Abstract
Abstract  3811
3811
A large number of genes have been found mutated in CMML including 18 encoding signaling molecules (CBL, N/KRAS, JAK2, FLT3, KIT), epigenetic regulators (TET2, IDH1/2, DNMT3A, ASXL1, EZH2) transcription (RUNX1, NPM1) and splicing (SRSF2, SF3B1, U2AF1, ZRSR2) factors. We report the genotypic patterns, clinical correlates and prognostic impact of mutations in those 18 genes in a large cohort of CMML patients (pts).
Bone marrow or peripheral CD14+ cells from 224 CMML pts from a non interventional study (n=186) or a phase II decitabine trial (n=38; Braun Blood 2011) were genotyped by mutation specific techniques and Sanger sequencing for up to 18 genes (depending on available material): TET2, IDH1, IDH2, DNMT3A, CBL, NRAS, KRAS, JAK2V617F, FLT3, KIT, NPM1, RUNX1, ASXL1, EZH2, SF3B1, SRSF2, U2AF1 and ZRSR2. The number of TET2 alleles with a functional Cystein Rich (CysR) domain (Delhommeau NEJM 2009) was predicted based on mutation type and zygosity, assuming that double mutations affect independent alleles. Overall (OS) and AML-free (AMLFS) survival were analyzed from the date of genotyping.
224 CMML pts (152M/72F, median age 75y) were genotyped at diagnosis (37%) or after a median of 7.2 months of evolution (none had received hypomethylating agents [HMA] before genotyping); WHO diagnosis was CMML-1/2 in 78%/22%, 70% pts had normal karyotype, 22% had extramedullary disease (EMD); 13 pts had autoimmune manifestations (AIM). The most frequently mutated genes were TET2 (58%), SRSF2 (47%) and ASXL1 (38%). Mutations in RUNX1, CBL and NRAS were found in 14%, 11% and 10% of pts, respectively (resp). All other genes were mutated in <10% of pts. Only 5% pts lacked any mutation, and 70% had ≥2 mutated genes; TET2, IDH1 and IDH2 mutations, present in 64% of pts, were mutually exclusive. Mutations in splice and signaling genes were present in 63% and 35% of pts, with 2 mutated genes within each group in 3% and 2% pts, resp. ASXL1 mutations were less frequent in the presence of TET2 mutations (P<.0001). Significant mutual associations included ASXL1/RUNX1, ASXL1/NRAS, TET2/SRSF2, RUNX1/SRSF2 and U2AF1/IDH2.
In multivariate analysis accounting for those interactions, TET2 status was the only independent predictor of hemoglobin values (median 10.2 vs 11.9 g/dL in wildtype [wt] vs mutated pts, P<.0001) with a gene dosage effect (P=.0003). Platelet counts were higher in JAK2V617F pts, and lower in pts with RUNX1, TET2 or SRSF2 mutations. WBC and monocyte counts were higher in pts with ASXL1 and NRAS mutations. EMD was associated to ASXL1, CBL, KRAS and JAK2 mutations. EMD, CMML-2 and abnormal karyotype were less frequent in TET2 mutated pts. All 13 pts with AIM had at least one mutated gene, with no specific genotype spectrum.
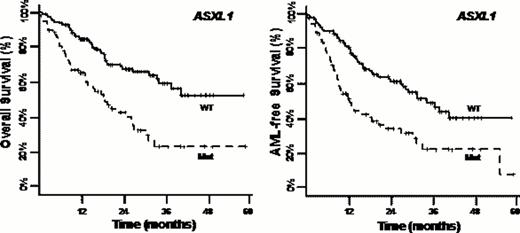
With a median follow-up of 25.4 months, median OS and AMLFS were 32.2 and 28.0 months resp. In univariate analysis, OS was decreased in pts with IDH2 mutations (P=.04), and AMLFS was shorter in pts with NRAS (P=.04), RUNX1 (P=.03) and SRSF2 (P=.04) mutations. ASXL1 mutations markedly reduced OS (median 18.5 vs 35.7 months in wt pts) and AMLFS (median 12.5 vs 34.7 months, both P<.0001), with similar results in the 74 pts who received HMA during follow-up. In the 224 pts, there was no significant effect of overall TET2 status (wt vs mutated) on OS or AMLFS (both P=.09) but median OS was 29.3, 35.7 months and not reached in pts with 2 (57%), 1 (30%) and 0 (13%) TET2 alleles with a putatively functional CysR domain (P=.01). Similar differences were noted for AMLFS (P=.008).
In multivariate analysis including peripheral blood counts, WHO classification, cytogenetics, disease evolution and therapy, ASXL1 was the only gene whose mutations independently predicted inferior OS (HR: 2.44, 95% CI: 1.36–4.37, P=.003)and AMLFS (HR: 2.54, 95% CI: 1.46–4.42, P=.001); SRSF2 mutations only predicted inferior AMLFS (HR: 2.05, 95% CI: 1.15–3.65, P=.02). The total number of mutated genes as a continuous variable, which was higher in ASXL1 mutated pts (mean 3.0 vs 1.8, P<.0001), was the only genetic variable to retain prognostic value when added to those models (OS and AMLFS both P<.0001).
TET2, SRSF2 and ASXL1 are the most frequent mutated genes in CMML. The number and location of TET2 mutations may impact CMML presentation and outcome. The total number of mutated genes has the strongest prognostic relevance, but ASXL1 mutational status provides a robust surrogate prognostic marker for daily practice.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal