Abstract
Abstract 1379
A large number of gene mutations have been recently detected in AML using novel sequencing technologies. We established a rapid, amplicon-based resequencing assay that allows efficient analysis of 16 of the most commonly mutated genes in AML and used it to study a cohort of AML patients (pts) carrying a translocation t(9;11)(p22;q23) (MLLT3-MLL; MLL-AF9). This genetic subgroup, accounting for ∼1% of adult AML, is associated with young age, treatment-related disease, FAB M4/M5 morphology, and an intermediate prognosis. There is limited information on the cooperating genetic lesions in adult AML with t(9;11). Importantly, several widely used murine AML models are based on MLLT3-MLL fusion transcript expression. Thus, a better understanding of the genetic basis of human MLLT3-MLL-rearranged AML is necessary to understand how well these animal models reflect their human counterpart and whether findings from MLLT3-MLL-induced disease are generalizable to other genetic subsets.
We studied 33 bone marrow samples from adult AML pts with t(9;11)(p22;q23) (age range, 20–71 years; median, 44 years; 21 de novo and 12 therapy-related AML). Mutations in ASXL1, CBL, DNMT3A, FLT3, IDH1, IDH2, KIT, KRAS, NRAS, NPM1, RUNX1, SF3B1, SRSF2, TET2, U2AF1 and WT1 were analyzed from 250ng of genomic DNA using a multiplexed, amplicon-based next-generation sequencing approach (Illumina TruSeq Custom Amplicon assay and MiSeq sequencer). KRAS mutations were independently verified using PCR followed by 454 sequencing (Roche), and NRAS and FLT3 mutations by PCR and melting curve analysis or Sanger sequencing.
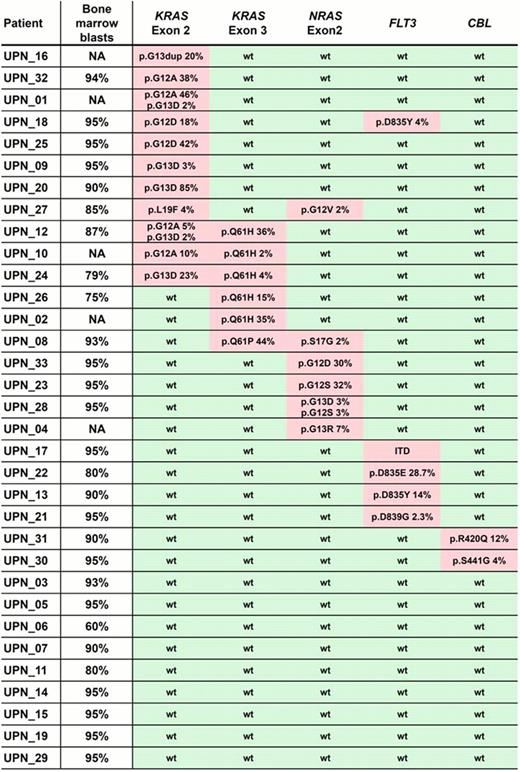
Per patient, we obtained between 96k and 235k paired-end reads (2×150bp) mapping to the regions of interest, resulting in median coverage depths of the target genes ranging from 180-fold (SRSF2) to >2500-fold (KRAS). Overall, mutations affecting growth factor signalling pathways were detected in 73% of MLLT3-MLL rearranged AML (24/33; Figure): Fourteen pts (42%) carried KRAS mutations mostly affecting the known hotspot codons 12, 13 and 61, 6 pts (18%) had NRAS mutations (mainly at codons 12 or 13), 5 pts had FLT3 mutations (4 tyrosine kinase domain mutations and 1 internal tandem duplication), and 2 pts had mutated CBL. The frequency of RAS gene mutations did not differ significantly between de novo AML and pts with treatment-related disease (P=.26).
More than one RAS mutation was found in 7 pts, including pts with 2 (n=3) or 3 (n=1) distinct KRAS mutations, 2 pts with mutations in both NRAS and KRAS, and one patient with 2 NRAS mutations. Interestingly, in some of these pts, one mutation was present in a relatively large proportion of sequencing reads (e.g., patient UPN12 showing a KRAS p.Q61H mutation in 36% of reads, consistent with a heterozygous mutation present in the majority of cells in the specimen), while other coexisting mutations affected a much smaller proportion of reads (in patient UPN12, two different KRAS exon 2 mutations in 5% and 2% of reads, respectively). These results suggest the presence of different subclones within the AML blast population, each carrying a different KRAS mutation. Analyses of follow-up samples are underway to assess changes of clonal architecture over time.
Other gene mutations were rarely found in this cytogenetic subgroup of AML: In our 33 pts, we detected 2 ASXL1 mutations, 1 mutation each in TET2, SRSF2 and U2AF1, and no mutations in the other 8 genes we studied.
Targeted resequencing using a multiplexed amplicon-based assay is a sensitive and rapid method to screen for mutations in a panel of genes commonly involved in AML pathogenesis. To our knowledge, our report is the first comprehensive analysis of cooperating gene mutations in adult AML with t(9;11)(p22;q23). We demonstrate that MLLT3-MLL-rearranged AML is characterized by frequent mutations in genes involved in growth factor signalling (particularly KRAS and NRAS, mutated in 40% and 18%, respectively, of our MLL-MLLT3 AML cohort compared to only about 5% of unselected AML pts), in the absence of other common AML-related gene mutations. Our results complement recent studies reporting RAS mutations in 45% of infant MLL-rearranged ALL, and functional data from mouse models showing that RAS mutations cooperate with the MLLT3-MLL fusion during leukemogenesis. Finally, our results provide evidence for clonal heterogeneity within MLLT3-MLL rearranged human AML.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal