;22)(q34;q11) (100) | No | PCgR | CCgR |
| 10 | Male | 47,XY,+8,t(9;22)(q34;q11) (14) | No | mCgR | Death |
| 11 | Male | 47,XY,+8,t(9;22)(q34;q11) (100) | No | PCgR | Failure |
| 12 | Male | 47,XY,+8,t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 13 | Male | 47,XY,t(9;22)(q34;q11),+19 (100) | No | PCgR | Drop-out |
| 14 | Male | 47,XY,t(9;22)(q34;q11),+19 (100) | No | CCgR | Failure |
| 15 | Male | 46,XY,t(9;22)(q34;q11),del(20)(q11q13) (100) | Yes | CCgR | CCgR |
| 16 | Male | 46,XY,del(7)(p21),t(9;22)(q34;q11) (100) | No | NE | Failure |
| 17 | Female | 46,XX,t(1;7)(p36;q31),t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 18 | Male | 46,XY,t(6;11)(q13;q13)t(9;22)(q34;q11) (100) | No | CCgR | CCgR |
| 19 | Female | 46,XX,t(7;19)(q21;p13),t(9;22;22)(q34;q11;q11) (100) | No | CCgR | CCgR |
| 20 | Male | 46,XY,t(9;22)(q34;q11),t(X;13)(q13;q32) (100) | Yes | CCgR | Failure |
| 21 | Female | 46,XX,t(2;14)(p13;q32),der(14)t(2;14)(p13;q32), t(9;22)(q34;q11) (100) | No | NE | Failure |
PCgR indicates partial CgR; mCgR, minor CgR, NE, not evaluable; Failure, according to 2009 European LeukemiaNet criteria, described under “Definition of progression, failure, abd events”; and Drop-out, imatinib permanent discontinuation for any reason different from failure.
Major route abnormalities (trisomies 8 and 19) occurred in 5 of 21 cases (24%).
One patient (no.19) showed a variant Ph chromosome: t(9;22;22)(q34;q11;q11) that was characterized by chromosome banding and FISH analysis.
Deletion of der(9) chromosome was detected by FISH analysis in 4 cases (19%; no. 1, 7, 15, and 20): 2 cases with loss of Y chromosome, 1 case with del(20)(q11q13), and 1 case with t(X;13)(q13;q32).
In most patients, ACAs have been found in all the observed cells; only 3 patients (no. 1, 8, and 10) had additional abnormalities in a subclone (in 65%, 33%, and 14% of the metaphases, respectively).
Response to imatinib mesylate therapy and survival
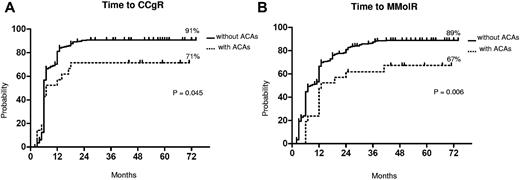
The median follow-up was 58 months (range, 25-75 months) and 60 months (range, 2-81 months) in patients with and without ACAs, respectively. The response rates to imatinib are shown in Table 3. The cytogenetic and molecular response rates were uniformly lower in patients with ACAs: no significant differences were observed at the 12th month, but the overall CCgR and MMolR rates were significantly lower in patients with ACAs: 71% versus 89% (P = .03) and 67% versus 86% (P = .03), respectively. The responses were significantly slower in the group of patients with ACAs (Figure 1). In a multivariate logistic regression, the presence of ACAs retained its prognostic significance, when adjusted for other relevant variables (Table 4).
Response to imatinib (patients were categorized by their response at the 12th month and by their best overall response)
| Response . | Patients with ACAs, N = 21 (%) . | Patients without ACAs, N = 357 (%) . | P . |
|---|---|---|---|
| CHR | |||
| Overall | 21 (100) | 350 (98) | 1.00 |
| CCgR | |||
| 12th mo | 12 (57) | 267 (75) | .12 |
| Overall | 15 (71) | 317 (89) | .03 |
| MMolR | |||
| 12th mo | 10 (48) | 205 (57) | .50 |
| Overall | 14 (67) | 306 (86) | .03 |
| Response . | Patients with ACAs, N = 21 (%) . | Patients without ACAs, N = 357 (%) . | P . |
|---|---|---|---|
| CHR | |||
| Overall | 21 (100) | 350 (98) | 1.00 |
| CCgR | |||
| 12th mo | 12 (57) | 267 (75) | .12 |
| Overall | 15 (71) | 317 (89) | .03 |
| MMolR | |||
| 12th mo | 10 (48) | 205 (57) | .50 |
| Overall | 14 (67) | 306 (86) | .03 |
Cytogenetic and molecular response. Kaplan-Meier analysis; estimates of time to CCgR (A) and of time to MMolR (B). Dotted line indicates presence of ACAs, and solid line indicates presence of only Ph chromosome.
Cytogenetic and molecular response. Kaplan-Meier analysis; estimates of time to CCgR (A) and of time to MMolR (B). Dotted line indicates presence of ACAs, and solid line indicates presence of only Ph chromosome.
Multivariate logistic regression estimating the association between the presence of ACAs and the CCgR, considering other covariates in the model
| CCgR . | Odds ratio . | SE . | z . | P > z . | 95% confidence interval . |
|---|---|---|---|---|---|
| ACAs | 0.2506491 | 0.1418693 | −2.44 | .014 | 0.082657-0.7600683 |
| Variant Ph | 0.5733353 | 0.3446664 | −0.93 | .355 | 0.1764819-1.86259 |
| del(9) | 2.417083 | 1.554654 | 1.37 | .170 | 0.6851781-8.526674 |
| Sex | 1.132707 | 0.3962182 | 0.36 | .722 | 0.5706497-2.24836 |
| Hemoglobin | 1.3555 | 0.1237197 | 3.33 | .001 | 1.133466-1.621029 |
| Eosinophils | 1.138051 | 0.100403 | 1.47 | .143 | 0.9573391-1.352876 |
| Basophils | 0.9109481 | 0.488844 | −1.74 | .082 | 0.8200029-1.01198 |
| Imatinib dose | 0.8333311 | 0.3054995 | −0.50 | .619 | 0.4062251-1.709497 |
| Sokal score | 0.9955652 | 0.2284137 | −0.02 | .985 | 0.6350055-1.560853 |
| CCgR . | Odds ratio . | SE . | z . | P > z . | 95% confidence interval . |
|---|---|---|---|---|---|
| ACAs | 0.2506491 | 0.1418693 | −2.44 | .014 | 0.082657-0.7600683 |
| Variant Ph | 0.5733353 | 0.3446664 | −0.93 | .355 | 0.1764819-1.86259 |
| del(9) | 2.417083 | 1.554654 | 1.37 | .170 | 0.6851781-8.526674 |
| Sex | 1.132707 | 0.3962182 | 0.36 | .722 | 0.5706497-2.24836 |
| Hemoglobin | 1.3555 | 0.1237197 | 3.33 | .001 | 1.133466-1.621029 |
| Eosinophils | 1.138051 | 0.100403 | 1.47 | .143 | 0.9573391-1.352876 |
| Basophils | 0.9109481 | 0.488844 | −1.74 | .082 | 0.8200029-1.01198 |
| Imatinib dose | 0.8333311 | 0.3054995 | −0.50 | .619 | 0.4062251-1.709497 |
| Sokal score | 0.9955652 | 0.2284137 | −0.02 | .985 | 0.6350055-1.560853 |
Variant Ph indicates variant Ph translocations; and del(9), deletion of derivative chromosome 9.
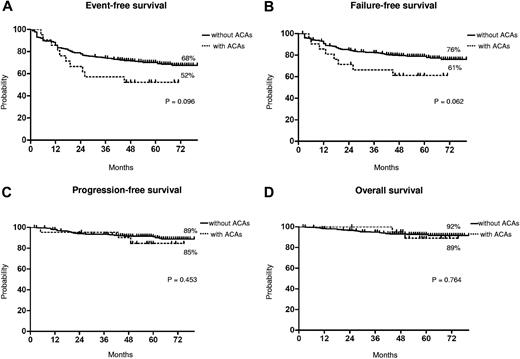
The median time to CCgR was 7 and 6 months in patients with and without ACAs, respectively (P = .045). The median duration of cytogenetic response has not been reached in both groups: 3 of the 15 (20%) CCgR patients with ACAs and 27 of the 317 (8.5%) CCgR patients without ACAs subsequently lost the previously achieved CCgR; the estimated probability of survival without cytogenetic relapse was 79% and 90% (P = .118) in patients with and without ACAs, respectively. The median time to MMolR was 13 and 8 months in patients with and without ACAs, respectively (P = .006). The long-term outcome is reported in Figure 2. The estimated overall probabilities of EFS, FFS, PFS, and OS were 52% versus 68% (P = .096), 61% versus 76% (P = .062), 85% versus 89% (P = .453), and 89% versus 92% (P = .764) for patients with and without clonal ACAs, respectively. Even if all outcomes were inferior for patients with ACAs baseline, no difference versus patients without ACAs baseline was statistically significant. According to the type of cytogenetic abnormality, 9 cases showed loss of Y chromosome, whereas in 5 cases a major route abnormality (trisomy 8 and 19) was reported, in 6 cases a single different abnormality was reported, and in the last case 2 ACAs (Table 2) were reported: 7 of the 9 cases with loss of Y chromosome (no. 1-9) retrieved and maintained a CCgR (6/7 MMolR either); 2 of 5 cases showing a major route abnormality obtained a CCgR by 12 months, but only 1 patient maintained the response with a longer follow-up; 4 of the 6 cases with different single abnormalities obtained and maintained either a CCgR and a MMolR (2/6 failed imatinib); and 1 case with 2 ACAs (no. 21) discontinued imatinib therapy because of treatment failure at 6 months. However, because of the small number of patients with ACAs, the different prognostic significance of each specific cytogenetic abnormality was not assessable.
Long-term outcome. Kaplan-Meier analysis; estimates of EFS (A), FFS (B), PFS (C), and OS (D). Dotted line indicates presence of ACAs, and solid line indicates presence of only Ph chromosome.
Long-term outcome. Kaplan-Meier analysis; estimates of EFS (A), FFS (B), PFS (C), and OS (D). Dotted line indicates presence of ACAs, and solid line indicates presence of only Ph chromosome.
Discussion
At diagnosis, the presence of clonal ACAs may be observed in ∼ 5% of CP CML patients4–6 ; however, their appearance is mostly associated with disease evolution and their frequency is higher in late CP, AP (∼ 30%), and blast crisis (∼ 80%). The current standard treatment approach for newly diagnosed CML patients is based on TKIs, imatinib mesylate, the former standard of care, and the second generation TKIs nilotinib and dasatinib, recently registered as frontline treatments. There is much debate about the “best” treatment strategy, between more or less conservative approaches, considering imatinib or not as still a good option for most patients as the frontline treatment. The prognostic significance of the presence of clonal ACAs at diagnosis has been discussed in previous studies, but data on the outcome of CP CML patients with ACAs after therapy with frontline imatinib are limited. The available studies are usually case reports, small series, or large series but involving patients in different phases of disease, more often in AP and/or treated with different drugs (eg, interferon and TKIs).4,29–34 Reports suggested that, within different disease stages, ACAs did not influence the response to therapy30,34 ; indeed after second TKI therapy, the hematologic and cytogenetic response rates, OS and EFS were no different between patients in CP with CE and patients in CP without CE; but CE had a significant impact when associated with other features of AP.34 These findings are partially supported by a very recent study, in which only major route ACAs were associated with a negative impact.35 On the other hand, another observation reported that in early CP CML the presence of ACAs was one of the independent adverse predictors for PFS in the 6-months analysis and ACAs were present in 4 of the 6 patients who progressed within 1 year.7 The European LeukemiaNet recommendations provide a warning for CP CML patients with ACAs treated frontline with imatinib, suggesting that ACAs have impact on the outcome of CML,14 as they predict significantly for shorter PFS and OS.7 The presence of ACAs has been considered a feature of AP in some classifications of CML; however, the World Health Organization classification suggested to include ACAs as a criterion for AP only if they are not present at diagnosis.13
In the present study, we report a large series of 559 early CP CML patients treated with only imatinib as frontline therapy within 3 clinical trials of the GIMEMA WP on CML.
Clonal ACAs occurred in 21/378 evaluable patients (5.6%), a frequency consistent with previous reports. We can confirm that ACAs are not frequently detected in early CP of CML.
None of the cytogenetic abnormalities observed was considered as constitutional, because of the type of abnormalities and/or the fact that such abnormalities disappeared in all patients who achieved a cytogenetic response. In our series the most frequent observed abnormality was the loss of Y chromosome (43%). The loss of Y chromosome is frequently associated with older age, and might be in some cases only a consequence of ageing. However, there are not univocal data on the significance of the loss of the Y chromosome in CP-CML.4,35–38 We considered this abnormality as ACA, differently from some other series published previously. In our cases, the loss of Y chromosome disappeared in all patients who finally reached a CgR, confirming that the abnormality was related to the hematologic disease.
The presenting features were substantially similar in the 2 groups with and without ACAs (Table 1), except for sex and the median percentage of peripheral blasts. The presence of ACAs seems more common in male patients, as in our series the loss of Y chromosome is the most frequent abnormality. As a matter of fact, the significant unbalance in favor of males patients with ACAs disappeared (P = .37) after exclusion of the 9 patients bearing the loss of Y chromosome. The median percentage of peripheral blasts was higher in the ACAs group: a slight difference most probably deprived of biologic and clinical meanings. Deletions of a sizable portion of the derivative chromosome 9 have been described in 10%-15% of CML patients and have been found more frequently in patients with variant Ph translocations,39–40 but no association has been described in patients with ACAs. However, a recent study41 asserted that deletions of der(9) do not influence the outcome of CML in early CP patients treated with imatinib as frontline therapy. In the present study, 4 of 21 patients (19% vs 10% without ACAs) showed deletions of der(9) by FISH, but the difference was not statistically significant.
Although the number of patients with clonal ACAs was relatively low, the presence of ACAs influences the response to imatinib therapy: the rates of CCgR and MMolR were significantly inferior to those of patients with only Ph translocation (Table 3) and the times to CCgR and MMolR were significantly longer in patients with ACAs (Figure 1).
Previous studies have suggested that achievement of CCgR on imatinib is a major prognostic factor for overall and progression free survival,11–12 and that early molecular response is predictive of long-term disease stability with lack of progression and durability of CCgR.42–44
Furthermore, we observed a higher rate of response loss in the patients with clonal ACAs, but the difference was not statistically significant (data not shown). These differences in terms of responses, rate and rapidity, did not translate into different outcomes. In fact, the number of negative events was higher in patients with ACAs, but the corresponding EFS and FFS curves were not significantly different (P = .09 and P = .06, respectively; Figure 2). Moreover, the survival without progression to AP/BP and the OS probabilities were superimposable, and the higher number of failures did not foresee a higher propensity to progress, either for the low number of events overall or because some of the patients who failed have been rescued to response using second generation TKIs.
We were not able to investigate the differences concerning response rate or survival related to the different kinds of abnormalities, as we found only 3, 2, and 1 patient with trisomy 8, trisomy 19, and 2 ACAs, respectively; moreover, 6 patients showed 6 different single abnormalities that are not frequent in CML. However, our data suggest a worse outcome in patients with major route abnormalities (trisomy 8 and 19; Table 1, case no. 10-14), in according to a recent report,35 even if we have not observed any i(17q) and +der(22q) in our patients, differently from their cases. The only patient with complex karyotype, without major route ACAs, failed the treatment and discontinued imatinib therapy. The increased genomic instability may facility the emergence of a clone with malignant phenotype.
In a recent paper, other authors reported that loss of Y chromosome had a poor impact in patients in imatinib therapy, in terms of cytogenetic and molecular response, EFS, and OS.38 Our data, in agreement with a recent observation,35 do not suggest a worse prognosis in 9 patients with loss of Y chromosome.
ACAs may be found in all the cells, or they may be acquired in a subclone, thus defining a CE with worse outcome.30–31 In our series, no correlation is possible, because only 3 patients (no. 1, 8, and 10; Table 2) had abnormalities in a subclone: 2 patients with loss of Y chromosome are still in CCgR and 1 patient with trisomy 8 died in mCgR.
In conclusion, this large series of patients suggests that ACAs at diagnosis, although not frequently detected, have a negative impact in early CP CML patients treated with imatinib as frontline therapy. Our data confirm that patients with ACAs constitute a “warning” category, in terms of responses, and they suggest that these cases could require a close monitoring and treatment with second generation TKIs as frontline therapy. In agreement with recent observations,35 more intensive therapy should be considered in the patients showing major route abnormalities and complex karyotype.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Katia Vecchi for valuable assistance.
This study was supported by Associazione Italiana contro le Leucemie, i linfomi e i mielomi Bologna (BolognaAIL); Progetti di Ricerca di Interesse Nazionale (PRIN2007), University of Bologna; Fondazione del Monte di Bologna e Ravenna; and European LeukemiaNet.
Authorship
Contribution: S.L., F.C., and N.T. analyzed the data and wrote the manuscript; S.L., G. Marzocchi, C.B., L.Z., M.M., M.G.G., A.G., G.A., G.P., and P.B. performed the molecular cytogenetic analysis; I.I. performed the molecular analysis; F.C., G.G., G. Specchia, G.R.-C., E.A., A.Z., M.A.C., and F. Palandri enrolled the study patients and collected clinical data; F. Pane, G. Saglio, G. Martinelli, G.R., and M.B. designed and supervised the trials; and G.R., M.B., and N.T. supervised the study and gave final approval for submission.
Conflict-of-interest disclosure: F. Pane has received research support from Novartis, served as a consultant for Novartis and Bristol-Myers Squibb, and served on the Novartis speaker's bureau; G. Saglio did consultancy work for Novartis and Bristol-Myers Squibb and served on the speakers' bureaus of Novartis and Bristol-Myers Squibb; G. Martinelli served on the speakers' bureaus of Novartis, Bristol-Myers Squibb, and Pfizer; M.B. received research support from Novartis, Bristol-Myers Squibb, and Pfizer; did consultancy work for Novartis, Bristol-Myers Squibb, and Pfizer; and served on the Novartis speaker's bureau; and G.R. did consultancy work for Novartis and served on the speakers' bureaus of Novartis and Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
A complete list of the members of the GIMEMA CML WP appears in the supplemental Appendix.
Correspondence: Nicoletta Testoni, Department of Hematology and Oncological Sciences “Sant'Orsola-Malpighi” Hospital, University of Bologna, Via Massarenti 9, 40138 Bologna, Italy; e-mail: nicoletta.testoni@unibo.it.
References
Author notes
S.L. and F.C. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal