Abstract
We studied the distribution of peripheral B-cell subsets in patients deficient for key factors of the TLR-signaling pathways (MyD88, TIRAP/MAL, IL-1 receptor–associated kinase 4 [IRAK-4], TLR3, UNC-93B, TRIF). All TLRs, except TLR3, which signals through the TRIF adaptor, require MyD88 and IRAK-4 to mediate their function. TLR4 and the TLR2 heterodimers (with TLR1, TLR6, and possibly TLR10) require in addition the adaptor TIRAP, whereas UNC-93B is needed for the proper localization of intracellular TLR3, TLR7, TLR8, and TLR9. We found that IgM+IgD+CD27+ but not switched B cells were strongly reduced in MyD88-, IRAK-4-, and TIRAP-deficient patients. This defect did not appear to be compensated with age. However, somatic hypermutation of Ig genes and heavy-chain CDR3 size distribution of IgM+IgD+CD27+ B cells were not affected in these patients. In contrast, the numbers of IgM+IgD+CD27+ B cells were normal in the absence of TLR3, TRIF, and UNC-93B, suggesting that UNC-93B–dependent TLRs, and notably TLR9, are dispensable for the presence of this subset in peripheral blood. Interestingly, TLR10 was found to be expressed at greater levels in IgM+IgD+CD27+ compared with switched B cells in healthy patients. Hence, we propose a role for TIRAP-dependent TLRs, possibly TLR10 in particular, in the development and/or maintenance of IgM+IgD+CD27+ B cells in humans.
Introduction
In humans, CD27+ blood B cells with mutated immunoglobulin (Ig) receptors comprise 2 major populations: isotype-switched memory cells (IgG+ or IgA+) and IgM+IgD+CD27+ cells. Whereas switched CD27+ cells are generated in germinal centers by T-dependent responses, the origin of IgM+IgD+CD27+ cells is still controversial. Because they display mutated Ig genes, they often are considered as “IgM memory” B cells that exited the germinal center reaction before isotype switch.1,2 However, data, including ours, support the view that these cells can develop and mutate along a germinal center–independent pathway and that they represent circulating marginal zone B cells3,4 involved in T-independent responses and in the protection against infections by encapsulated bacteria, notably Streptococcus pneumoniae.5,6 Moreover, our recent data provide evidence for a developmental diversification of IgM+IgD+CD27+ B cells, at least in very young children, outside of T-dependent and -independent immune responses.7
In mice, the marginal zone lineage branches at the transitional/immature stage of B-cell development.8 Previous investigators have shown that inflammation and/or Toll-like receptor (TLR) signals can drive a T-independent proliferation and accumulation of somatic mutations in transitional B cells.9,10 A similar observation was made recently for human B cells: in response to TLR9 engagement in vitro, a fraction of immature B cells from cord blood proliferated and differentiated into IgM-secreting plasma cells or into cells expressing a marginal zone–like B-cell phenotype with a low level of somatic mutation.11,12 It was proposed that these marginal zone–like B cells obtained in vitro after CpG stimulation could correspond in vivo to a first line of defense that would provide protection against invading bacteria. TLR9, as well as other TLRs, may thus play an important role in the development and/or maintenance of IgM+IgD+CD27+ cells in vivo, but also of other B-cell subsets, because TLR engagement on mature B cells (especially TLR7 and TLR9) triggers a range of functional responses, including proliferation, cytokine/chemokine production, isotype switching, terminal differentiation, and Ig secretion.13,14
To test this hypothesis, we studied the peripheral blood B-cell subsets of patients either deficient for key mediators of TLR-signaling pathways (MyD88, TIRAP/MAL, IRAK-4, UNC-93B, and TRIF) or for specific TLRs (TLR3). In humans, 10 TLRs have been identified (TLR1-TR10) and their triggering, with the exception of TLR3, induces the recruitment to their Toll/IL-1 receptor (TIR) domain of the adaptor protein MyD88 and the IL-1 receptor–associated kinase 4 (IRAK-4) kinase complex that are essential for mediating signaling of these receptors.15-18 This MyD88/IRAK-4–dependent signaling pathway is also used by several IL-1R family members, including IL-1R, IL-18R, and IL-33R.19 If TLR5, TLR7, TLR8, and TLR9 interact directly with MyD88 after ligand recognition, TLR4, and also TLR2, which can heterodimerize with either TLR1 or TLR6 (and possibly TLR10, see “Discussion”), bind to the bridging adaptor MAL/TIRAP to recruit MyD88.20,21 UNC-93B has been shown in the mouse to be essential for TLR3, TLR7, and TLR9 signaling through its requirement for their intracellular trafficking from the endoplasmic reticulum to the endolysosomes from which they signal,22,23 and data obtained in humans have confirmed the UNC-93B dependence of TLR3, TLR7, TLR8, and TLR9 function.24-26 TLR3 signaling relies on another TIR-adaptor molecule named TRIF, also used in the MyD88-independent, TLR4 signaling pathway (a schematic description of TLR signaling pathways is included in supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).27,28
By studying the B-cell compartment of patients with different inborn errors affecting the signaling of most TLRs (MyD88, IRAK-4) or only some of them (TIRAP, TLR3, UNC-93B, TRIF), we observed that IgM+IgD+CD27+ but not switched cells are strongly dependent on a few specific TLRs for their maintenance, and that, surprisingly, the role of TLR9 appears dispensable.
Methods
Patients and healthy controls
This study was conducted in accordance with the Declaration of Helsinki, with informed consent obtained from each patient or the patient's family. Spleen and blood samples were obtained from young patients undergoing splenectomy because of a nonimmunologic disease (spherocytosis). Adult spleen samples were obtained from organ donors with the authorization of the French Agence de la Biomédecine. Blood samples from healthy adult controls were obtained from the blood bank (Etablissement Français du Sang). Leftover blood samples (taken for blood counts) from healthy children undergoing orthopedic surgery at the Necker Hospital were obtained from the Service d'Hématologie Biologique. The patients studied are described in Table 1. A detailed report of clinical and biologic data has been published elsewhere,29 except for the TIRAP-deficient patients (L.I. and J.-L.C., unpublished manuscript data, 2012). Approval was obtained from the institutional review board of Necker Hospital for this study.
Genotype, age, and proportion of B-cell subsets in the cohort of patients deficient for specific TLRs or molecules that impair the TLR/IL1R-signaling pathways
| Deficiency . | Patient . | Mutation . | Reference . | Age, y* . | % CD19+ . | IgD+CD27+ (% of B cells) . | IgD−CD27+ (% of B cells) . | IgM-only CD27+ (% of IgD-CD27+)† . | IgA+CD27+ (% of B cells) . | DN IgD−CD27− (% of B cells) . | Transitional 19+24++38++ (% of B cells) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IRAK-4 | P30 | M1V/1188 + 520A > G | 29 | 2 | 20 | 2.5 | 6.2 | 3.1 | 1 | 2.7 | 7 |
| 2.5 | 28 | 0.8 | 2.9 | nd | 0.4 | 7.7 | nd | ||||
| IRAK-4 | P8 | 1189-1G > T/ 1188 + 520A > G | 29 | 4 | 22 | 4 | 11 | 5.3 | nd | 4.5 | nd |
| 8.5 | 21 | 1.4 | 5.8 | nd | 1.65 | 4 | nd | ||||
| 9 | 11 | 2.4 | 8 | 1.8 | 2.44 | 2.8 | 2.43 | ||||
| IRAK-4 | P15 | 1-1096_40 + 23del / 1-1096_40 + 23del | 29 | 7 | 28 | 1.3 | 8.9 | nd | 1.65 | 8.2 | nd |
| 9.5 | nd | 3.3 | 16.3 | 4 | 2.4 | 3 | 7 | ||||
| IRAK-4 | P13 | E402X/E402X | 29 | 10 | 8 | 2.4 | 7.3 | 5.8 | nd | 2.1 | 6 |
| IRAK-4 | P2 | Q293X/ BAC210N13del | 29 | 12 | nd | 3 | 11 | 5.1 | nd | 4.2 | nd |
| 16.5 | 15 | 1.4 | 3.7 | nd | nd | 7.5 | nd | ||||
| IRAK-4 | P48 | Q293X/Q293X | 29 | 12 | nd | 6 | 11 | nd | nd | 3.7 | 8 |
| IRAK-4 | P24 | 1240insA/9421481_ 1125 + 547del | 29 | 16 | nd | 6.6 | 7.4 | nd | 1.8 | 1.5 | 10 |
| IRAK-4 | P31 | R12C/831 + 5G > T | 29 | 17 | nd | 3.7 | 16 | nd | 5.2 | 5.6 | 5 |
| IRAK-4 | P17 | Q293X/Q293X | 29 | 25 | 20 | 1.8 | 7.7 | nd | 1 | 8 | nd |
| MyD88 | P2 | R209C/L106P | 29 | 2 | 18 | 2.8 | 6.7 | 4.8 | 1.9 | 4 | 7 |
| MyD88 | P6 | E65del/E65del | 29 | 4.5 | 19 | 4.6 | 7.7 | 6.7 | 1.7 | 2.3 | 12.4 |
| MyD88 | P5 | E65del/E65del | 29 | 7 | 17 | 5.4 | 9.2 | 4.3 | 3 | 2.7 | 22 |
| MyD88 | P3 | R209C/R209C | 29 | 9 | 8 | 9 | 15.5 | 2.3 | 2.5 | 5 | 5.7 |
| 9.5 | 11 | 5.8 | 12 | nd | 2.8 | 11.4 | 5 | ||||
| 10.5 | 5.5 | 5.6 | 7.6 | 1.7 | 1.5 | 3.3 | 13 | ||||
| MyD88 | P4 | R209C/R209C | 29 | 16 | 13 | 6 | 21 | nd | 7.2 | 9.6 | 5 |
| UNC-93B | P1 | 1034 del4/1034 del4 | 24 | 16 | 23 | 8.4 | 7.7 | 7.4 | 2.5 | 2.5 | 4.4 |
| UNC93-B | P2 | G781A/G781A | 24 | 15 | 23 | 15 | 14 | 4.7 | 5.1 | 5.6 | 4.4 |
| TLR3 | P1 | P554S/wt‡ | 30 | 15 | 17 | 14 | 9 | 4.5 | 2.5 | 1.7 | 5.6 |
| TLR3 | P2 | E746X/P554S | 31 | 19 | 12 | 11.5 | 6.5 | nd | 2 | 7 | 1.7 |
| TRIF | P1 | R141X/R141X | 27 | 3.5 | 22 | 4.6 | 5.5 | nd | 0.6 | 15 | 35 |
| TIRAP | R2 | R121W/R121W | § | 15 | 6.3 | 3.8 | 18.4 | 2.1 | 6 | 10 | 4.5 |
| TIRAP | R3 | R121W/R121W | § | 20 | 7.5 | 3 | 17.5 | 1.8 | 5.8 | 6.5 | 5.2 |
| TIRAP | R1 | R121W/R121W | § | 22 | 17 | 1.8 | 6.5 | nd | 1.6 | 5.6 | 7 |
| TIRAP | R4 | R121W/R121W | § | 23 | 11 | 2.6 | 10.5 | 1.8 | 3.7 | 7.8 | 4.6 |
| Controls | Normal values¶ | 2-3 | 20.8 | 5.4 | 2.6 | nd | nd | 2.5 | 8.7 | ||
| 4-5 | 16.1 | 6.5 | 5.6 | nd | nd | 4.5 | 7.3 | ||||
| 6-10 | 12.2 | 10 | 6.5 | nd | nd | 5 | 6 | ||||
| 11-18 | 13.3 | 7.3 | 5.4 | nd | nd | 3.7 | 5.6 | ||||
| 19-25 | 9.1 | 11.7 | 9.4 | nd | nd | 3.2 | 4.7 | ||||
| Normal values# | 2-5 | 1.31 | |||||||||
| 6-10 | 2.44 | ||||||||||
| 11-15 | 2.93 | ||||||||||
| > 16 | 3.11 |
| Deficiency . | Patient . | Mutation . | Reference . | Age, y* . | % CD19+ . | IgD+CD27+ (% of B cells) . | IgD−CD27+ (% of B cells) . | IgM-only CD27+ (% of IgD-CD27+)† . | IgA+CD27+ (% of B cells) . | DN IgD−CD27− (% of B cells) . | Transitional 19+24++38++ (% of B cells) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IRAK-4 | P30 | M1V/1188 + 520A > G | 29 | 2 | 20 | 2.5 | 6.2 | 3.1 | 1 | 2.7 | 7 |
| 2.5 | 28 | 0.8 | 2.9 | nd | 0.4 | 7.7 | nd | ||||
| IRAK-4 | P8 | 1189-1G > T/ 1188 + 520A > G | 29 | 4 | 22 | 4 | 11 | 5.3 | nd | 4.5 | nd |
| 8.5 | 21 | 1.4 | 5.8 | nd | 1.65 | 4 | nd | ||||
| 9 | 11 | 2.4 | 8 | 1.8 | 2.44 | 2.8 | 2.43 | ||||
| IRAK-4 | P15 | 1-1096_40 + 23del / 1-1096_40 + 23del | 29 | 7 | 28 | 1.3 | 8.9 | nd | 1.65 | 8.2 | nd |
| 9.5 | nd | 3.3 | 16.3 | 4 | 2.4 | 3 | 7 | ||||
| IRAK-4 | P13 | E402X/E402X | 29 | 10 | 8 | 2.4 | 7.3 | 5.8 | nd | 2.1 | 6 |
| IRAK-4 | P2 | Q293X/ BAC210N13del | 29 | 12 | nd | 3 | 11 | 5.1 | nd | 4.2 | nd |
| 16.5 | 15 | 1.4 | 3.7 | nd | nd | 7.5 | nd | ||||
| IRAK-4 | P48 | Q293X/Q293X | 29 | 12 | nd | 6 | 11 | nd | nd | 3.7 | 8 |
| IRAK-4 | P24 | 1240insA/9421481_ 1125 + 547del | 29 | 16 | nd | 6.6 | 7.4 | nd | 1.8 | 1.5 | 10 |
| IRAK-4 | P31 | R12C/831 + 5G > T | 29 | 17 | nd | 3.7 | 16 | nd | 5.2 | 5.6 | 5 |
| IRAK-4 | P17 | Q293X/Q293X | 29 | 25 | 20 | 1.8 | 7.7 | nd | 1 | 8 | nd |
| MyD88 | P2 | R209C/L106P | 29 | 2 | 18 | 2.8 | 6.7 | 4.8 | 1.9 | 4 | 7 |
| MyD88 | P6 | E65del/E65del | 29 | 4.5 | 19 | 4.6 | 7.7 | 6.7 | 1.7 | 2.3 | 12.4 |
| MyD88 | P5 | E65del/E65del | 29 | 7 | 17 | 5.4 | 9.2 | 4.3 | 3 | 2.7 | 22 |
| MyD88 | P3 | R209C/R209C | 29 | 9 | 8 | 9 | 15.5 | 2.3 | 2.5 | 5 | 5.7 |
| 9.5 | 11 | 5.8 | 12 | nd | 2.8 | 11.4 | 5 | ||||
| 10.5 | 5.5 | 5.6 | 7.6 | 1.7 | 1.5 | 3.3 | 13 | ||||
| MyD88 | P4 | R209C/R209C | 29 | 16 | 13 | 6 | 21 | nd | 7.2 | 9.6 | 5 |
| UNC-93B | P1 | 1034 del4/1034 del4 | 24 | 16 | 23 | 8.4 | 7.7 | 7.4 | 2.5 | 2.5 | 4.4 |
| UNC93-B | P2 | G781A/G781A | 24 | 15 | 23 | 15 | 14 | 4.7 | 5.1 | 5.6 | 4.4 |
| TLR3 | P1 | P554S/wt‡ | 30 | 15 | 17 | 14 | 9 | 4.5 | 2.5 | 1.7 | 5.6 |
| TLR3 | P2 | E746X/P554S | 31 | 19 | 12 | 11.5 | 6.5 | nd | 2 | 7 | 1.7 |
| TRIF | P1 | R141X/R141X | 27 | 3.5 | 22 | 4.6 | 5.5 | nd | 0.6 | 15 | 35 |
| TIRAP | R2 | R121W/R121W | § | 15 | 6.3 | 3.8 | 18.4 | 2.1 | 6 | 10 | 4.5 |
| TIRAP | R3 | R121W/R121W | § | 20 | 7.5 | 3 | 17.5 | 1.8 | 5.8 | 6.5 | 5.2 |
| TIRAP | R1 | R121W/R121W | § | 22 | 17 | 1.8 | 6.5 | nd | 1.6 | 5.6 | 7 |
| TIRAP | R4 | R121W/R121W | § | 23 | 11 | 2.6 | 10.5 | 1.8 | 3.7 | 7.8 | 4.6 |
| Controls | Normal values¶ | 2-3 | 20.8 | 5.4 | 2.6 | nd | nd | 2.5 | 8.7 | ||
| 4-5 | 16.1 | 6.5 | 5.6 | nd | nd | 4.5 | 7.3 | ||||
| 6-10 | 12.2 | 10 | 6.5 | nd | nd | 5 | 6 | ||||
| 11-18 | 13.3 | 7.3 | 5.4 | nd | nd | 3.7 | 5.6 | ||||
| 19-25 | 9.1 | 11.7 | 9.4 | nd | nd | 3.2 | 4.7 | ||||
| Normal values# | 2-5 | 1.31 | |||||||||
| 6-10 | 2.44 | ||||||||||
| 11-15 | 2.93 | ||||||||||
| > 16 | 3.11 |
DN indicates double negative; IRAK-4, IL-1 receptor–associated kinase 4; and nd, not determined.
Age at analysis.
IgM-only CD27+ cells represent on average 7.9% ± 1.6% of the IgD−CD27+ subset in healthy adult controls.4
The P554S allele is loss-of-function and dominant negative.
R1 to R4 are relatives of a TIRAP deficient patient (proband); they have a confirmed TIRAP deficiency but no clinical phenotype (L.I. and J.-L.C., unpublished data, 2012)
Reference values (median) are taken from Morbach et al.32
Reference values for the IgA+CD27+ B-cell subset (mean) are from Dr Menno Van Zelm, Netherlands, (Erasmus Medical Center, Rotterdam, The Netherlands; personal written communication, January 2012).
Flow cytometry analysis and cell sorting of blood and splenic B-cell subsets
Antibodies used for cell staining are described in supplemental Table 1. PBMCs were isolated by centrifugation on Ficoll-Paque PLUS (GE Healthcare), with, in some cases, previous B-cell enrichment by negative selection with the RosetteSep B-cell enrichment cocktail (StemCell Technologies). Splenocytes were obtained from splenic samples by mechanical disruption followed by a Ficoll gradient centrifugation. Then, splenic B cells were enriched with the Dynabeads Untouched Human B Cells kit (Invitrogen). PBMCs or enriched blood or splenic B cells were then incubated with different combinations of antibodies. Flow cytometric data acquisition was performed on a FACSCalibur or a FACSCantoII, and data were analyzed either with the CellQuest, Diva (BD Biosciences), or FlowJo (Tree Star) software. For the analysis of somatic hypermutation frequencies or for VH3-spectratyping, cells were stained with anti-CD19-PC5, IgD-FITC, and CD27-PE antibodies and separated into CD19+IgD+CD27+, CD19+IgD+CD27−, and CD19+IgD−CD27+ fractions, respectively, by the use of a FACSAria Cell Sorter (BD Biosciences). For the cytokine/chemokine secretion assays, B cells were labeled with anti-CD19-APC-H7 and CD27-APC antibodies, together with either anti-IgA-FITC and -IgG-PE or with -IgD-FITC antibodies. Sorting was then performed by gating on CD19+IgD+CD27+, CD19+IgD+CD27−, and CD19+IgG+ or IgA+CD27+ cells. Purity of sorted cell fractions was > 98%.
B-cell culture with TLR agonists
Sorted B cells were incubated in 96-well flat-bottom plates (BD Biosciences) at a concentration of 2 × 105 cells/200μL in RPMI1640/Glutamax/PenStrep (Gibco-Invitrogen) and 10% FCS (FetalClone I; HyClone) and were stimulated with different TLR agonists, used at the following final concentrations: CpG ODN 2006 (TLR9 agonist, Sigma-Aldrich) at 6 μg/mL; flagellin (TLR5 agonist) at 0.5 μg/mL; resiquimod or R848 (TLR7 and 8 agonist), Pam2CSK4 (TLR2/6 agonist), and Pam3CSK4 (TLR1/2 agonist; all from Invivogen) at 1 μg/mL; PamCys(Pam)-SK4 (a putative TLR10 agonist,33 EMC Microcollections) at 1 μg/mL; and poly-IC (polyinosine-polycytidic acid, a TLR3 agonist, GE Healthcare) at 25 ng/μL. The quantitative determination of secreted cytokines/chemokines was performed on 96-well supernatants collected after 4 days in culture via the use of multiplex cytokine assays (Human Cytokine 25-plex panel from Invitrogen or Bio-Plex Pro Human Cytokine 27-Plex Panel from Bio-Rad) read on a Bio-Plex 200 system (Bio-Rad). Alternatively, the concentration of IL-6, IL-8, MIP-1α, and MIP-1β was assessed by ELISA by use of the following kits: human IL-6 and IL-8 (Sanquin), human MIP-1α (Peprotech), and MIP-1β (Mabtech) ELISA kits.
Analysis of the mutation frequencies in JH4-JH5 introns flanking rearranged VHDJH4
Starting from 2500-3000 sorted IgD+CD27+ and IgD−CD27+ cells, total genomic DNA was extracted by proteinase K digestion. The JH4 segment is used in 50% of rearranged VH genes, and the adjacent JH4-JH5 intron can be studied when limited numbers of cells are available. The JH4-JH5 intronic region was amplified with Phusion DNA polymerase (Finnzymes) by the use of a mixture of 6 FR3 primers (FR3-mix) designed to amplify all VH gene sequences and a primer binding 5′ to the JH5 exon, as previously described.7 PCR conditions were as follows: 40 cycles of 98°C for 10 seconds, 60°C for 20 seconds, and 72°C for 20 seconds. The resulting JH4-JH5 PCR products were gel purified and cloned with the Zero Blunt PCR Cloning Kit (Invitrogen). Sequences were run in an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Mutation frequencies were calculated by comparing the sequences obtained with germline intronic JH4-JH5 sequences over 341 bp, starting at the 3′ border of JH4.
VH3 spectratyping
Total RNA was isolated from 10 000-30 000 cells using the Allprep DNA/RNA Mini Kit (QIAGEN) and reverse transcribed by random priming with the ProSTAR First Strand Reverse Transcription-PCR Kit (Stratagene). VH3-μ transcripts were then amplified with GoTaq polymerase (Promega) for 40 cycles (45 seconds at 94°C, 60 seconds at 59°C, and 90 seconds at 72°C) using a VH3–consensus primer (5′-GGGGTCCCTGAGACTCTCCTGTGCAG-3′) and a μ-CH1 region primer (5′-GGGAATTCTCACAGGAGACGA-3′). PCR products of the expected size (∼ 400 bp) were gel purified with the Gel Extraction Kit (QIAGEN) and fluorescently labeled using a run-off reaction consisting of 10 cycles of 30 seconds at 94°C, 30 seconds at 58°C, and 1 minute at 72°C, with the mix of FR3 primers coupled in 5′ to the fluorescent dye HEX (Sigma-Aldrich). The run-off product were subjected to electrophoresis on an ABI PRISM 3100 genetic analyzer, with GeneScan 400HD ROX Size Standard and were analyzed with Genemapper version 4 (both from Applied Biosystems) as described previously.7
Statistical analysis
The comparison of the proportions of IgD+CD27+ and IgD−CD27+ B cells between IRAK-4–, MyD88-, and TIRAP-deficient patients and controls or between IRAK-4– and MyD88- or between MyD88- and TIRAP-deficient patients was conducted my means of generalized estimating equations to account for the correlation arising from repeated measurements and to avoid the multivariate normality assumption. The generalized estimating equation analysis was performed by use of the GENMOD procedure of the SAS Version 9.2 software (SAS institute). The comparisons of mean fluorescence intensities of TLR10 between IgD+CD27+ and IgD−CD27+ B cells from blood and spleen were performed by paired t test as implemented in the TTEST procedure of the SAS Version 9.2 software.
Results
IRAK-4–, MyD88-, and TIRAP-deficient patients show a specific reduction of the IgM+IgD+CD27+ B-cell subset
We studied the peripheral blood B-cell subsets of patients deficient for IRAK-4 (n = 9), MyD88 (n = 5), TIRAP (n = 4), UNC-93B (n = 2), TLR3 (n = 2), and TRIF (n = 1; Figure 1 and supplemental Figure 2). For some patients, 2 or 3 blood samples could be obtained at different time points (Table 1). All the patients have been previously described,17,18,24,27,29-31 except the 4 TIRAP-deficient patients (L.I. and J.-L.C., unpublished data, 2012) who carry a homozygous missense mutation in the TIR domain of TIRAP (Table 1), resulting in a selective impairment of the signaling downstream of TLR2/TLR6. All patients had B-cell numbers within normal values (Table 1).17,18,24,27,29-31 The age of the patients ranged from 2 to 25 years, with a mean age of 10.7 and 8.4 years for the IRAK-4– and MyD88-deficient patients, respectively. Our control group consisted of healthy patients with a similar distribution of age (2-25 years; mean, 10.7 years).
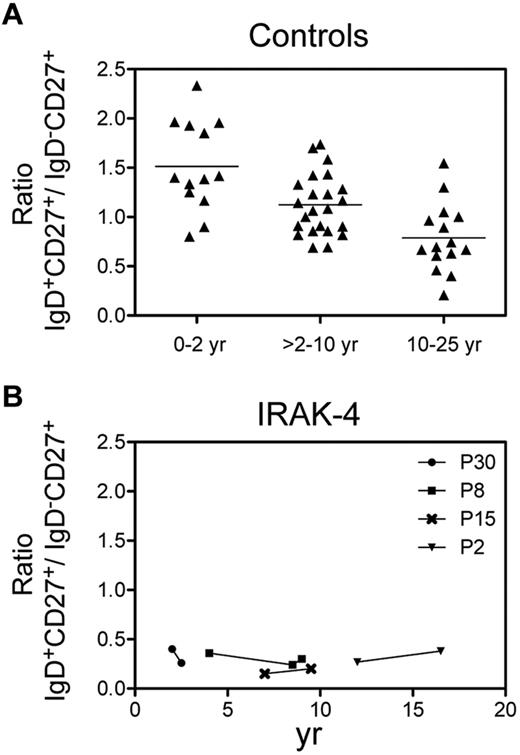
IgM+IgD+CD27+ cells are significantly decreased in IRAK-4–, MyD88-, and TIRAP-deficient patients. (A) Relative frequencies of blood IgD+CD27+ and IgD−CD27+ cells (expressed as percentages of CD19+ B cells) in IRAK-4–, MyD88-, TIRAP-, UNC-93B–, and TLR3-deficient patients and controls were determined by flow cytometry after IgD, CD27, and CD19 staining. Only values from patients or control patients older than 4 years of age were plotted (ie, excluding the 3.5-year-old TRIF-deficient patient, as well as patients P30 [IRAK-4, 2 years] and P2 [MyD88, 2 years]). Mean values (expressed as percentage of total B cells) are indicated below the graph for IRAK-4–, MyD88-, and TIRAP-deficient patients and controls. *P < .05, **P < .01, ***P < .001 (see “Methods” for statistics). (B) Ratio of the IgM+IgD+CD27+ versus IgD−CD27+ cell proportions for all the patients and controls (from 2 to 25 years of age).
IgM+IgD+CD27+ cells are significantly decreased in IRAK-4–, MyD88-, and TIRAP-deficient patients. (A) Relative frequencies of blood IgD+CD27+ and IgD−CD27+ cells (expressed as percentages of CD19+ B cells) in IRAK-4–, MyD88-, TIRAP-, UNC-93B–, and TLR3-deficient patients and controls were determined by flow cytometry after IgD, CD27, and CD19 staining. Only values from patients or control patients older than 4 years of age were plotted (ie, excluding the 3.5-year-old TRIF-deficient patient, as well as patients P30 [IRAK-4, 2 years] and P2 [MyD88, 2 years]). Mean values (expressed as percentage of total B cells) are indicated below the graph for IRAK-4–, MyD88-, and TIRAP-deficient patients and controls. *P < .05, **P < .01, ***P < .001 (see “Methods” for statistics). (B) Ratio of the IgM+IgD+CD27+ versus IgD−CD27+ cell proportions for all the patients and controls (from 2 to 25 years of age).
IgM+IgD+CD27+ and switched B cells emerge in parallel in the blood of healthy children younger than 2 years of age and represent an increasing fraction of total B cells with age.4 In very young patients, the proportion of IgM+IgD+CD27+ cells is generally slightly greater than switched cells, with a mean ratio of 1.5. Later in life, the ratio of IgM+IgD+CD27+ versus switched cells is generally close to 1.0, with mean ratios of 1.1 from 2 to 10 years and 0.8 from 10 to 25 years (Figure 2A). Thus, to evaluate whether the absence of signaling from one or several TLRs may affect the development and/or homeostasis of IgM+IgD+CD27+ and switched B cells in the patients studied, 2 different values were considered and compared with age-matched controls: first, their proportions relative to total B cells, for patients older than 4 years of age to avoid the inclusion of low values typical of young ages (Figure 1A); and second, the IgM+IgD+CD27+ versus switched cell ratio for all of them (Figure 1B). In patients for whom IgM-only B cells were analyzed, they represented a minor subset and accounted for 1.7%-7.4% of IgD−CD27+ B cells (Table 1), making percentages of IgD−CD27+ cells a valid surrogate for isotype-switched B cells.
The defect of IgM+IgD+CD27+ cells is stable with age. Ratio of the IgD+CD27+ versus IgD−CD27+ cell proportions as a function of age in control patients (A) and in several patients deficient for IRAK-4 who were analyzed at different ages (B). Control patients are divided into 3 different age groups.
The defect of IgM+IgD+CD27+ cells is stable with age. Ratio of the IgD+CD27+ versus IgD−CD27+ cell proportions as a function of age in control patients (A) and in several patients deficient for IRAK-4 who were analyzed at different ages (B). Control patients are divided into 3 different age groups.
We observed a significant reduction in the proportion of IgM+IgD+CD27+ B cells for IRAK-4– and MyD88-deficient patients in comparison with control patients, and the reduction was surprisingly slightly more pronounced for IRAK-4– than for MyD88-deficient patients. In contrast, the proportion of switched B cells was only slightly or marginally reduced in IRAK-4– or MyD88-deficient patients, respectively, in comparison with switched cells of controls. Accordingly, the mean ratios of IgM+IgD+CD27+ versus switched cells in IRAK-4– and MyD88-deficient patients were significantly lower than for controls (0.34 ± 0.19 and 0.53 ± 0.15 vs 1 ± 0.31).
Similar results were observed for the 4 adult TIRAP-deficient patients (15-23 years): their proportions of IgD+CD27+ cells were severely reduced, whereas switched cells were clearly unaffected, and the mean ratio of IgM+IgD+CD27+ versus switched cells was approximately 4 times lower than for the controls (0.23 ± 0.05 vs 1 ± 0.31). One should mention as a caveat that the 4 TIRAP-deficient patients are brothers and sisters of a consanguineous family. However, the very similar phenotype observed for all of them, together with the rarity of a deficiency limited to the sole IgM+IgD+CD27+ subset (see “Discussion”), makes the possibility of a phenotype contributed by the fortuitous coinheritance of a second genetic lesion by all members of the family somehow unlikely.
In contrast, in the TLR3- and the UNC-93B–deficient patients, the proportions of IgM+IgD+CD27+ and switched cells were within normal ranges, and the ratios of IgM+IgD+CD27+ versus switched cells were always superior to 1. At last, in the young TRIF-deficient patient, the proportion of IgM+IgD+CD27+ and switched cells were similar and within the range of normal age-matched values. Altogether, these results suggest a minor contribution of TLR3, TLR7, TLR8, and TLR9 for the generation and/or maintenance of IgM+IgD+CD27+ cells in humans but points toward a role of other receptors that rely on TIRAP, MyD88, and IRAK-4 for their signaling.
The reduction of IgM+IgD+CD27+ B cells in IRAK-4–deficient patients is not compensated with age
For 4 IRAK-4–deficient patients, the relative proportions of IgM+IgD+CD27+ and IgD−CD27+ cells could be followed at different ages (Figure 2B). The ratio of IgM+IgD+CD27+ versus switched CD27+ cells did not appear to vary significantly with time and was consistently lower for IRAK-4–deficient patients than the mean ratio observed for the age-matched control groups. This finding indicates that the absence of IRAK-4–mediated signaling in IgM+IgD+CD27+ cells is not overcome by homeostatic mechanisms as patients grow older and suggests a possible role for this pathway in the maintenance of these cells.
Normal somatic diversification and heavy-chain CDR3 size selection of IgM+IgD+CD27+ cells in IRAK-4– and MyD88-deficient patients
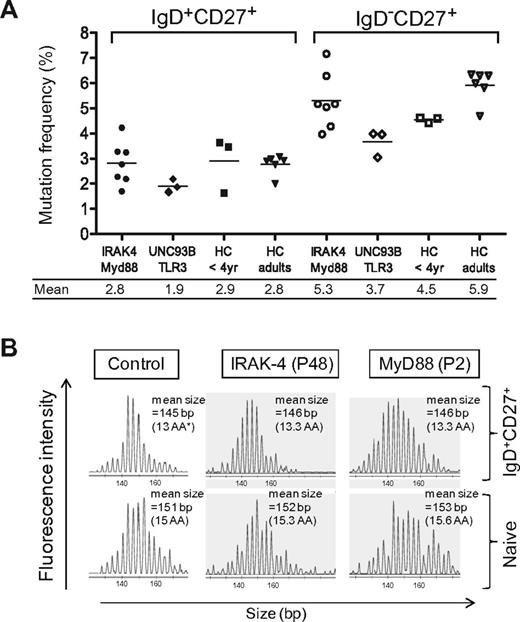
To study the role of TLR signaling in the somatic hypermutation process, mutation frequencies of Ig genes were determined for sorted blood IgD+CD27+ and IgD−CD27+ subsets of several patients and age-matched controls (Figure 3 and supplemental Table 2). The mutation frequencies of IgD+CD27+ and IgD−CD27+ cells of the 2 MyD88- and the 5 IRAK-4–deficient patients studied were within the range of the control values. Mutation characteristics, like transitions/transversions ratio or hotspot targeting, according to Longo et al34 also were comparable with control patients. These results suggest that the diversification of IgM+IgD+CD27+ cells does not rely on a IRAK-4– and MyD88-mediated signaling, whether triggered directly on B cells or on other cell types, like the recently described B-cell helper neutrophils that were proposed to stimulate the diversification of marginal zone B cells.35 Mutation frequencies for the 2 Unc93-B and the single TLR3-deficient patients were somewhat in the lower range of controls, notably for the TLR3 switched subset, but conclusions are obviously difficult for this single case.
Normal somatic mutation frequencies and heavy-chain CDR3 repertoire in sorted IgD+CD27+ cells from IRAK-4– and MyD88-deficient patients. (A) Somatic mutations frequencies in the JH4-JH5 intron flanking rearranged VHDJH4 sequences from sorted blood IgD+CD27+ and IgD−CD27+ cells of IRAK-4–, MyD88-, UNC-93B-, and TLR3-deficient patients (3.5 years) and young (< 4 years) or adult healthy controls (HC; see also supplemental Table 2). Mean values (expressed as mutations per 100 bp of total sequences) are indicated below the graph for each B-cell subset. IRAK-4– or MyD88-deficient patients and those deficient for UNC-93B or TLR3 were grouped. In control JH4-JH5 sequences, mutation frequencies gradually increased with age, reaching adult values for blood IgD+CD27+ B cells by approximately 4 years of age (S.W., J.-C. W and C.-A.R., unpublished data, 2009). (B) H-CDR3 spectratypes of the VH3 transcripts expressed by sorted blood IgD+CD27+ and naive B cells of a healthy control and a MyD88- and an IRAK-4–deficient patient. VH3-μ transcripts were prepared from 10 000 to 30 000 sorted cells, reverse transcribed, and amplified by PCR (see “Methods”). The PCR products were labeled by a run-off reaction with specific fluorescent VH-FR3 consensus primers and subjected to capillary gel electrophoresis. As observed for naive cells, control IgD+CD27+ cells display a regular distribution of heavy-chain CDR3 sizes but showed a shift of 6 bp on average toward shorter CDR3 lengths. IgD+CD27+ cells from the IRAK-4– and the MyD88-deficient patient (P48 and P2) display a similar Gaussian distribution and average heavy-chain CDR3 size compared with control IgD+CD27+ cells (ie, a 6- or 7-bp mean difference in comparison with autologous naive cells). *The mean heavy-chain (H)–CDR3 length in amino acids (AA) inferred from the mean size of the PCR products is indicated. (H-CDR3 being defined as the region included between the invariant Cys and Trp residues of VH and JH genes, respectively.)
Normal somatic mutation frequencies and heavy-chain CDR3 repertoire in sorted IgD+CD27+ cells from IRAK-4– and MyD88-deficient patients. (A) Somatic mutations frequencies in the JH4-JH5 intron flanking rearranged VHDJH4 sequences from sorted blood IgD+CD27+ and IgD−CD27+ cells of IRAK-4–, MyD88-, UNC-93B-, and TLR3-deficient patients (3.5 years) and young (< 4 years) or adult healthy controls (HC; see also supplemental Table 2). Mean values (expressed as mutations per 100 bp of total sequences) are indicated below the graph for each B-cell subset. IRAK-4– or MyD88-deficient patients and those deficient for UNC-93B or TLR3 were grouped. In control JH4-JH5 sequences, mutation frequencies gradually increased with age, reaching adult values for blood IgD+CD27+ B cells by approximately 4 years of age (S.W., J.-C. W and C.-A.R., unpublished data, 2009). (B) H-CDR3 spectratypes of the VH3 transcripts expressed by sorted blood IgD+CD27+ and naive B cells of a healthy control and a MyD88- and an IRAK-4–deficient patient. VH3-μ transcripts were prepared from 10 000 to 30 000 sorted cells, reverse transcribed, and amplified by PCR (see “Methods”). The PCR products were labeled by a run-off reaction with specific fluorescent VH-FR3 consensus primers and subjected to capillary gel electrophoresis. As observed for naive cells, control IgD+CD27+ cells display a regular distribution of heavy-chain CDR3 sizes but showed a shift of 6 bp on average toward shorter CDR3 lengths. IgD+CD27+ cells from the IRAK-4– and the MyD88-deficient patient (P48 and P2) display a similar Gaussian distribution and average heavy-chain CDR3 size compared with control IgD+CD27+ cells (ie, a 6- or 7-bp mean difference in comparison with autologous naive cells). *The mean heavy-chain (H)–CDR3 length in amino acids (AA) inferred from the mean size of the PCR products is indicated. (H-CDR3 being defined as the region included between the invariant Cys and Trp residues of VH and JH genes, respectively.)
Blood IgM+IgD+CD27+ cells as well as splenic IgM+IgD+CD27+ cells of infants harbor shorter Ig heavy-chain CDR3s than naive cells, the hallmark of a unclear selection process that has been described as well for marginal zone B cells in the mouse.4,36 To address the role of TLR signaling in this selection, we performed H-CDR3 spectratyping of VH3 transcripts expressed by sorted blood IgD+CD27+ and naive B cells from a MyD88- and an IRAK-4–deficient patient (Figure 3B). In the control, a regular distribution of heavy-chain CDR3 sizes was observed for IgD+CD27+ cells, the mean difference in CDR3 sizes between IgD+CD27+ and naive cells being of 6 bp on the basis of mean peak height values. As for the control, IgD+CD27+ B cells from the IRAK-4– and Myd88-deficient patients P48 and P2 harbored shorter H-CDR3 lengths, with a shift of 6 and 7 bp, respectively, compared with naive B cells. Thus, the absence of IRAK-4– and MyD88-mediated signaling does not appear to impact the selection that is applied to IgM+IgD+CD27+ cells during their development.
Blood and splenic B-cell subsets reveal that IgM+IgD+CD27+ cells display greater TLR10 expression
Human B cells have been shown to express various TLRs, but depending on their tissue origins (tonsils, blood), their state of maturation (naive vs memory), or activation (after a BCR stimulation or under inflammatory conditions), the patterns of mRNA expression showed some differences.37-41 In agreement with previous observations, and by mean of a gene expression profiling study with Affymetrix Human Genome U133 Plus2 arrays, we found that different ex vivo blood and splenic B-cell subsets expressed very high levels of TLR10 and TLR1; high-to-intermediate levels of TLR6, TLR7, and TLR9; barely detectable levels of TLR2 and TLR4; and undetectable levels of TLR3, TLR5, and TLR8 transcripts (supplemental Figure 3). Notably, mRNA transcript levels of TLR1, TLR6, and TLR10 were greater on average in the splenic compared with blood subsets, a difference that was, however, not observed at the protein level.
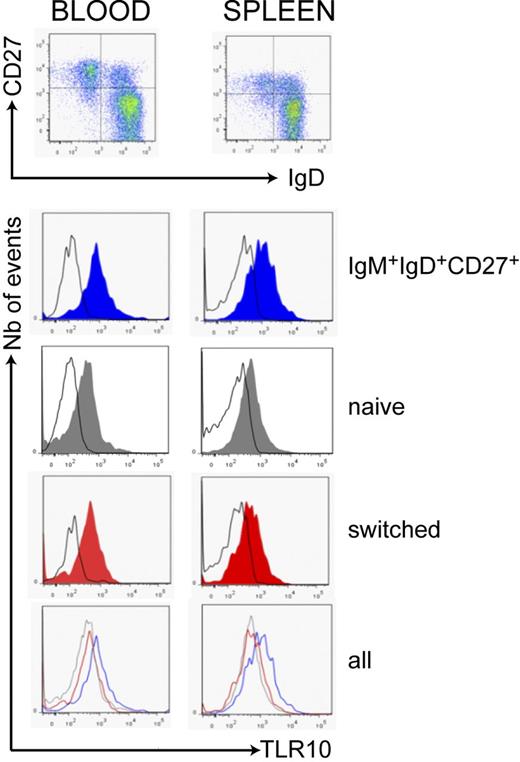
To confirm the protein expression of TIRAP-dependent TLRs at the cell surface (TLR1, TLR2, TLR4, TLR6, and TLR10), enriched B cells from blood and spleen were analyzed by flow cytometry. Consistent with the absence or very low level of transcripts, no staining for TLR2 or TLR4 was detected in the blood or splenic B-cell subsets, but, surprisingly, surface TLR6 was hardly detected (not shown). Splenic IgM+IgD+CD27+ and switched cells were nevertheless able to proliferate and to secrete cytokines/chemokines in response to a TLR2/6 agonist (see below). TLR1 and TLR10 were expressed by all B-cell subsets, TLR1 being expressed at a similar level on IgM+IgD+CD27+ and switched cells (supplemental Figure 4), whereas TLR10 was reproducibly and significantly greater on IgM+IgD+CD27+ cells (Figure 4) in comparison with switched cells (a mean 2-fold [P < .002] or 1.8-fold [P < .004]) difference in mean fluorescence intensity was observed in blood and spleen, respectively). This may confer to IgM+IgD+CD27+ cells a specific responsiveness after TLR10 engagement with yet-unknown TLR10 ligands.
Greater TLR10 expression in IgM+IgD+CD27+ cells from blood and spleen. B cells from blood and spleen were analyzed for TLR10 surface expression by flow cytometry after gating on different CD19+ subsets distinguished by IgD and CD27 labeling. Expression of TLR10 was greater on IgD+CD27+ cells (blue) in comparison with naive IgD+CD27− (gray) and IgD−CD27+ cells (red); the black line represents the isotype-matched control antibody. Data are representative of 6 blood samples and 10 splenic samples. An average 2- and 1.8-fold difference in mean fluorescence intensity was observed for IgM+IgD+CD27+ versus switched B cells from blood and spleen, respectively (P < .002 and P < .004, see “Methods”).
Greater TLR10 expression in IgM+IgD+CD27+ cells from blood and spleen. B cells from blood and spleen were analyzed for TLR10 surface expression by flow cytometry after gating on different CD19+ subsets distinguished by IgD and CD27 labeling. Expression of TLR10 was greater on IgD+CD27+ cells (blue) in comparison with naive IgD+CD27− (gray) and IgD−CD27+ cells (red); the black line represents the isotype-matched control antibody. Data are representative of 6 blood samples and 10 splenic samples. An average 2- and 1.8-fold difference in mean fluorescence intensity was observed for IgM+IgD+CD27+ versus switched B cells from blood and spleen, respectively (P < .002 and P < .004, see “Methods”).
The MyD88/ IRAK-4 pathway is also used by IL-1R family members, including IL-1R, IL-18R, and IL-33R. We therefore looked for mRNA expression of the corresponding genes (IL1R1, IL18R1, and IL1RL1) in different ex vivo blood and splenic B-cell subsets (supplemental Figure 3). IL1R1 and IL1RL1 transcripts were not detected, whereas IL18R1 transcripts were present at various but low levels in splenic subsets and were barely detectable in blood samples. We looked for the protein expression of these genes on the surface of blood and splenic subsets by flow cytometry. IL-1R4 (also known as ST2, the product of the IL1RL1 gene), which associates with IL-1RAcP to form the receptor complex for IL-33, could not be detected on blood or splenic B cells (not shown). IL-1R1, the biologically active form of the IL-1R, was undetectable on the blood B cells of 5 different adult donors (not shown) and only very weakly expressed on splenic B cells of 4 of 5 individuals (supplemental Figure 4). In contrast, most of the blood and splenic B cells stained for IL-18Rα (the product of the IL18R1 gene) with a comparable level of expression on both IgM+IgD+CD27+ and switched B cells.
IgM+IgD+CD27+ and switched B cells show distinct cytokine responses to TLR agonists
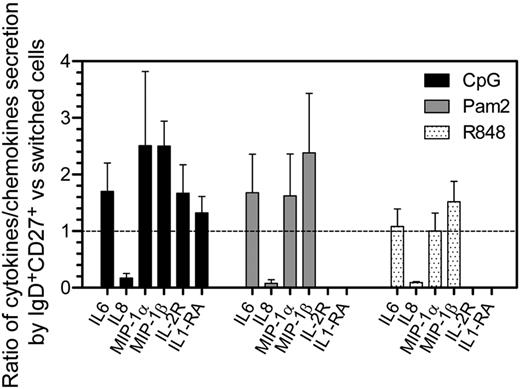
To test both their cytokine expression profiles and proliferative response after TLR engagement, splenic switched (IgG+ and IgA+CD27+), IgD+CD27+, and naive B cells were sorted and activated with a panel of TLR ligands. Among those ligands, only CpG (TLR9), Pam2CSK4 (TLR2/6), and R848 (TLR7/TLR8) induced a reproducible secretion above the background level of more than 3 cytokines/chemokines among the 25 or 27 that were measured in the culture supernatants by multiplex assays (Figure 5 and supplemental Figure 5). IgD+CD27+ cells secreted moderately but reproducibly greater levels of IL-6, MIP-1α, and MIP-1β than switched cells in response to CpG and Pam2CSK4. A greater amount of IL-2R and IL-1RA was also produced by IgM+IgD+CD27+ compared with switched cells, in response to CpG stimulation. To note, IL-8, which is a potent neutrophil attractant, was mainly secreted by switched cells. Interestingly, among the large panel of cytokines/chemokines tested, some that are usually secreted by non–B-cell types were induced (eg, GM-CSF, IP-10, and VEGF; see supplemental Table 3). Such a secretion profile is unlikely to be because of contaminations by other cell types, eg, macrophages, given the cell sorting purity, and may therefore reflect the presence of subpopulations with yet-unknown functions, an observation that obviously warrants further studies.
Differences in cytokine secretion levels between IgM+IgD+CD27+ and switched B cells in response to different TLR agonists. Switched (IgG+ and IgA+) CD27+ cells and IgD+CD27+ were sorted and activated with different agonists, and cytokine secretion in culture supernatants after 4 days was evaluated by multiplex assays (see “Methods”). CpG (TRL9 agonist), R848 (TLR7 and TLR8 agonist), and Pam2CSK4 (Pam2, TLR2/6 agonist) did induce a reproducible secretion of IL-6, IL-8, MIP-1-α, and MIP-1-β (and IL-2R and IL-1RA for CpG stimulation). Because of a high variability of the amount of cytokines secreted by B-cell subsets from one splenic sample to another, the mean ratio of cytokine secretion ± SD between IgD+CD27+ and switched cells from the same sample is represented. The data were obtained from 3 or 4 independent experiments. The dashed line indicates a ratio equal to 1.
Differences in cytokine secretion levels between IgM+IgD+CD27+ and switched B cells in response to different TLR agonists. Switched (IgG+ and IgA+) CD27+ cells and IgD+CD27+ were sorted and activated with different agonists, and cytokine secretion in culture supernatants after 4 days was evaluated by multiplex assays (see “Methods”). CpG (TRL9 agonist), R848 (TLR7 and TLR8 agonist), and Pam2CSK4 (Pam2, TLR2/6 agonist) did induce a reproducible secretion of IL-6, IL-8, MIP-1-α, and MIP-1-β (and IL-2R and IL-1RA for CpG stimulation). Because of a high variability of the amount of cytokines secreted by B-cell subsets from one splenic sample to another, the mean ratio of cytokine secretion ± SD between IgD+CD27+ and switched cells from the same sample is represented. The data were obtained from 3 or 4 independent experiments. The dashed line indicates a ratio equal to 1.
Two attempts at inducing TLR10 signaling were made: first, by the use of PamCys(Pam)–SK4, which was proposed as a potential specific ligand of TLR10 on the basis of modeling studies and might activate human TLR10/1 hetero- and TLR10 homodimers,33 and second, through TLR10 cross-linking via the use of anti-TLR10 antibodies. However, both approaches were unable to trigger cytokine/chemokine secretion (or cell proliferation) from CD27− or CD27+ B-cell subsets.
Among the tested TLR agonists, CpG, and to a lesser extent R848, Pam2CSK4, and Pam3CSK4 (that triggers TLR1/2), were able to induce the proliferation (as well as a plasma cell differentiation) of both IgM+IgD+CD27+ and switched B cells. Because of great variability between samples, no significant differences emerged between the 2 subsets regarding their proliferative responses (not shown). On CpG stimulation, switched cells showed nevertheless a much greater differentiation level into CD27high plasmacytic cells (not shown). Altogether, IgM+IgD+CD27+ and switched cells show differences in their effector profile through stimulation of TLR9 that they express at the same level.39 One may thus envision that more pronounced differences could be generated through activation of TLR10 that is differentially expressed.
Discussion
We addressed in this report the role of TLRs in the ontogeny and homeostasis of human IgM+IgD+CD27+ cells in vivo through the analysis of patients bearing genetic defects that impair the TLR signaling pathways, as well as through the study of the expression profile of TLR/IL-1R of blood and splenic B-cell subsets and of their chemokine/cytokine secretion after stimulation by TLR ligands.
At first, and to evaluate which TLRs may be responsible for possible B-cell–intrinsic effects, we studied TLRs expression on IgM+IgD+CD27+, switched, and naive B cells from blood and spleen of healthy individuals. Several studies have described the TLR expression patterns of B cells or B-cell subsets, mostly from peripheral or mucosal B cells and at the mRNA level (reviewed in Bekeredjian-Ding and Jego13 ). In close agreement with these previous reports, we observed that naive, IgM+IgD+CD27+ and switched cells from blood or spleen express TLR1, TLR6, TLR7, TLR9, and TLR10 transcripts. Compared with their blood counterparts, splenic subsets express greater levels of these transcripts, which might probably endow them with a greater and/or faster responsiveness to TLR engagement. No expression of TLR3, TLR5, and TLR8 was found, whereas TLR2 and TLR4 transcripts were barely detectable. Because TIRAP-deficient patients showed a specific reduction of IgM+IgD+CD27+ cells (see next paragraph), we focused, for cell-surface expression, on TLRs dependent on this pathway. Among those, only TLR10, which was shown to be specifically restricted to B cells and plasmacytoid dendritic cells,42 was differentially expressed, being constantly greater in IgM+IgD+CD27+ cells in comparison with switched and naive B cells, which suggests that TLR10, whose function remains elusive, may play a specific role for this subset. We also analyzed the expression of the IL-1R family that includes receptors to IL-1, IL-18, and IL-33 and uses the same MyD88-IRAK-4 signaling pathway. A clear IL-18R expression was detected in blood and splenic samples, which did not differ between switched and unswitched B-cell subsets.
We then studied the peripheral B-cell compartment of IRAK-4–, MyD88-, and TIRAP-deficient patients and reported here a marked reduction of IgM+IgD+CD27+ B cells in presence of a normal or slightly decreased switched subset. Surprisingly, we observed only a minor impact of MyD88 deficiency on the peripheral pool of IgG+ and IgA+ memory B cells in vivo, despite the recent implication of MyD88 in a TACI-mediated isotype switch process.43 To our knowledge, a deficit restricted to the sole IgM+IgD+CD27+ population is a rather rare situation, because in most of the primary immunodeficiencies in which a severe reduction of circulating CD27+ memory B cells was reported, this reduction was caused either by the preferential decrease (or absence) of switched cells, or of both unswitched (IgD+) and switched subsets: this is indeed the case in common variable immunodeficiency; in the X-linked lymphoproliferative syndrome type 1; in the warts, hypogammaglobulinemia, infections, myelokathexis (WHIM) syndrome; or in DOCK8 (Dedicator of cytokinesis 8) deficiency.44-47 For patients whom we were able to study at different ages (up to 5 years apart), the deficit in IgM+IgD+CD27+ cells appeared stable with time, indicating that homeostatic mechanisms were unable to normalize it as patients grow older. Other functional characteristics, like the specific shift toward smaller CDR3 sizes observed in this population, or the mutation frequency of its Ig genes, appeared as well unaffected. We would therefore favor a role of IRAK-4/MyD88 signaling in the maintenance/survival of the IgM+IgD+CD27+ subset rather than its development, considering its otherwise-normal characteristics, notably in terms of repertoire diversification, and its persistent reduction with age. Accordingly, whether the residual IgM+IgD+CD27+ population observed is because of a partial TLR dependence for its survival, or whether it represents the persistence of a subset with different maintenance requirements, is an open question that pertains to the more general issue of a possible heterogeneity of the IgM+IgD+CD27+ B-cell population.
There was a significantly more pronounced deficit of the IgM+IgD+CD27+ subset in IRAK-4– compared with MyD88-deficient patients. This result was surprising because no signal transduction function has been uniquely assigned to IRAK-4 so far. Moreover, both IRAK-4 and MyD88 deficiencies correspond to diseases that are clinical phenocopies, with patients displaying a similar sensitivity to pathogens, mainly encapsulated bacteria.18,29 The invasive infections they suffer occur essentially at very young ages, at which time T-independent responses are nonfunctional, thus being likely the consequence of a deficient innate immune response, rather than of a deficit in IgM+IgD+CD27+ B cells. Nevertheless, approximately one-half of the IRAK-4 patients show an impaired antipneumococcal Ig response at later ages,29 the main function attributed to this B-cell subset.
TLR3- and TRIF-deficient patients showed no apparent alterations of their B-cell compartment, suggesting that TLR3 is not crucial for the development and/or maintenance of IgM+IgD+CD27+ B cells. To our surprise, the 2 UNC-93B–deficient patients behaved like control patients, suggesting that, among TLRs that rely both on MyD88/IRAK-4 and UNC-93B to signal, TLR7, TLR8 (which is not detectable in human B cells), and TLR9 are dispensable for IgM+IgD+CD27+ B cells. Alternatively, TLR7 and TLR9 signaling in human B cells may be partly independent from UNC-93B; however, this seems unlikely because TLR7 and TLR9 agonists were shown to require UNC-93B expression to activate human B cells.25
The IRAK-MyD88 pathway transmits signals not only from TLRs but also from the family of IL-1 receptors (including receptors to IL-1, IL-18, and IL-33). Moreover, the involvement of MyD88 in other types of signal transduction (like TACI) has been described, thus leaving open whether the deficit observed could be mediated by actors other than TLRs.43 However, the TIRAP-deficient patients, for whom the dissociation between the 2 CD27+ subsets was the most important, allowed us to limit further the number and nature of receptors involved. It has been shown that TIRAP-deficient mice respond normally to IL-1 and IL-18 but are impaired in their responses to ligands for TLR4 and for TLR2 heterodimers (and TLR1 and 6 that heterodimerize with TLR2), for which TIRAP constitutes a “sorting adaptor,” allowing the recruitment of MyD88 to the TLR complex at the cell membrane.48 Accordingly, for the 4 TIRAP-deficient patients studied, who belong to the same family and carry a homozygous missense mutation in the TIR domain of TIRAP, the response to IL-1β was normal in whole blood cells, whereas the response to Pam2CSK4 (a TLR2/6 agonist) was markedly impaired (L.I. and J.-L.C., unpublished data, 2012). The phenotype of TIRAP patients would thus restrict the possible B-cell–specific TLRs involved to those associating with TLR2, ie, TLR6, and possibly TLR1.
But what about TLR10? TLR10 is a highly N-glycosylated protein that can associate with TLR2 or TLR1, or constitutes a homodimer.42 However, it is controversial whether TLR10 needs TIRAP to mediate MyD88 recruitment. Nevertheless, in a recent structural study of TLR4-TIRAP interaction, Bovijn et al identified a TIRAP binding site within the TIR domain of TLR4, generated on its dimerization. On the basis of the sequence conservation between the TIR domains of TLR4 and 10 (whose crystallized structure was used to model the interaction), these authors concluded that TIRAP interaction sites are likely to be present in TLR10.49 One has to emphasize that the search for TLR10 agonists has been greatly hindered by the absence of a functional homologue in mice and also because TLR10, alone or in cooperation with TLR2, fails to activate typical TLR-induced signaling, including NF-κB reporters.50 Moreover, the analysis of chimeric receptors in which the intracellular part of TLR10 was replaced with that of TLR1 revealed that TLR10 may be able to sense triacylated lipopeptides and a wide variety of other microbial-derived agonists shared by TLR1 but not TLR6. In the reverse swapping experiment, however, a receptor with the extracellular and transmembrane domain of TLR1 fused to the signaling intracellular domain of TLR10 did not respond.50 However, none of these experiments has been performed in B-cell lines in which lineage specific posttranslational modifications of TLR10 could possibly occur and allow for its signaling.
TLRs are expressed by many actors of the immune response, and it is obviously unsettled whether the impact of the TLR signaling deficiency observed is B-cell intrinsic. Along this line, we addressed whether the 2 CD27+ subsets have different effector profiles in response to various TLR agonists, including PamCys(Pam)SK4, a proposed TLR10-specific ligand based on modeling studies.33 Unfortunately, no response either in terms of cytokine/chemokine secretion or cellular proliferation was observed for this agonist, thus precluding to get insight as to whether TLR10 may have a unique function for IgM+IgD+CD27+ cells. We found that TLR2/6, TLR7, and TLR9 triggering activated all splenic subsets to produce cytokines, chemokines, and hematopoietic growth factors. Although TLR9 was expressed with similar intensity on both CD27+ subsets,39 its engagement resulted in reproducibly higher levels of secretion for IL-6, MIP-1α (CCL3) and MIP-1β (CCL4) for IgM+IgD+CD27+ compared with switched cells (an average 2-fold difference), and a much lower secretion of IL-8 (CCL8), a chemokine attracting neutrophils at sites of inflammation (a 10-fold difference). Whether these differences in effector profile may have functional consequences remains obviously an open question at that stage.
Several reports have emphasized the integration of BCR and TLRs signals in the outcome of the immune response.14 We would like to propose to extend further this notion to the survival/maintenance signals delivered to specific B-cell subsets, in particular to the IgM+IgD+CD27+ population that harbors features of both innate-like and adaptive activation. Accordingly, recent data on diffuse large B-cell lymphomas of the activated B-cell type (which represent CD27+IgM+ transformed cells) have highlighted a survival/proliferative pathway mediated by both MyD88 and BCR signals.51 A gain-of-function mutant of MyD88 was described in this study that triggered a survival signal to which lymphomas became “addicted.” Most strikingly, in lymphomas that harbor instead a mutation in the BCR signaling complex leading to its constitutive activation, MyD88 and BCR signaling were shown to provide nonredundant survival signals.51 Along the same line, the inefficient central tolerance checkpoint observed for developing B cells from IRAK-4– and MyD88-deficient patients could be viewed as the consequence of a greater signalization threshold for the BCR in the absence of cooperative MyD88/IRAK-4 signals.25 We thus would like to propose a B-cell intrinsic role of TLRs in the specific homeostatic maintenance of IgM+IgD+CD27+ cells, possibly through microbial induced proliferation and/or through provision of survival signals that may cooperate with those delivered specifically through the IgM and not the IgG or IgA receptor.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jérome Mégret and Corinne Cordier for help with the cell sorter, Nicolas Cagnard for microarray data analysis, Dr Chantal Brouzes (Necker Hospital, Paris) for providing children blood samples, and Damiana Lecoeuche for her technical assistance. Microarray data were obtained within the frame of the LYMPH-IMM-MARGE INCA project. The authors thank Dr Menno van Zelm for sharing data about B-cell subsets control values. The authors are very much indebted to the patients and their families.
Inserm U783 is supported by the Ligue contre le Cancer (équipe labellisée), an ERC Advanced Grant, and the Fondation Princesse Grace. J.-L.C. is supported in part by the St Giles Foundation and The Rockefeller University grant UL1 TR000043 from the National Center for Research Resources and the National Center for Advancing Sciences (NCATS), National Institutes of Health. E.M. is supported by grants from the National Institutes of Health-National Institute of Allergy and Infectious Diseases (grants AI061093, AI071087, AI082713, and AI095848) and L.M. by TÁMOP 4.2.1/B-09/1/KONV-2010-0007.
National Institutes of Health
Authorship
Contribution: S.W. performed experiments, analyzed and discussed data, and wrote the manuscript; M.B., H.D., and M.C. performed experiments and analyzed experimental data. L.M., C.R.-G., B.-Z.G., C.R., A.C.I., S.E.Z., C.H., Y.C., J.V., C.R., and P.D.A. provided samples and participated in the clinical care of the patients; L.I., E.M., S.-Y.Z., A.P., J.-L.C., and C.P. provided samples from patients, discussed data, and edited the manuscript; A.A. performed statistical analysis; A.C. discussed data; and J.-C.W. and C.-A.R. share senior authorship.
Conflict-of-interest disclosure: The authors have no conflicting financial interests.
Correspondence: Jean-Claude Weill and Claude-Agnès Reynaud, Inserm U783, Développement du Système Immunitaire, Faculté de Médecine-Site Necker-Enfants Malades, Université Paris Descartes, 156, rue de Vaugirard 75730 Paris Ceder 15, France; e-mail: claude-agnes.reynaud@inserm.fr or jean-claude.weill@inserm.fr.

![Figure 1. IgM+IgD+CD27+ cells are significantly decreased in IRAK-4–, MyD88-, and TIRAP-deficient patients. (A) Relative frequencies of blood IgD+CD27+ and IgD−CD27+ cells (expressed as percentages of CD19+ B cells) in IRAK-4–, MyD88-, TIRAP-, UNC-93B–, and TLR3-deficient patients and controls were determined by flow cytometry after IgD, CD27, and CD19 staining. Only values from patients or control patients older than 4 years of age were plotted (ie, excluding the 3.5-year-old TRIF-deficient patient, as well as patients P30 [IRAK-4, 2 years] and P2 [MyD88, 2 years]). Mean values (expressed as percentage of total B cells) are indicated below the graph for IRAK-4–, MyD88-, and TIRAP-deficient patients and controls. *P < .05, **P < .01, ***P < .001 (see “Methods” for statistics). (B) Ratio of the IgM+IgD+CD27+ versus IgD−CD27+ cell proportions for all the patients and controls (from 2 to 25 years of age).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/25/10.1182_blood-2012-07-440776/4/m_zh89991299010001.jpeg?Expires=1769113710&Signature=chrp6iS2MM1IU~4lw-r78Hc2q~6Bl4MEuJMs1QMNI5TGKXKHqZkPQUT1teTm3F8bW5ISnPE6GbVbFRc9ZthNJECfMSuIcUJmODmWhVFs0WaAVCuFWywRlegGpdwJ5OBRomHTmWiujyl-h-5nT8NKRfPA0pbqBfUjOjqOZNG4PRDM365aGsH5P3wuCNDT8cjjgydt6SVtlaG~NjJ~JqbVmzbtyWV5K8N3HwLl92lM-SFwaxYJDKFMoVZQWtv59Srm-DceHgoBBrEm4zXa7ioX2Gg4Eq7lB0105TrhnmQ9g8CMlL0jyGgIaJ6YhaBcWuF0iHPIO6iSrX6K2V3zv27g1Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal