Abstract
Abstract 784
Whole Genome Sequencing of Therapy-Related Acute Myeloid Leukemia
Giridharan Ramsingh, Dong Shen, Tamara L. Lamprecht, Sharon E. Heath, Robert S. Fulton, Elaine Mardis, Li Ding, Peter Westervelt, John Welch, Matthew J. Walter, Timothy A. Graubert, John F. DiPersio, Timothy J. Ley, Richard K. Wilson, and Daniel C. Link.
Therapy related therapy-related acute myeloid leukemia (t-AML) accounts for 10–20% of all new cases of AML, and its incidence is rising. A fundamental difference in the pathogenesis of de novo AML and t-AML is prior treatment with chemotherapy and/or radiotherapy. The exposure of hematopoietic stem/progenitors cells (HSPCs) to this genotoxic stress is hypothesized to alter the number and spectrum of mutations that arise in t-AML. Moreover, the genotoxic stress may exert selective pressure to expand those HSPC clones that are inherently resistant to chemotherapy, a common feature in t-AML. To test these hypotheses, we sequenced the genomes of 23 cases of t-AML and compared them to the genomes of 24 cases of de novo AML, which we recently reported (Welch et al., Cell, July 2012). We choose to focus our initial studies on the subset of t-AML with normal cytogenetics or simple balanced translocations. Specifically, MLL gene rearrangements were observed in 22% of cases, other balanced translocations in 22%, trisomy 8 in 22%, normal karyotype in 31%, and a complex karyotype in a single case. All patients had received prior alkylator chemotherapy (62%), topoisomerase inhibitor chemotherapy (65%), or radiotherapy (77%).
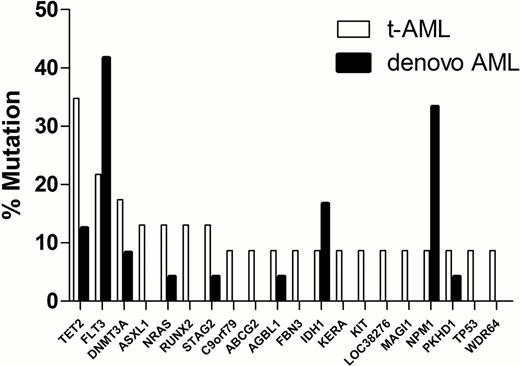
To identify somatic mutations, whole genome sequencing was performed on leukemic bone marrow (average 65% blasts) and skin (normal) DNA. Average haploid coverage was 37.5X and 34.7X for the leukemia and skin genomes, respectively. All somatic mutations were verified using patient-specific custom NimbleGen capture arrays, followed by Illumina sequencing. Although the total number of somatic single nucleotide variants in older patients (>50 years) with t-AML was similar to that observed in de novo AML (484 ± 68 vs. 506 ± 45, respectively), significantly more mutations were present in younger (≤ 50 years) patients with t-AML (743 ± 228) compared with de novo AML (336 ± 179, P=0.04). Exposure to chemotherapy is associated with an increased rate of transversions in relapsed AML (Ding et al., Nature 2012). However, the percentage of somatic mutations that were transversions in t-AML (35.8 ± 1.91%) was similar to that seen in de novo AML (33.5 ± 0.93%), regardless of age. In the 23 t-AML genomes, we identified recurring mutations (present in at least 2 cases) in 20 genes. Many of these mutations were also observed in de novo AML genomes (Figure 1). The most commonly mutated gene in t-AML was TET2, which was mutated in 35% of cases. Of interest, missense mutations of the ABC transporter gene ABCG2 were significantly enriched in t-AML (2/23, 8.7%) compared with de novo AML (0 in 200 cases, P=0.01). ABCG2 (also known as breast cancer resistance protein, BCRP) has been implicated in chemotherapy resistance. ABCG2 is expressed at high levels in hematopoietic stem cells, where it is known to function as a key drug transporter. Studies are underway to define the frequency of ABCG2 mutations (and other ABC transporter genes) in a larger cohort of t-AML, including cases with alterations in chromosome 5 or 7 or with complex cytogenetic abnormalities. In summary, in younger patients with t-AML, the mutational burden is higher than that of de novo AML patients, possibly reflecting prior exposure to chemoradiotherapy, though no increase in transversions was observed. Mutations of ABCG2 may contribute to chemotherapy resistance in a subset of t-AML.
Recurring mutations in t-AML (n = 23) compared with de novo AML (n = 24).
Ley:Washington University: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal